

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147405.9 + phase: 0
(589 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
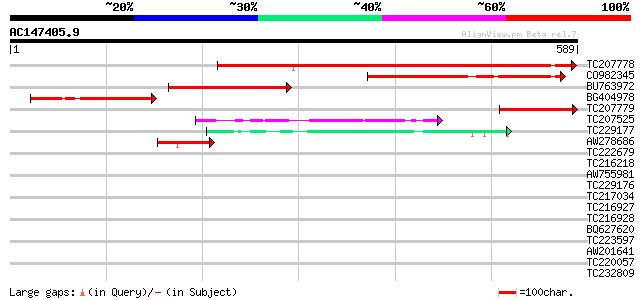
Score E
Sequences producing significant alignments: (bits) Value
TC207778 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (41%) 549 e-156
CO982345 256 3e-68
BU763972 197 1e-50
BG404978 148 8e-36
TC207779 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (6%) 113 3e-25
TC207525 weakly similar to UP|NGP1_HUMAN (Q13823) Autoantigen NG... 84 2e-16
TC229177 weakly similar to UP|Q21086 (Q21086) C. elegans NST-1 p... 82 7e-16
AW278686 44 2e-04
TC222679 similar to UP|Q9LH58 (Q9LH58) GTP-binding protein-like,... 38 0.014
TC216218 homologue to UP|Q9M7P3 (Q9M7P3) GTP-binding protein, co... 35 0.071
AW755981 35 0.093
TC229176 35 0.12
TC217034 similar to UP|Q8L732 (Q8L732) Expressed protein, partia... 35 0.12
TC216927 homologue to UP|Q9SVA6 (Q9SVA6) GTP-binding-like protei... 35 0.12
TC216928 similar to UP|Q9SVA6 (Q9SVA6) GTP-binding-like protein,... 33 0.27
BQ627620 similar to GP|10176988|db GTP-binding protein-like {Ara... 33 0.27
TC223597 homologue to UP|Q9SGT3 (Q9SGT3) T6H22.14 protein, parti... 33 0.35
AW201641 33 0.35
TC220057 similar to GB|AAP41843.1|32396052|AY254472 short integu... 32 0.60
TC232809 similar to UP|ENGC_ECOLI (P39286) Probable GTPase engC ... 32 1.0
>TC207778 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (41%)
Length = 1511
Score = 549 bits (1415), Expect = e-156
Identities = 286/382 (74%), Positives = 315/382 (81%), Gaps = 10/382 (2%)
Frame = +1
Query: 217 REKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSS-QADNMASADNPDTKIYGRDELLAR 275
REKWAEYFRAHDILFIFWSAKAATA LEGKKLGSS + DNM ++PDTKIYGRDELLAR
Sbjct: 13 REKWAEYFRAHDILFIFWSAKAATAALEGKKLGSSWEDDNMGRTNSPDTKIYGRDELLAR 192
Query: 276 LQSEAEAIVDMRRSSGSS-------KSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINAL 328
LQSEAE IVD RR+SGSS KS +N + SSSSS+V+VGFVGYPNVGKSSTINAL
Sbjct: 193 LQSEAEEIVDRRRNSGSSDAGPSNIKSPAENTAGSSSSSNVIVGFVGYPNVGKSSTINAL 372
Query: 329 VGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPIDRMTQ 388
VGQK+TGVTSTPGKTKHFQTLIIS++L LCDCPGLVFPSFSSSRYEMI CGVLPIDRMT+
Sbjct: 373 VGQKRTGVTSTPGKTKHFQTLIISDELTLCDCPGLVFPSFSSSRYEMIACGVLPIDRMTE 552
Query: 389 HRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLP 448
RE VQVVA+RVPRHVIEEIY I LPKPKSYESQSRPPLASELLR YC SRG SSGLP
Sbjct: 553 QRESVQVVADRVPRHVIEEIYKIRLPKPKSYESQSRPPLASELLRAYCTSRGYVASSGLP 732
Query: 449 DETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVE 508
DETRA+R +LKDY DGKLPHY+ PPG S +E A E+ HD V+ SD+S E+SS +
Sbjct: 733 DETRAARLMLKDYTDGKLPHYQKPPGASDEEQALEEPVGHDLVDLDASDSSGTEESSDDK 912
Query: 509 TELAPKLEHVLDDLSSFDMANGLA-SNKVAPKKTKESQK-HHRKPPRTKNRSWRAGNAGK 566
+ELAP LEHVLDDL+SFDMANGLA N KK K S K HH+KP RTKNRSWRAGN
Sbjct: 913 SELAPNLEHVLDDLNSFDMANGLAPKNITTVKKPKASHKHHHKKPQRTKNRSWRAGN--- 1083
Query: 567 DDTDGMPIARFHQKPVNSGPLK 588
+D DGMP+ARF QK N+GP+K
Sbjct: 1084EDADGMPVARFFQKAANTGPMK 1149
>CO982345
Length = 804
Score = 256 bits (653), Expect = 3e-68
Identities = 133/206 (64%), Positives = 156/206 (75%)
Frame = -2
Query: 372 RYEMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASEL 431
RY+MI CGVLPIDRMT+HRE VQVVA++VPR VIEEIY ISLPKPK YE QSRPPLASEL
Sbjct: 803 RYKMIACGVLPIDRMTEHREAVQVVADKVPRQVIEEIYKISLPKPKPYEPQSRPPLASEL 624
Query: 432 LRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQV 491
LR YCASRG T+SGLPDETRA+RQILKDYIDGKLPH+EMPPG+S + E D V
Sbjct: 623 LRAYCASRGYVTASGLPDETRAARQILKDYIDGKLPHHEMPPGMS--------NVEPDPV 468
Query: 492 NPHVSDASDIEDSSVVETELAPKLEHVLDDLSSFDMANGLASNKVAPKKTKESQKHHRKP 551
N H S + ED V++++AP E VLDDL+SFD+ANGL SN+V KK+ S HH+KP
Sbjct: 467 NLHGSVSYGTED---VDSKVAPDFEQVLDDLNSFDLANGLVSNEVTIKKSDASHNHHKKP 297
Query: 552 PRTKNRSWRAGNAGKDDTDGMPIARF 577
K+RS R N +D DGMP+ +F
Sbjct: 296 QGKKDRSQRTEN---NDADGMPVVKF 228
>BU763972
Length = 409
Score = 197 bits (501), Expect = 1e-50
Identities = 100/128 (78%), Positives = 111/128 (86%), Gaps = 1/128 (0%)
Frame = +1
Query: 166 VERSDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFR 225
VERSDLLVMVVDSRDPLFYRCPDLEAYA+EVD HK TLLLVNKADLLPASVREKW EYF
Sbjct: 1 VERSDLLVMVVDSRDPLFYRCPDLEAYAREVDEHKRTLLLVNKADLLPASVREKWVEYFH 180
Query: 226 AHDILFIFWSAKAATAVLEGKKLGSS-QADNMASADNPDTKIYGRDELLARLQSEAEAIV 284
AH+IL+IFWSAKAA+A +EGK S +AD+ +NPDT+IY RDELLARLQSEAE IV
Sbjct: 181 AHNILYIFWSAKAASAAVEGKMFRSPLEADDSGKNNNPDTEIYDRDELLARLQSEAEEIV 360
Query: 285 DMRRSSGS 292
+MRR+S S
Sbjct: 361 EMRRNSSS 384
>BG404978
Length = 384
Score = 148 bits (373), Expect = 8e-36
Identities = 82/131 (62%), Positives = 95/131 (71%)
Frame = +2
Query: 22 QQTKEKGKIYKKKFLESFTEVSDIDAIIEQADEDVDEQQLLDIPLPPPTALINLDHASGS 81
QQ+KE G++ KK LESFTE+SDIDAIIEQA+ +Q L IP P INLD +S
Sbjct: 8 QQSKENGRLQKKNVLESFTEISDIDAIIEQAEHP--QQPLFSIPQRP----INLDLSSSI 169
Query: 82 DFNGLTVEEMKKEQKIEEALHASSLRVPRRPFWSAEMSADELHANETQHFLTWRRSLARL 141
N +T+EEM+K+Q EEALHASSLRVPRRP W A MS +EL NE Q FL WRR LARL
Sbjct: 170 GKNEMTLEEMRKQQTREEALHASSLRVPRRPPWRAYMSVEELDDNERQAFLIWRRRLARL 349
Query: 142 EENKKLVLTPF 152
EEN+ LVLTPF
Sbjct: 350 EENENLVLTPF 382
>TC207779 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (6%)
Length = 599
Score = 113 bits (282), Expect = 3e-25
Identities = 57/82 (69%), Positives = 63/82 (76%), Gaps = 2/82 (2%)
Frame = +3
Query: 510 ELAPKLEHVLDDLSSFDMANGLA-SNKVAPKKTKESQKHH-RKPPRTKNRSWRAGNAGKD 567
ELAP LEHVL DL+SFDMANGLA N KK K S KHH +KP R K+RSWRAG AG +
Sbjct: 3 ELAPNLEHVLVDLNSFDMANGLAPKNTTTVKKPKASHKHHHKKPQRKKDRSWRAGKAGNE 182
Query: 568 DTDGMPIARFHQKPVNSGPLKV 589
D DGMP+ARF QKP N+GPLKV
Sbjct: 183 DADGMPVARFFQKPANTGPLKV 248
>TC207525 weakly similar to UP|NGP1_HUMAN (Q13823) Autoantigen NGP-1, partial
(26%)
Length = 603
Score = 83.6 bits (205), Expect = 2e-16
Identities = 71/256 (27%), Positives = 114/256 (43%)
Frame = +2
Query: 194 KEVDVHKNTLLLVNKADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQA 253
KE HK+ +LL+NK DL+PA W+ K VL S +
Sbjct: 5 KENCKHKHMVLLLNKCDLVPA-------------------WATKGWLRVL------SKEF 109
Query: 254 DNMASADNPDTKIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGF 313
+A N + K +G+ LL+ L+ A D + S VGF
Sbjct: 110 PTLAFHANIN-KSFGKGSLLSVLRQFARLKRDKQAIS--------------------VGF 226
Query: 314 VGYPNVGKSSTINALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRY 373
VGYPNVGKSS IN L + V PG+TK +Q + +++++ L DCPG+V+ + + +
Sbjct: 227 VGYPNVGKSSVINTLRTKNVCKVAPIPGETKVWQYITLTKRIFLIDCPGVVYHN-NDTET 403
Query: 374 EMITCGVLPIDRMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLR 433
+++ GV+ + + + + V RV + +E Y I K ++ + ++ L
Sbjct: 404 DVVLKGVVRVTNLKDAADHIGEVLKRVKKEHLERAYKI-----KEWDDE------NDFLL 550
Query: 434 TYCASRGQTTSSGLPD 449
C S G+ G PD
Sbjct: 551 QLCKSTGKLLKGGEPD 598
>TC229177 weakly similar to UP|Q21086 (Q21086) C. elegans NST-1 protein
(Corresponding sequence K01C8.9), partial (21%)
Length = 1353
Score = 82.0 bits (201), Expect = 7e-16
Identities = 83/331 (25%), Positives = 131/331 (39%), Gaps = 14/331 (4%)
Frame = +2
Query: 205 LVNKADLLPASVREKWAEYFRAHDILFIFWSAKAATAVLEGKKLGSSQADNMASADNPDT 264
L+NK DL+P EKW +Y R F K +T Q N++ DT
Sbjct: 2 LLNKIDLVPKEALEKWLKYLREELPTVAF---KCST---------QQQRSNLSDCLGADT 145
Query: 265 KIYGRDELLARLQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSST 324
I + + + RS KS + VG +G PNVGKSS
Sbjct: 146 LI--------------KLLKNYSRSHEIKKS-------------ITVGLIGLPNVGKSSL 244
Query: 325 INALVGQKKTGVTSTPGKTKHFQTLIISEKLILCDCPGLVFPSFSSSRYEMITCGVLPID 384
IN+L V STPG T+ Q + + + + L DCPG+V + + I+
Sbjct: 245 INSLKRSHVVNVGSTPGLTRSMQEVQLDKNVKLLDCPGVVMLKSQENDASVALLNCKRIE 424
Query: 385 RMTQHRECVQVVANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTS 444
++ V+ + P + Y I K + + L RG+
Sbjct: 425 KLDDPISPVKEIFKLCPPEQLVTHYKIGTFKFGDVD---------DFLLKIATVRGKLKK 577
Query: 445 SGLPDETRASRQILKDYIDGKLPHYEMPPGLSTQE-----LASEDSNEH--DQV----NP 493
G+ D A+R +L D+ +GK+ +Y +PP E + SE S E D+V +
Sbjct: 578 GGIVDINAAARIVLHDWNEGKIIYYTIPPNRDQGEPAEVKIVSEFSKEFNIDEVYKSESS 757
Query: 494 HVSDASDIEDSSVVETELAPKL---EHVLDD 521
++ ++D + VE + L E +L+D
Sbjct: 758 YIGSLKSVDDLNAVEVPSSRPLNLDETMLED 850
>AW278686
Length = 310
Score = 44.3 bits (103), Expect = 2e-04
Identities = 23/64 (35%), Positives = 40/64 (61%), Gaps = 5/64 (7%)
Frame = +2
Query: 154 KNLDIWRQLWRVVERSDLL-----VMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNK 208
+++ +W +L+ V+ SD++ MV+D+RDP RC L+ + K+ H+ +LL+NK
Sbjct: 119 QSVRLWGELYTVIHSSDVVDQAAESMVLDARDPPGTRCYHLKKHLKDNCQHERMVLLLNK 298
Query: 209 ADLL 212
DLL
Sbjct: 299 CDLL 310
>TC222679 similar to UP|Q9LH58 (Q9LH58) GTP-binding protein-like, partial
(18%)
Length = 653
Score = 37.7 bits (86), Expect = 0.014
Identities = 23/66 (34%), Positives = 30/66 (44%)
Frame = +3
Query: 276 LQSEAEAIVDMRRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTG 335
L E E D + S+ S + S + + V VG PNVGKS+ N LVG +
Sbjct: 282 LSIEDEEKSDRKESAQSRRKSPRRTKIIPDNLLPRVAIVGRPNVGKSALFNRLVGGNRAI 461
Query: 336 VTSTPG 341
V PG
Sbjct: 462 VVDEPG 479
>TC216218 homologue to UP|Q9M7P3 (Q9M7P3) GTP-binding protein, complete
Length = 1545
Score = 35.4 bits (80), Expect = 0.071
Identities = 16/41 (39%), Positives = 25/41 (60%)
Frame = +3
Query: 288 RSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINAL 328
+S+ S ++ + + SSH+ +G VG PNVGKS+ N L
Sbjct: 81 KSAKSKEAPAERPILGRFSSHLKIGIVGLPNVGKSTLFNTL 203
>AW755981
Length = 451
Score = 35.0 bits (79), Expect = 0.093
Identities = 23/56 (41%), Positives = 27/56 (48%), Gaps = 1/56 (1%)
Frame = +3
Query: 311 VGFVGYPNVGKSSTINALVGQKKTGVTS-TPGKTKHFQTLIISEKLILCDCPGLVF 365
+ F G NVGKSS +NAL Q TS PG T+ + K L D PG F
Sbjct: 117 IAFAGRSNVGKSSLLNALTRQWGVVRTSDKPGLTQTINFFNLGTKHCLVDLPGYGF 284
>TC229176
Length = 887
Score = 34.7 bits (78), Expect = 0.12
Identities = 23/78 (29%), Positives = 38/78 (48%), Gaps = 5/78 (6%)
Frame = +3
Query: 429 SELLRTYCASRGQTTSSGLPDETRASRQILKDYIDGKLPHYEMPPGL-----STQELASE 483
S+ L RG+ G+ D A+R +L D+ +GK+ +Y MPP S + S+
Sbjct: 15 SDFLLKVGTVRGKLKKGGIVDINAAARIVLHDWNEGKIIYYTMPPNRDQGEPSEANIVSD 194
Query: 484 DSNEHDQVNPHVSDASDI 501
S E + + S++S I
Sbjct: 195 FSKEFNIDEVYSSESSYI 248
>TC217034 similar to UP|Q8L732 (Q8L732) Expressed protein, partial (24%)
Length = 1633
Score = 34.7 bits (78), Expect = 0.12
Identities = 27/115 (23%), Positives = 45/115 (38%)
Frame = +2
Query: 396 VANRVPRHVIEEIYNISLPKPKSYESQSRPPLASELLRTYCASRGQTTSSGLPDETRASR 455
+A+ V R +E I N LP P++ RPP +G P
Sbjct: 578 LASLVARKSLEHIGNCDLPHPQTKHVSPRPPY----------PKGVDHHDKTPPSLNRKT 727
Query: 456 QILKDYIDGKLPHYEMPPGLSTQELASEDSNEHDQVNPHVSDASDIEDSSVVETE 510
+ Y DG G+ST + + +DS++H + H D+S + + +E
Sbjct: 728 ETGSSYADG------YTTGISTSDCSFQDSSKHSS-SSHSKDSSSSTEGCQINSE 871
>TC216927 homologue to UP|Q9SVA6 (Q9SVA6) GTP-binding-like protein, complete
Length = 1771
Score = 34.7 bits (78), Expect = 0.12
Identities = 36/140 (25%), Positives = 57/140 (40%), Gaps = 8/140 (5%)
Frame = +2
Query: 233 FWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQS-EAEAIVDMRRSSG 291
+ S+K AT + + K + D MA + L A+L E + + +G
Sbjct: 194 YLSSKMATVMQKIKDI----EDEMAKTQKNKATAHHLGLLKAKLAKLRRELLTPSSKGAG 361
Query: 292 SSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVG-------QKKTGVTSTPGKTK 344
+ D S VG VG+P+VGKS+ +N L G + T +T PG
Sbjct: 362 GAGEGFDVTKSGDSR----VGLVGFPSVGKSTLLNKLTGTFSEVASYEFTTLTCIPGVIT 529
Query: 345 HFQTLIISEKLILCDCPGLV 364
+ K+ L D PG++
Sbjct: 530 Y-----RGAKIQLLDLPGII 574
>TC216928 similar to UP|Q9SVA6 (Q9SVA6) GTP-binding-like protein, partial
(25%)
Length = 354
Score = 33.5 bits (75), Expect = 0.27
Identities = 27/101 (26%), Positives = 44/101 (42%), Gaps = 1/101 (0%)
Frame = +1
Query: 231 FIFWSAKAATAVLEGKKLGSSQADNMASADNPDTKIYGRDELLARLQS-EAEAIVDMRRS 289
+ ++S+K AT + + K + D MA + L A+L E + +
Sbjct: 61 YYYFSSKMATVMQKIKDI----EDEMARTQKNKATAHHLGLLKAKLAKLRRELLTPSSKG 228
Query: 290 SGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVG 330
+G + D S VG VG+P+VGKS+ +N L G
Sbjct: 229 AGGAGEGFDVTKSGDSR----VGLVGFPSVGKSTLLNKLTG 339
>BQ627620 similar to GP|10176988|db GTP-binding protein-like {Arabidopsis
thaliana}, partial (21%)
Length = 431
Score = 33.5 bits (75), Expect = 0.27
Identities = 18/58 (31%), Positives = 29/58 (49%)
Frame = +1
Query: 287 RRSSGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTPGKTK 344
R +S + + V S + + VG PNVGKS+ +NAL+ + + V G T+
Sbjct: 55 RENSHNEVEDSSDLDVDKSKLPLQLAIVGRPNVGKSTLLNALLQEDRVLVGPEAGLTR 228
>TC223597 homologue to UP|Q9SGT3 (Q9SGT3) T6H22.14 protein, partial (19%)
Length = 441
Score = 33.1 bits (74), Expect = 0.35
Identities = 17/42 (40%), Positives = 23/42 (54%)
Frame = +1
Query: 299 NASVSSSSSHVVVGFVGYPNVGKSSTINALVGQKKTGVTSTP 340
++S S S + G VG PNVGKS+ NA+V K + P
Sbjct: 196 SSSSSKISMSLKAGIVGLPNVGKSTLFNAVVENGKAQAANFP 321
>AW201641
Length = 425
Score = 33.1 bits (74), Expect = 0.35
Identities = 18/41 (43%), Positives = 23/41 (55%)
Frame = +3
Query: 290 SGSSKSSDDNASVSSSSSHVVVGFVGYPNVGKSSTINALVG 330
SG K + N ++ + VG PNVGKSS +NALVG
Sbjct: 306 SGLQKIEESN-NLDEEDYVPAISIVGRPNVGKSSILNALVG 425
>TC220057 similar to GB|AAP41843.1|32396052|AY254472 short integuments 2
{Arabidopsis thaliana;} , partial (35%)
Length = 556
Score = 32.3 bits (72), Expect = 0.60
Identities = 17/58 (29%), Positives = 29/58 (49%)
Frame = +2
Query: 169 SDLLVMVVDSRDPLFYRCPDLEAYAKEVDVHKNTLLLVNKADLLPASVREKWAEYFRA 226
+DL++ V D+R P DL+ + K ++ +NK DL ++ KW YF +
Sbjct: 200 ADLVIEVRDARIPFSSANADLQPHLSA----KRRVVALNKKDLANPNIMHKWTHYFES 361
>TC232809 similar to UP|ENGC_ECOLI (P39286) Probable GTPase engC precursor ,
partial (5%)
Length = 699
Score = 31.6 bits (70), Expect = 1.0
Identities = 15/25 (60%), Positives = 19/25 (76%)
Frame = +2
Query: 314 VGYPNVGKSSTINALVGQKKTGVTS 338
VG NVGKSS +N+LV +KK +TS
Sbjct: 248 VGRSNVGKSSLLNSLVRRKKLALTS 322
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.130 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,199,473
Number of Sequences: 63676
Number of extensions: 344760
Number of successful extensions: 1631
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 1602
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1616
length of query: 589
length of database: 12,639,632
effective HSP length: 103
effective length of query: 486
effective length of database: 6,081,004
effective search space: 2955367944
effective search space used: 2955367944
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC147405.9


