

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146865.12 - phase: 0
(516 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
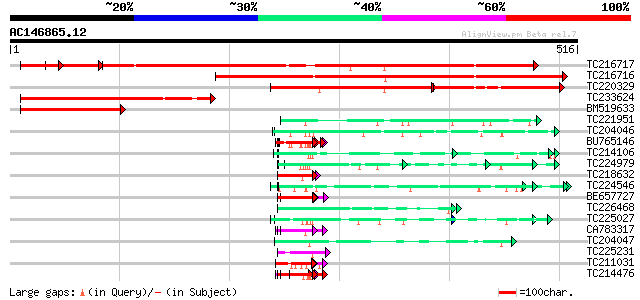
Score E
Sequences producing significant alignments: (bits) Value
TC216717 similar to UP|Q8H3E3 (Q8H3E3) Transcription factor (PWW... 578 e-178
TC216716 similar to UP|Q9ZS47 (Q9ZS47) Arbuscular mycorrhiza pro... 491 e-139
TC220329 weakly similar to UP|Q9ZS47 (Q9ZS47) Arbuscular mycorrh... 165 6e-81
TC233624 similar to UP|Q8H3E3 (Q8H3E3) Transcription factor (PWW... 260 9e-70
BM519633 119 4e-27
TC221951 UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75... 67 3e-11
TC204046 UP|Q39835 (Q39835) Extensin, complete 64 2e-10
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 64 2e-10
TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 62 8e-10
TC224979 similar to UP|Q09083 (Q09083) Hydroxyproline-rich glyco... 58 9e-09
TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wal... 58 1e-08
TC224546 similar to UP|Q41707 (Q41707) Extensin class 1 protein ... 57 2e-08
BE657727 similar to SP|P10495|GRP1_ Glycine-rich cell wall struc... 57 2e-08
TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich prote... 57 2e-08
TC225027 similar to UP|Q39835 (Q39835) Extensin, partial (49%) 57 3e-08
CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 56 3e-08
TC204047 hydroxyproline-rich glycoprotein 56 4e-08
TC225231 homologue to UP|PGKH_TOBAC (Q42961) Phosphoglycerate ki... 55 6e-08
TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 55 1e-07
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 55 1e-07
>TC216717 similar to UP|Q8H3E3 (Q8H3E3) Transcription factor (PWWP domain
protein)-like protein, partial (10%)
Length = 1582
Score = 578 bits (1490), Expect(2) = e-178
Identities = 280/401 (69%), Positives = 328/401 (80%), Gaps = 4/401 (0%)
Frame = +1
Query: 85 KVLRLWLERKILPEPMVRHHIRELDLYSSVSAGVYSRRSLRTERALDDPIREMEGMHVDE 144
+VLRLWLER+ILPE ++R HIRELDLYSS S G+Y RRS+RTERALDDP+REMEGM VDE
Sbjct: 409 QVLRLWLERRILPESIIRRHIRELDLYSS-SGGIYLRRSMRTERALDDPVREMEGMLVDE 585
Query: 145 YGSNSSLQLPGFCMPRMLKDEDDNEESDSDGGNFEAVTPEHNSEVHEMTSIIDKHRHILE 204
YGSNS+ QLPGFCMP+MLKDEDD E SDSDGGNFEAVTPEH SE++E+TS I+KHRHILE
Sbjct: 586 YGSNSTFQLPGFCMPQMLKDEDDGEGSDSDGGNFEAVTPEHTSEIYEITSAIEKHRHILE 765
Query: 205 DVDGELEMEDVSPSRDVEMNSFSNVDRGNATQFENNIHLPSAPPHQLVPQSSVPPPLAPP 264
DVDGELEMEDV+PS +VEMNS NVDR NA Q E N+ L AP HQ + +SS PPPL+
Sbjct: 766 DVDGELEMEDVAPSNEVEMNSICNVDRENAKQCEKNLPLFFAPLHQDM-RSSSPPPLSFL 942
Query: 265 PPPPPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHTESQCVKDN--PLHPMDRPLAAPRS 322
PPPPPP +PH + STSDP TV NS+G T SQ +K+N PLH + + +AAPR
Sbjct: 943 PPPPPP-------SIPHHMPSTSDPYNTVVNSKGCTVSQTLKENHHPLHSVAQLMAAPRH 1101
Query: 323 SQPISNAVHHHAPEYRE--AHISESDRSFNSFPVPHPVNYRHSDGVTMHDRGHSIRPPRH 380
SQPI +AVHH PEYRE H+ ES SFNSFPVP P N+RH+DGVT H++G+SIRPP+H
Sbjct: 1102SQPICDAVHHQVPEYREMQMHMPESTCSFNSFPVPPPENFRHTDGVTTHNKGYSIRPPQH 1281
Query: 381 VPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMEREHFYHNNHERLKPPPYDYRER 440
VP NQFSFV+GEQH +HRREVPPP PYS+RQHFV+N+ERE+FY NNHERL+PPPYDY+ER
Sbjct: 1282VPCNQFSFVNGEQHVKHRREVPPPLPYSSRQHFVQNIERENFY-NNHERLRPPPYDYQER 1458
Query: 441 WDVPPPYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRFPPR 481
W+ P PYPGP Y ++ +P PYGCHPCE RIPDHGWRFPP+
Sbjct: 1459WNGPAPYPGPWYQEKGVPPPYGCHPCESSRIPDHGWRFPPQ 1581
Score = 67.0 bits (162), Expect(2) = e-178
Identities = 34/39 (87%), Positives = 36/39 (92%)
Frame = +2
Query: 11 QVMEILADNLETESSLHRRVDLFFLVDSIAQFSRGLKGD 49
+VMEILA LE ESS+HRRVDLFFLVDSIAQFSRGLKGD
Sbjct: 182 KVMEILAHCLEMESSVHRRVDLFFLVDSIAQFSRGLKGD 298
Score = 64.7 bits (156), Expect = 1e-10
Identities = 35/53 (66%), Positives = 37/53 (69%)
Frame = +3
Query: 33 FFLVDSIAQFSRGLKGDVCLVYSSAIQAVLPRLLSAAVPTGNAAQENRRQCLK 85
FFL+ + F K VC VYS AIQAVLPRLLSAA P GN QENRRQCLK
Sbjct: 249 FFLLTLLHSFLVV*KAMVCGVYSFAIQAVLPRLLSAAAPPGNTGQENRRQCLK 407
>TC216716 similar to UP|Q9ZS47 (Q9ZS47) Arbuscular mycorrhiza protein,
partial (73%)
Length = 1385
Score = 491 bits (1265), Expect = e-139
Identities = 231/322 (71%), Positives = 263/322 (80%), Gaps = 2/322 (0%)
Frame = +3
Query: 188 EVHEMTSIIDKHRHILEDVDGELEMEDVSPSRDVEMNSFSNVDRGNATQFENNIHLPSAP 247
EV+EMTS I+KHRHILEDVDGELEMEDV+PS VEMNS NVD GNA Q E N+ L AP
Sbjct: 9 EVYEMTSAIEKHRHILEDVDGELEMEDVAPSNAVEMNSICNVDTGNAKQCEKNLPLSFAP 188
Query: 248 PHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHTESQCVKD 307
HQ V SS PPP PPPPPPP PPPPP PM H + STSDP TV NS+G T SQ +KD
Sbjct: 189 LHQDVRSSSPPPPSFLPPPPPPPRPPPPP-PMSHHMPSTSDPYDTVVNSKGCTVSQTLKD 365
Query: 308 NPLHPMDRPLAAPRSSQPISNAVHHHAPEYREA--HISESDRSFNSFPVPHPVNYRHSDG 365
NPLH + +P+AAPR SQPIS+AVHH PEYRE H+ ES FNSFPVP P N+RH+DG
Sbjct: 366 NPLHSVAQPMAAPRHSQPISDAVHHLVPEYREMQMHMPESTCCFNSFPVPPPDNFRHTDG 545
Query: 366 VTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMEREHFYHN 425
VTMH++G+SIRPP+HVPSNQFSFV+GEQH +H+REVPPPPPYS+ QHFV+NMERE+FY N
Sbjct: 546 VTMHNKGYSIRPPQHVPSNQFSFVNGEQHVKHQREVPPPPPYSSSQHFVQNMERENFY-N 722
Query: 426 NHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRFPPRSMNH 485
NHERL+PPPY Y +RW+ P PYPGPRY ++ +P PYGCHPCE RIPDHGWRFPP+SMN
Sbjct: 723 NHERLRPPPYVYEDRWNGPAPYPGPRYQEKGVPPPYGCHPCESSRIPDHGWRFPPQSMNQ 902
Query: 486 RNSMPFRPPFEDAIPVTNRGSS 507
RNSMPFRPPFEDAIPV+NRG S
Sbjct: 903 RNSMPFRPPFEDAIPVSNRGPS 968
>TC220329 weakly similar to UP|Q9ZS47 (Q9ZS47) Arbuscular mycorrhiza protein,
partial (54%)
Length = 918
Score = 165 bits (418), Expect(2) = 6e-81
Identities = 76/120 (63%), Positives = 92/120 (76%)
Frame = +2
Query: 386 FSFVHGEQHARHRREVPPPPPYSNRQHFVENMEREHFYHNNHERLKPPPYDYRERWDVPP 445
F FV+GE + +REVPPPP YSN HF+ +M RE+ Y +NHERL+PP YDY+ERW+VPP
Sbjct: 476 FLFVNGEHQMKPQREVPPPPSYSNGHHFMPSMTREYGY-DNHERLRPP-YDYQERWNVPP 649
Query: 446 PYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRFPPRSMNHRNSMPFRPPFEDAIPVTNRG 505
GPRYHD +P+PYGCHP E P HGWRFPP SMN+R+S+ FRP FEDAIPV +RG
Sbjct: 650 -CSGPRYHDRGVPAPYGCHPSESFSFPGHGWRFPPPSMNYRDSLHFRPHFEDAIPVADRG 826
Score = 154 bits (389), Expect(2) = 6e-81
Identities = 83/161 (51%), Positives = 102/161 (62%), Gaps = 10/161 (6%)
Frame = +1
Query: 238 ENNIHLPSAPP-HQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP-------HLVSSTSDP 289
E N+ + PP Q +P SS PPP PPPPPP PPP LP+P H S+TSD
Sbjct: 1 EKNLPVSFGPPLPQDLPPSSPPPPSYRSPPPPPPTPPPSSLPIPPPPPATLHFKSATSDQ 180
Query: 290 CRTVFNSRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYRE--AHISESDR 347
+S+G +S VK N LHPM PLAAPR+SQPIS+AV + PE R+ + ES
Sbjct: 181 YHVAVDSKGFEDSLTVKANVLHPMAEPLAAPRNSQPISDAVQYTVPECRDMPMQMPESTC 360
Query: 348 SFNSFPVPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSF 388
SFN+FPV N R++DG TMH++G+SI PP HVPSNQFSF
Sbjct: 361 SFNTFPVQPTDNSRNTDGATMHNKGYSIPPPHHVPSNQFSF 483
>TC233624 similar to UP|Q8H3E3 (Q8H3E3) Transcription factor (PWWP domain
protein)-like protein, partial (10%)
Length = 677
Score = 260 bits (665), Expect = 9e-70
Identities = 136/178 (76%), Positives = 152/178 (84%), Gaps = 1/178 (0%)
Frame = +3
Query: 11 QVMEILADNLETESSLHRRVDLFFLVDSIAQFSRGLKGDVCLVYSSAIQAVLPRLLSAAV 70
+VMEI+ NLE ESSLHRRVDLFFLVDSIAQ SRGLKGD+ VY S ++AVLPRLLSAA
Sbjct: 159 KVMEIVVHNLEIESSLHRRVDLFFLVDSIAQCSRGLKGDIGGVYPSTMKAVLPRLLSAAA 338
Query: 71 PTGNAAQENRRQCLKVLRLWLERKILPEPMVRHHIRELDLY-SSVSAGVYSRRSLRTERA 129
P GNAA+ENRRQCLKVLRLWLERKILPEP++RHH++ELD Y SSVSAGV+S RSLR +R
Sbjct: 339 PPGNAAKENRRQCLKVLRLWLERKILPEPIIRHHMQELDSYSSSVSAGVHSHRSLRRDRP 518
Query: 130 LDDPIREMEGMHVDEYGSNSSLQLPGFCMPRMLKDEDDNEESDSDGGNFEAVTPEHNS 187
DDP+R+MEGM +DEYGSNSS QLPGFCMPRML D SDSDGG FEAVTPEH+S
Sbjct: 519 FDDPVRDMEGM-LDEYGSNSSFQLPGFCMPRMLGDGG----SDSDGGEFEAVTPEHDS 677
>BM519633
Length = 429
Score = 119 bits (297), Expect = 4e-27
Identities = 59/95 (62%), Positives = 71/95 (74%)
Frame = +1
Query: 11 QVMEILADNLETESSLHRRVDLFFLVDSIAQFSRGLKGDVCLVYSSAIQAVLPRLLSAAV 70
+V+E+L LETE+S HR+VDLFFLVDSI Q S KG Y +QA LPRLL AA
Sbjct: 142 EVVELLIRKLETETSFHRKVDLFFLVDSITQCSHNQKGIAGASYIPTVQAALPRLLGAAA 321
Query: 71 PTGNAAQENRRQCLKVLRLWLERKILPEPMVRHHI 105
P G +A+ENRRQCLKVLRLWLERKI PE ++RH++
Sbjct: 322 PPGASARENRRQCLKVLRLWLERKIFPESVLRHYM 426
>TC221951 UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75) (NGM-75),
complete
Length = 1179
Score = 66.6 bits (161), Expect = 3e-11
Identities = 69/274 (25%), Positives = 94/274 (34%), Gaps = 36/274 (13%)
Frame = +1
Query: 247 PPHQLVPQSSVPPPLAPPPPP--PPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHTESQC 304
PPH+ P +PPP PPP PP PPP PH
Sbjct: 202 PPHEKTPPEYLPPPHEKPPPEYLPPHEKPPPEYQPPH----------------------- 312
Query: 305 VKDNPLHPMDRPLAAPRSSQPISNAVHHH--APEYREAHISESDRSFNSFPVPH---PVN 359
+ P H P P +P + H PEY H ++ + PH P
Sbjct: 313 --EKPPHENPPPEHQPPHEKPPEHQPPHEKPPPEYEPPH----EKPPPEYQPPHEKPPPE 474
Query: 360 YR--HSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREV-----PPPPPYSNRQH 412
Y+ H + H PP H P ++ H H + E PPP Y Q
Sbjct: 475 YQPPHEKPPPEYQPPHEKPPPEHQPPHEKPPEHQPPHEKPPPEYQPPHEKPPPEYQPPQE 654
Query: 413 FVENMEREHFYHNNHER--------------LKPPPYD-----YRERWDVPPPYPGPRYH 453
+ + Y HE+ + PPPY+ Y ++ PPP P H
Sbjct: 655 KPPHEKPPPEYQPPHEKPPPEHQPPHEKPPPVYPPPYEKPPPVYEPPYEKPPPVVYPPPH 834
Query: 454 DEDMPSPYGCHPCEPPRI---PDHGWRFPPRSMN 484
++ P Y P E P + P +G R+PP N
Sbjct: 835 EK--PPIYEPPPLEKPPVYNPPPYG-RYPPSKKN 927
>TC204046 UP|Q39835 (Q39835) Extensin, complete
Length = 1627
Score = 63.9 bits (154), Expect = 2e-10
Identities = 72/279 (25%), Positives = 101/279 (35%), Gaps = 18/279 (6%)
Frame = +1
Query: 240 NIHLPSAPPHQLVPQSSVPPPLAPPPPP---PPPPPPPPPLPMPHLVSSTSDPCRTVFNS 296
++ LPS SS PPP PPPP PPPP P P+ S P +
Sbjct: 52 SLTLPSQTLADNYIYSSPPPPKHSPPPPYYYHSPPPPKHSPPPPYYYHSPPPPKHSPPPP 231
Query: 297 RGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPH 356
+ K +P P P P +H P + H ++S P P
Sbjct: 232 YYYHSPPPPKHSPPPPYYYHSPPPPKHSPPPPYYYHSPPPPK--HSPPPPYYYHSPPPPK 405
Query: 357 ----PVNYRHSDGVTMHDRG-----HSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPY 407
P Y HS H HS PP+H P + + H +H PPPP Y
Sbjct: 406 HSPPPPYYYHSPPPPKHSPPPPYYYHSPPPPKHSPPPPY-YYHSPPPPKHS---PPPPYY 573
Query: 408 SNRQHFVENMEREHFYHNNHERLKPPPYDYRERWDVPPP------YPGPRYHDEDMPSPY 461
+ ++ +Y+ + PPP ++ PPP P P Y P P
Sbjct: 574 YHSPPPPKHSPPPPYYYKS----PPPPPKKPYKYPSPPPPVYKYKSPPPPYKYPSPPPPP 741
Query: 462 GCHPCEPPRIPDHGWRFPPRSMNHRNSMPFRPPFEDAIP 500
+P PP P + ++ PP + S P PP++ P
Sbjct: 742 YKYPSPPP--PVYKYKSPPPPVYKYKSPP--PPYKYPSP 846
Score = 57.4 bits (137), Expect = 2e-08
Identities = 75/288 (26%), Positives = 95/288 (32%), Gaps = 29/288 (10%)
Frame = +1
Query: 242 HLPSAPPHQLVPQ---SSVPPPLAPPPPP-----PPPP---PPPP-----PLPMPHLVSS 285
H P P H P S PPP PPPP PPPP PPPP P P P
Sbjct: 481 HSPPPPKHSPPPPYYYHSPPPPKHSPPPPYYYHSPPPPKHSPPPPYYYKSPPPPPKKPYK 660
Query: 286 TSDPCRTVFNSRGHTESQCVKDNPLHPMDRPLAAP---RSSQPISNAVHHHAPEYREAHI 342
P V+ + P P P P + P + +P +
Sbjct: 661 YPSPPPPVYKYKSPPPPYKYPSPPPPPYKYPSPPPPVYKYKSPPPPVYKYKSPPPPYKYP 840
Query: 343 SESDRSFNSFPVPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVP 402
S + +P P P Y++ + S PP PS P
Sbjct: 841 SPPPPPYK-YPSPPPPVYKYKSPPPPVYKYKSPPPPYKYPS------------------P 963
Query: 403 PPPPYSNRQHFVENMEREHFYHNNHE-----RLKPPPYDYRERWDVPPPY-----PGPRY 452
PPPPY + + Y + + PPPY Y PPPY P P Y
Sbjct: 964 PPPPYK----YPSPPPPVYKYKSPPPPVYKYKSPPPPYKYPS--PPPPPYKYPSPPPPVY 1125
Query: 453 HDEDMPSPYGCHPCEPPRIPDHGWRFPPRSMNHRNSMPFRPPFEDAIP 500
+ P PY +P PP P + + PP + S P PP P
Sbjct: 1126KYKSPPPPYK-YPSPPP--PPYKYPSPPPPVYKYKSPP--PPVHSPPP 1254
Score = 53.5 bits (127), Expect = 2e-07
Identities = 69/273 (25%), Positives = 89/273 (32%), Gaps = 14/273 (5%)
Frame = +1
Query: 242 HLPSAPPHQLVPQ---SSVPPPLAPPPPP-----PPPP--PPPPPLPMPHLVSSTSDPCR 291
H P P H P S PPP PPPP PPPP PPP P+ S P
Sbjct: 289 HSPPPPKHSPPPPYYYHSPPPPKHSPPPPYYYHSPPPPKHSPPP----PYYYHSPPPPKH 456
Query: 292 TVFNSRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNS 351
+ + K +P P P P +H P + H + S
Sbjct: 457 SPPPPYYYHSPPPPKHSPPPPYYYHSPPPPKHSPPPPYYYHSPPPPK--HSPPPPYYYKS 630
Query: 352 FPVPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQ 411
P P Y++ + S PP PS PPPPPY
Sbjct: 631 PPPPPKKPYKYPSPPPPVYKYKSPPPPYKYPS------------------PPPPPY---- 744
Query: 412 HFVENMEREHFYHNNHERLKPPPYDYRERWDVPPPY----PGPRYHDEDMPSPYGCHPCE 467
+ PPP Y+ + PP Y P P Y P P +P
Sbjct: 745 ----------------KYPSPPPPVYKYKSPPPPVYKYKSPPPPYKYPSPPPPPYKYPSP 876
Query: 468 PPRIPDHGWRFPPRSMNHRNSMPFRPPFEDAIP 500
PP P + ++ PP + S P PP++ P
Sbjct: 877 PP--PVYKYKSPPPPVYKYKSPP--PPYKYPSP 963
Score = 40.4 bits (93), Expect = 0.002
Identities = 21/60 (35%), Positives = 27/60 (45%), Gaps = 16/60 (26%)
Frame = +1
Query: 246 APPHQLVPQSSVPPPLAPPPPPPPP---PPPPPPL-------------PMPHLVSSTSDP 289
+PP + S PPP P PPPPP P PPPP+ P PH + ++ P
Sbjct: 1105 SPPPPVYKYKSPPPPYKYPSPPPPPYKYPSPPPPVYKYKSPPPPVHSPPPPHYIYASPPP 1284
Score = 32.7 bits (73), Expect = 0.40
Identities = 18/41 (43%), Positives = 18/41 (43%), Gaps = 9/41 (21%)
Frame = +1
Query: 245 SAPPHQLVPQSSVPP---PLAPPP------PPPPPPPPPPP 276
S PP P PP P PPP PPPP PPPP
Sbjct: 1135 SPPPPYKYPSPPPPPYKYPSPPPPVYKYKSPPPPVHSPPPP 1257
Score = 32.0 bits (71), Expect = 0.69
Identities = 18/43 (41%), Positives = 22/43 (50%), Gaps = 10/43 (23%)
Frame = +1
Query: 244 PSAPPHQLVPQSSVPPPL----APPPPPPPPPPP------PPP 276
P PP++ S PPP+ +PPPP PPPP PPP
Sbjct: 1165 PPPPPYKY---PSPPPPVYKYKSPPPPVHSPPPPHYIYASPPP 1284
Score = 31.2 bits (69), Expect = 1.2
Identities = 15/33 (45%), Positives = 17/33 (51%)
Frame = +1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPP 276
PS PP +S PP +PPPP PPPP
Sbjct: 1189 PSPPPPVYKYKSPPPPVHSPPPPHYIYASPPPP 1287
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 63.5 bits (153), Expect = 2e-10
Identities = 27/44 (61%), Positives = 28/44 (63%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTS 287
P PP P S PPP PPPPPPPPPPPPPP P+P L S S
Sbjct: 357 PPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPPPPPLPPLPXSLS 226
Score = 60.1 bits (144), Expect = 2e-09
Identities = 24/37 (64%), Positives = 24/37 (64%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPPL PPPPPPPPPPPPPP P P
Sbjct: 153 PPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPP 43
Score = 60.1 bits (144), Expect = 2e-09
Identities = 24/37 (64%), Positives = 24/37 (64%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPPL PPPPPPPPPPPPPP P P
Sbjct: 208 PPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPP 98
Score = 59.3 bits (142), Expect = 4e-09
Identities = 26/46 (56%), Positives = 27/46 (58%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDP 289
P PP P +PPP PPPPPPPPPPPPPP P P TS P
Sbjct: 131 PPPPPPPPPPSPPLPPPPPPPPPPPPPPPPPPPPPPP---GQTSPP 3
Score = 59.3 bits (142), Expect = 4e-09
Identities = 24/37 (64%), Positives = 24/37 (64%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
PS PP PPP PPPPPPPPPPPPPPLP P
Sbjct: 256 PSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPLPPP 146
Score = 59.3 bits (142), Expect = 4e-09
Identities = 24/38 (63%), Positives = 24/38 (63%)
Frame = -3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
LP PP P PPP PPPPPPPPPPPPPP P P
Sbjct: 375 LPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPPP 262
Score = 58.9 bits (141), Expect = 5e-09
Identities = 24/39 (61%), Positives = 24/39 (61%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHL 282
P PP P PPP PPPPPPPPPPPPPP P P L
Sbjct: 206 PPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPSPPL 90
Score = 58.9 bits (141), Expect = 5e-09
Identities = 23/37 (62%), Positives = 24/37 (64%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P +PPP PPPPPPPPPPPPPP P P
Sbjct: 196 PPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSP 86
Score = 58.9 bits (141), Expect = 5e-09
Identities = 24/37 (64%), Positives = 24/37 (64%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P S PPP PPPPPPPPPPP PPLP P
Sbjct: 191 PPPPPPPPPPPPSPPPPPPPPPPPPPPPPPSPPLPPP 81
Score = 58.5 bits (140), Expect = 7e-09
Identities = 23/37 (62%), Positives = 24/37 (64%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P +PPP PPPPPPPPPPPPPP P P
Sbjct: 141 PPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPPP 31
Score = 58.5 bits (140), Expect = 7e-09
Identities = 23/37 (62%), Positives = 24/37 (64%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P S+PP PPPPPPPPPPPPPP P P
Sbjct: 268 PPPPPSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPP 158
Score = 58.2 bits (139), Expect = 9e-09
Identities = 23/37 (62%), Positives = 24/37 (64%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP +PPPPPPPPPPPPPP P P
Sbjct: 145 PPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPPP 35
Score = 58.2 bits (139), Expect = 9e-09
Identities = 25/46 (54%), Positives = 27/46 (58%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDP 289
P PP P PPP PPPPPPPPPPPPPP P P ++ S P
Sbjct: 138 PPPPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPPPPPPRANLSPP 1
Score = 57.4 bits (137), Expect = 2e-08
Identities = 23/37 (62%), Positives = 23/37 (62%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPPPPPPPPPPP P P
Sbjct: 211 PPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPPP 101
Score = 57.4 bits (137), Expect = 2e-08
Identities = 24/39 (61%), Positives = 25/39 (63%), Gaps = 1/39 (2%)
Frame = -1
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPP-PPPPPPPPLPMP 280
LP PP +P PPP PPPPPP PPPPPPPP P P
Sbjct: 407 LPPPPPPSSLPSPPPPPPPPPPPPPPLPPPPPPPPXPPP 291
Score = 57.0 bits (136), Expect = 2e-08
Identities = 23/37 (62%), Positives = 23/37 (62%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP L P PPP PPPPPPPPPP PPP P P
Sbjct: 178 PPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPP 68
Score = 56.6 bits (135), Expect = 3e-08
Identities = 25/38 (65%), Positives = 25/38 (65%)
Frame = -1
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
LPS PP P PPPL PPPPPPP PPPPPP P P
Sbjct: 380 LPSPPPPP-PPPPPPPPPLPPPPPPPPXPPPPPPPPPP 270
Score = 56.6 bits (135), Expect = 3e-08
Identities = 24/38 (63%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Frame = -1
Query: 244 PSAPPH-QLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P +PP L P PPP PPPPPPPPPPPPPP P P
Sbjct: 257 PLSPPSLXLSPP*KTPPPPPPPPPPPPPPPPPPPSPPP 144
Score = 56.2 bits (134), Expect = 3e-08
Identities = 24/38 (63%), Positives = 25/38 (65%), Gaps = 1/38 (2%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAP-PPPPPPPPPPPPPLPMP 280
P PP P S +PPP P PPPPPPPPPPPPP P P
Sbjct: 361 PPPPPPPPPPPSPLPPPPPPXPPPPPPPPPPPPPPPSP 248
Score = 55.8 bits (133), Expect = 4e-08
Identities = 23/38 (60%), Positives = 23/38 (60%)
Frame = -2
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
LP PP P PPP PP PPPPPPPPPPP P P
Sbjct: 157 LPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPP 44
Score = 55.8 bits (133), Expect = 4e-08
Identities = 23/39 (58%), Positives = 24/39 (60%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHL 282
P PP P +PPP PPP PPPPPPPPPP P P L
Sbjct: 368 PPPPPPPPPPPPPLPPPPPPPPXPPPPPPPPPPPPPPPL 252
Score = 55.8 bits (133), Expect = 4e-08
Identities = 23/39 (58%), Positives = 23/39 (58%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHL 282
P PP P PP PPPPPPPPPPPPPP P P L
Sbjct: 271 PPPPPPSPPPPXXSLPPKKPPPPPPPPPPPPPPPPPPPL 155
Score = 55.8 bits (133), Expect = 4e-08
Identities = 23/38 (60%), Positives = 23/38 (60%)
Frame = -3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
LP PP P PPP P PPPPPPPPPPPP P P
Sbjct: 384 LPPLPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPP 271
Score = 55.5 bits (132), Expect = 6e-08
Identities = 24/41 (58%), Positives = 25/41 (60%), Gaps = 4/41 (9%)
Frame = -3
Query: 244 PSAPPHQLVPQS----SVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP +P S PPP PPPPPPPPPPPPPP P P
Sbjct: 267 PPPPPLPPLPXSLSPLKNPPPPPPPPPPPPPPPPPPPFPPP 145
Score = 55.5 bits (132), Expect = 6e-08
Identities = 22/37 (59%), Positives = 23/37 (61%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP +P PPP PPPPPPPPPPP PP P P
Sbjct: 181 PPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPP 71
Score = 55.1 bits (131), Expect = 8e-08
Identities = 24/46 (52%), Positives = 27/46 (58%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDP 289
P PP P PPP PPPPPPPPPPPPP P+P +S +P
Sbjct: 342 PPPPPPPSPPPPPPPPP--PPPPPPPPPPPPPLPPLPXSLSPLKNP 211
Score = 54.3 bits (129), Expect = 1e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPPPPPPPPPP P P
Sbjct: 148 PPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPPP 38
Score = 54.3 bits (129), Expect = 1e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP P PPPPPPPPPPPP P P
Sbjct: 151 PPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPPPPP 41
Score = 54.3 bits (129), Expect = 1e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPP PPPPPPPP P P
Sbjct: 163 PPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPP 53
Score = 53.9 bits (128), Expect = 2e-07
Identities = 22/36 (61%), Positives = 23/36 (63%)
Frame = -2
Query: 245 SAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
S PP + P PPP PPPPPPP PPPPPP P P
Sbjct: 232 SLPPKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPP 125
Score = 53.9 bits (128), Expect = 2e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPP PPPPPPPPP P P
Sbjct: 160 PLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPPPPPP 50
Score = 53.9 bits (128), Expect = 2e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPP PPPPPPPPPP P P
Sbjct: 223 PKKPPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPP 113
Score = 53.9 bits (128), Expect = 2e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPPPPP PPPPP P P
Sbjct: 172 PPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPPPPPPP 62
Score = 53.9 bits (128), Expect = 2e-07
Identities = 23/38 (60%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPP-PPPPPPPPLPMP 280
P PP P PPP PPPPPP PPPPPPPP P P
Sbjct: 177 PPPPPPPFPPPPPPPPPPPPPPPPPLPPPPPPPPPPPP 64
Score = 53.9 bits (128), Expect = 2e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPPPPPPPPPP P P
Sbjct: 187 PPPPPPPPPPLPPPPPPPPPPPPPPPPPPPPPSPPPP 77
Score = 53.5 bits (127), Expect = 2e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPPPPPPPPPP P P
Sbjct: 214 PPPPPPPPPPPPPPPPPPPLPPPPPPPPPPPPPPPPP 104
Score = 53.5 bits (127), Expect = 2e-07
Identities = 24/41 (58%), Positives = 24/41 (58%), Gaps = 4/41 (9%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPP----PPPPLPMP 280
P PP P S PPP PPPPPPPPPP PPPP P P
Sbjct: 188 PPPPPPPPPPPSPPPPPPPPPPPPPPPPPSPPLPPPPPPPP 66
Score = 53.1 bits (126), Expect = 3e-07
Identities = 23/38 (60%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAP-PPPPPPPPPPPPPLPMP 280
P PP P PPP P PPPPPPPPPPPPP P P
Sbjct: 152 PPPPPPPPPPPPPPPPPSPPLPPPPPPPPPPPPPPPPP 39
Score = 53.1 bits (126), Expect = 3e-07
Identities = 22/37 (59%), Positives = 22/37 (59%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P PPP PPPPPPPPPPP PP P P
Sbjct: 192 PPPPPPPPPPPPFPPPPPPPPPPPPPPPPPLPPPPPP 82
Score = 52.8 bits (125), Expect = 4e-07
Identities = 24/41 (58%), Positives = 25/41 (60%), Gaps = 1/41 (2%)
Frame = -3
Query: 243 LPSAPPHQLVPQSSVPPPLAPP-PPPPPPPPPPPPLPMPHL 282
L + PP P PPP PP PPPPPPPPPPPP P P L
Sbjct: 222 LKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPPPPPPPPPPL 100
Score = 52.8 bits (125), Expect = 4e-07
Identities = 24/48 (50%), Positives = 25/48 (52%), Gaps = 11/48 (22%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPP-----------PPPPPPPPPPPPLPMP 280
P PP P PPPL+PP PPPPPPPPPPPP P P
Sbjct: 305 PXPPPPPPPPPPPPPPPLSPPSLXLSPP*KTPPPPPPPPPPPPPPPPP 162
Score = 52.4 bits (124), Expect = 5e-07
Identities = 23/38 (60%), Positives = 23/38 (60%), Gaps = 1/38 (2%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPP-PPPPPPPPPLPMP 280
P PP P P PL PPPPP PPPPPPPPP P P
Sbjct: 373 PPPPPPPPPPPPPPPSPLPPPPPPXPPPPPPPPPPPPP 260
Score = 52.0 bits (123), Expect = 6e-07
Identities = 23/41 (56%), Positives = 23/41 (56%), Gaps = 4/41 (9%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPP----PPPPPPPPPLPMP 280
P PP P PPP PPPPP PPPPPPPPP P P
Sbjct: 173 PPPPPPSPPPPPPPPPPPPPPPPPSPPLPPPPPPPPPPPPP 51
Score = 52.0 bits (123), Expect = 6e-07
Identities = 22/37 (59%), Positives = 23/37 (61%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
PS+ L P PPP PPPPPP PPPPPPP P P
Sbjct: 396 PSSLLPPLPPPPPPPPPPPPPPPPPSPPPPPPPPPPP 286
Score = 51.6 bits (122), Expect = 8e-07
Identities = 23/39 (58%), Positives = 24/39 (60%), Gaps = 2/39 (5%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAP--PPPPPPPPPPPPPLPMP 280
PS PP P PPP +P PPPPP PPPPPPP P P
Sbjct: 382 PSPPPPPPPPPPPPPPPPSPLPPPPPPXPPPPPPPPPPP 266
Score = 50.1 bits (118), Expect = 2e-06
Identities = 23/45 (51%), Positives = 25/45 (55%), Gaps = 8/45 (17%)
Frame = -3
Query: 244 PSAPPHQLVP--------QSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP L P ++ PPP PPPPPPPPPPPP P P P
Sbjct: 273 PPPPPPPLPPLPXSLSPLKNPPPPPPPPPPPPPPPPPPPFPPPPP 139
Score = 49.7 bits (117), Expect = 3e-06
Identities = 21/36 (58%), Positives = 22/36 (60%)
Frame = -3
Query: 245 SAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
S P + P PPP PPPPPPPP PPPPP P P
Sbjct: 234 SLSPLKNPPPPPPPPPPPPPPPPPPPFPPPPPPPPP 127
Score = 49.3 bits (116), Expect = 4e-06
Identities = 23/43 (53%), Positives = 24/43 (55%), Gaps = 8/43 (18%)
Frame = -2
Query: 246 APPHQLVPQSSVPPPLAPPPPPPPP--------PPPPPPLPMP 280
+PP L P S PPP PPPPPPPP PP PPP P P
Sbjct: 406 SPPPLLPPPSPPPPPPPPPPPPPPPPSPLPPPPPPXPPPPPPP 278
Score = 49.3 bits (116), Expect = 4e-06
Identities = 23/47 (48%), Positives = 23/47 (48%), Gaps = 10/47 (21%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPP----------PPPPPPPPPPPPLPMP 280
P PP P PPP PP PPPPPPPPPPPP P P
Sbjct: 306 PPPPPPPPPPPPPPPPPPLPPLPXSLSPLKNPPPPPPPPPPPPPPPP 166
Score = 48.5 bits (114), Expect = 7e-06
Identities = 23/50 (46%), Positives = 23/50 (46%), Gaps = 13/50 (26%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPP-------------PPPPPLPMP 280
P PP P PPP PPPPPPPPP PPPPP P P
Sbjct: 336 PPPPPSPPPPPPPPPPPPPPPPPPPPPPLPPLPXSLSPLKNPPPPPPPPP 187
Score = 48.1 bits (113), Expect = 9e-06
Identities = 23/47 (48%), Positives = 23/47 (48%), Gaps = 9/47 (19%)
Frame = -2
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPP---------PPPPPPLPMP 280
LP PP P PPP PPP PPPP PPPPPP P P
Sbjct: 322 LPPPPPPXPPPPPPPPPPPPPPPSPPPPXXSLPPKKPPPPPPPPPPP 182
Score = 47.4 bits (111), Expect = 2e-05
Identities = 23/51 (45%), Positives = 23/51 (45%), Gaps = 14/51 (27%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPP--------------PPPPPPPLPMP 280
P PP P PPP PPPPPPP PPPPPPP P P
Sbjct: 332 PLPPPPPPPPXPPPPPPPPPPPPPPPLSPPSLXLSPP*KTPPPPPPPPPPP 180
Score = 32.0 bits (71), Expect = 0.69
Identities = 14/30 (46%), Positives = 14/30 (46%)
Frame = -1
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPPP 272
LP PP P PPP PPPP PP
Sbjct: 92 LPPPPPPPPPPPPPPPPPPPPPPPGQTSPP 3
>TC214106 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (22%)
Length = 1296
Score = 61.6 bits (148), Expect = 8e-10
Identities = 72/256 (28%), Positives = 84/256 (32%), Gaps = 4/256 (1%)
Frame = +1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPP---PPLPMPHLVSSTSDPCRTVFNSRGHT 300
PS PP PPP +PPPP PPPPPPP PP P+ + SS P + H+
Sbjct: 70 PSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVFSPPPPVQYYYSSPPPPQHSPPPPPPHS 249
Query: 301 ESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPE-YREAHISESDRSFNSFPVPHPVN 359
P P P P S P VH P Y S P P P
Sbjct: 250 PP------PPPPSPPPPVYPYLSPPPPPPVHSPPPPVYSPPPPPPSPPPCIEPPPPPPP- 408
Query: 360 YRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMER 419
I PP PS E+H+ PPP P+ H +
Sbjct: 409 --------------CIEPPPPPPSPPPC----EEHSP-----PPPSPHPAPYHPPPSPSP 519
Query: 420 EHFYHNNHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRFP 479
PPP Y PP P P YH P P PP P + P
Sbjct: 520 P-----------PPPVQYNSPPPPSPPPPTPVYHYNSPPP-----PSFPPPTPVYEGPLP 651
Query: 480 PRSMNHRNSMPFRPPF 495
P + + P PPF
Sbjct: 652 P-VIGVSYASPPPPPF 696
Score = 58.2 bits (139), Expect = 9e-09
Identities = 76/278 (27%), Positives = 89/278 (31%), Gaps = 22/278 (7%)
Frame = +1
Query: 245 SAPPHQLVPQSSVPPPLAPPPPPPPPPP---------PPPPLPMPHLVSSTSDPCRTVFN 295
S PP P S PP +PPPPPP PPP PPPP P P S P VF
Sbjct: 1 SPPPPSPPPPSPPPPVYSPPPPPPSPPPPSPTYCVRSPPPPSPPP---PSPPPPPPPVF- 168
Query: 296 SRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVP 355
+P P+ ++P Q H+P + S P P
Sbjct: 169 ------------SPPPPVQYYYSSPPPPQHSPPPPPPHSPPPPPPSPPPPVYPYLSPPPP 312
Query: 356 HPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVE 415
PV H + P P + + E PPPPP
Sbjct: 313 PPV----------HSPPPPVYSPPPPPPSPPPCIEPPPPPPPCIEPPPPPP--------- 435
Query: 416 NMEREHFYHNNHERLKPPPYDYRERWDVPPPYPGPR-YHDEDMPSP------YGCHPCEP 468
PPP E PPP P P YH PSP Y P P
Sbjct: 436 ---------------SPPPC---EEHSPPPPSPHPAPYHPPPSPSPPPPPVQYNSPP--P 555
Query: 469 PRIPDHGWRFPPRSMNHRNSMP---FRPP---FEDAIP 500
P P PP + H NS P F PP +E +P
Sbjct: 556 PSPP------PPTPVYHYNSPPPPSFPPPTPVYEGPLP 651
Score = 55.1 bits (131), Expect = 8e-08
Identities = 50/170 (29%), Positives = 59/170 (34%), Gaps = 3/170 (1%)
Frame = +1
Query: 241 IHLPSAPPHQLVPQSSVPPPLAPPPPPPPP---PPPPPPLPMPHLVSSTSDPCRTVFNSR 297
+H P P + P PPP PPPPPPP PPPPPP P P S P
Sbjct: 319 VHSPPPPVYSPPPPPPSPPPCIEPPPPPPPCIEPPPPPPSPPP--CEEHSPP-------- 468
Query: 298 GHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHP 357
P P P P S P V +++P S P P P
Sbjct: 469 -----------PPSPHPAPYHPPPSPSPPPPPVQYNSPPP------------PSPPPPTP 579
Query: 358 VNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPY 407
V + +S S PP V V G +A PPPPP+
Sbjct: 580 VYHYNSPPPP------SFPPPTPVYEGPLPPVIGVSYAS-----PPPPPF 696
Score = 30.0 bits (66), Expect = 2.6
Identities = 24/67 (35%), Positives = 26/67 (37%), Gaps = 2/67 (2%)
Frame = +1
Query: 444 PPPYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRFPPRSMNHRNSMPFRPP--FEDAIPV 501
PPP P P + SP P PP P + R PP S P PP F PV
Sbjct: 7 PPPSPPPPSPPPPVYSPPPPPPSPPPPSPTYCVRSPPPPSPPPPSPPPPPPPVFSPPPPV 186
Query: 502 TNRGSSP 508
SSP
Sbjct: 187 QYYYSSP 207
>TC224979 similar to UP|Q09083 (Q09083) Hydroxyproline-rich glycoprotein
precursor, partial (32%)
Length = 693
Score = 58.2 bits (139), Expect = 9e-09
Identities = 70/271 (25%), Positives = 93/271 (33%), Gaps = 15/271 (5%)
Frame = +1
Query: 245 SAPPHQLVPQSSVPPPLAPPPP-------PPPPPPPPPPLPMPHLVSSTSDPCRTVFNSR 297
S PP + PPP +PPPP PPPP PPP P+ S P + +
Sbjct: 73 SPPPPKKPYYYHSPPPPSPPPPYKKPYYYHSPPPPSPPPPKKPYYYHSPPPPPKKPYYYH 252
Query: 298 GHTESQCVKDNPLHPMDRPL----AAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFP 353
+P P +P P S P ++H+P S P
Sbjct: 253 SPP-----PPSPPPPYKKPYYYHSPPPPSPPPPKKPYYYHSPPP------------PSPP 381
Query: 354 VPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHF 413
P P Y HS PP P + + H PPPPP
Sbjct: 382 PPKPYYY------------HSPPPP---PKKKPYYYHSP---------PPPPP------- 468
Query: 414 VENMEREHFYHNNHERLKPPPYDYRERWDVPP----PYPGPRYHDEDMPSPYGCHPCEPP 469
++ ++YH+ PPP ++ PP PYP P H P P HP PP
Sbjct: 469 ----KKPYYYHS-----PPPPPKKPYKYPSPPPAVHPYPDPHPH----PHP---HP-HPP 597
Query: 470 RIPDHGWRFPPRSMNHRNSMPFRPPFEDAIP 500
IP + PP P++ P+ A P
Sbjct: 598 YIPHPVYHSPP-------PPPYKKPYIYASP 669
Score = 57.4 bits (137), Expect = 2e-08
Identities = 63/258 (24%), Positives = 88/258 (33%), Gaps = 28/258 (10%)
Frame = +1
Query: 251 LVPQSSVPPPLAP-----PPPP--------PPPPPPPPPLPMPHLVSSTSDPCRTVFNSR 297
L+P S PPP P PPPP PPPP PPPP P+ S P S
Sbjct: 22 LLPAPSPPPPKKPYYYHSPPPPKKPYYYHSPPPPSPPPPYKKPYYYHSPPPP------SP 183
Query: 298 GHTESQCVKDNPLHPMDRPL-----AAPRSSQPISNAVHHHAPEYREAHISESDRSFNSF 352
+ +P P +P P P ++H+P + ++S
Sbjct: 184 PPPKKPYYYHSPPPPPKKPYYYHSPPPPSPPPPYKKPYYYHSPPPPSPPPPKKPYYYHSP 363
Query: 353 PVPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQH 412
P P P + HS PP P + + H PPPPP
Sbjct: 364 PPPSPPPPKPY-------YYHSPPPP---PKKKPYYYHSP---------PPPPP------ 468
Query: 413 FVENMEREHFYHNNHERLKPPPYDYRERWDVPPP----------YPGPRYHDEDMPSPYG 462
++ ++YH+ PPP ++ PPP +P P H +P P
Sbjct: 469 -----KKPYYYHS-----PPPPPKKPYKYPSPPPAVHPYPDPHPHPHPHPHPPYIPHPVY 618
Query: 463 CHPCEPPRIPDHGWRFPP 480
P PP + + PP
Sbjct: 619 HSPPPPPYKKPYIYASPP 672
Score = 50.1 bits (118), Expect = 2e-06
Identities = 51/200 (25%), Positives = 69/200 (34%), Gaps = 6/200 (3%)
Frame = +1
Query: 244 PSAPPHQLVPQS-SVPPPLAPPPPPPP-----PPPPPPPLPMPHLVSSTSDPCRTVFNSR 297
PS PP P PPP +PPPP P PPPP PP P P+ S P
Sbjct: 265 PSPPPPYKKPYYYHSPPPPSPPPPKKPYYYHSPPPPSPPPPKPYYYHSPPPP-------- 420
Query: 298 GHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHP 357
K P + P P+ ++H+P + +P P P
Sbjct: 421 -------PKKKPYYYHSPPPPPPKKPY------YYHSPP-------PPPKKPYKYPSPPP 540
Query: 358 VNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENM 417
+ + D H H P ++P H H+ PPPPPY
Sbjct: 541 AVHPYPDP---HPHPHPHPHPPYIP-------HPVYHS------PPPPPY---------- 642
Query: 418 EREHFYHNNHERLKPPPYDY 437
++ + Y + PPPY Y
Sbjct: 643 KKPYIYAS-----PPPPYHY 687
Score = 45.1 bits (105), Expect = 8e-05
Identities = 40/137 (29%), Positives = 53/137 (38%), Gaps = 18/137 (13%)
Frame = +1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPP-------PPPPP---------PPPPPLPMPHLVSSTS 287
PS PP + PPP +PPPP PPPPP PPPPP P+ S
Sbjct: 319 PSPPPPKKPYYYHSPPPPSPPPPKPYYYHSPPPPPKKKPYYYHSPPPPPPKKPYYYHSPP 498
Query: 288 DPCRTVFNSRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHH--APEYREAHISES 345
P + + + +P HP P P I + V+H P Y++ +I S
Sbjct: 499 PPPKKPYKYPSPPPAVHPYPDP-HPHPHPHPHP---PYIPHPVYHSPPPPPYKKPYIYAS 666
Query: 346 DRSFNSFPVPHPVNYRH 362
P P +Y H
Sbjct: 667 P--------PPPYHY*H 693
Score = 36.6 bits (83), Expect = 0.028
Identities = 20/64 (31%), Positives = 25/64 (38%), Gaps = 16/64 (25%)
Frame = +1
Query: 242 HLPSAPPHQLVPQSSVPPPLAPPPPPPPPP----------------PPPPPLPMPHLVSS 285
H P PP + S PP + P P P P P PPPPP P++ +S
Sbjct: 487 HSPPPPPKKPYKYPSPPPAVHPYPDPHPHPHPHPHPPYIPHPVYHSPPPPPYKKPYIYAS 666
Query: 286 TSDP 289
P
Sbjct: 667 PPPP 678
>TC218632 similar to UP|GRP1_PHAVU (P10495) Glycine-rich cell wall structural
protein 1.0 precursor (GRP 1.0), partial (27%)
Length = 447
Score = 57.8 bits (138), Expect = 1e-08
Identities = 23/37 (62%), Positives = 24/37 (64%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP L P + PPP APPPP PPPPPPPPP P
Sbjct: 196 PWPPPSPLPPPAPYPPPRAPPPPEPPPPPPPPPTAPP 86
Score = 43.1 bits (100), Expect = 3e-04
Identities = 23/49 (46%), Positives = 23/49 (46%), Gaps = 9/49 (18%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPP---------PPPPPPPPPPPLPMPHLV 283
P APP P PPP APPP P PPP PPP P P LV
Sbjct: 148 PRAPPPPEPPPPPPPPPTAPPPYPPPAPCPAPSPPPYPPPCAPPAPPLV 2
Score = 36.2 bits (82), Expect = 0.036
Identities = 14/23 (60%), Positives = 14/23 (60%)
Frame = -3
Query: 258 PPPLAPPPPPPPPPPPPPPLPMP 280
PPP PP P PPP P PPP P
Sbjct: 202 PPPWPPPSPLPPPAPYPPPRAPP 134
Score = 28.9 bits (63), Expect = 5.8
Identities = 22/74 (29%), Positives = 23/74 (30%), Gaps = 2/74 (2%)
Frame = -3
Query: 400 EVPPPPPYSNRQHFVENMEREHFYHNNHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPS 459
E PPPPP PPP PPPYP P P
Sbjct: 127 EPPPPPP-------------------------PPP-------TAPPPYPPPAPCPAPSPP 44
Query: 460 PYG--CHPCEPPRI 471
PY C P PP +
Sbjct: 43 PYPPPCAPPAPPLV 2
>TC224546 similar to UP|Q41707 (Q41707) Extensin class 1 protein precursor,
partial (67%)
Length = 1240
Score = 57.4 bits (137), Expect = 2e-08
Identities = 70/278 (25%), Positives = 96/278 (34%), Gaps = 14/278 (5%)
Frame = +3
Query: 245 SAPPHQLVPQSSV----PPPLAPPPPPP-----PPPPPPPPLPMPHLVSSTSDPCRTVFN 295
S PP VP+ PPP +P PPPP PPPP P P P P+ S P +
Sbjct: 204 SPPPPSPVPKPPYYYHSPPPPSPSPPPPYYYKSPPPPSPVPKP-PYYYHSPPPPSPSPPP 380
Query: 296 SRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVP 355
+ +P P P S P ++H+P + H+
Sbjct: 381 PYYYKSPPPPSPSPPPPYYYKSPPPPSPSP-PPPYYYHSPPPPKEHV------------- 518
Query: 356 HPVNYRHS---DGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQH 412
HP Y HS + + PP PS + + H+ PPPPY
Sbjct: 519 HPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYY---HSPPPPSPSPPPPY----- 674
Query: 413 FVENMEREHFYHN--NHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCEPPR 470
+YH+ PPPY Y+ P P P P Y+ + P P P
Sbjct: 675 ---------YYHSPPPPSPSPPPPYYYKSP-PPPSPSPPPPYYYQSPPPP------SPTP 806
Query: 471 IPDHGWRFPPRSMNHRNSMPFRPPFEDAIPVTNRGSSP 508
P + ++ PP +H PP+ P S P
Sbjct: 807 HPPYYYKSPPPPTSH------PPPYHYVSPPPPSPSPP 902
Score = 54.7 bits (130), Expect = 1e-07
Identities = 70/273 (25%), Positives = 87/273 (31%), Gaps = 5/273 (1%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPP---PPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHT 300
PS PP S PPP PPPP PPPP P P P+ S P V +
Sbjct: 366 PSPPPPYYY--KSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPPPKEHVHPPYYYH 539
Query: 301 ESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHPVNY 360
+P P P S P +H P P P P Y
Sbjct: 540 SPPPPSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPP--------------PSPSPPPPYY 677
Query: 361 RHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMERE 420
HS PP PS + + + PPPP Y
Sbjct: 678 YHS-------------PPPPSPSPPPPYYY--KSPPPPSPSPPPPYYYQSPPPPSPTPHP 812
Query: 421 HFYHNN--HERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRF 478
+Y+ + PPPY Y P P P P YH P P P P + ++
Sbjct: 813 PYYYKSPPPPTSHPPPYHYVSP-PPPSPSPPPPYHYTSPPPP------SPAPAPKYIYKS 971
Query: 479 PPRSMNHRNSMPFRPPFEDAIPVTNRGSSPFTI 511
PP + S P PP + + +PFTI
Sbjct: 972 PPPPVYIYASPP--PPIYK*VRI---AEAPFTI 1055
Score = 51.2 bits (121), Expect = 1e-06
Identities = 71/262 (27%), Positives = 95/262 (36%), Gaps = 26/262 (9%)
Frame = +3
Query: 245 SAPPHQLVPQSSV----PPPLAPPPPPP-----PPPPPPPPLPMPHLVSSTSDPCRTVFN 295
S PP VP+ PPP +P P PP PPPP P P P P+ S P +
Sbjct: 12 SPPPPSPVPKPPYYYQSPPPPSPIPKPPYYYHSPPPPSPSP-PPPYYYKSPPPP-----S 173
Query: 296 SRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVP 355
P P+ +P S P S + P Y ++ S PVP
Sbjct: 174 PSPPPPYYYKSPPPPSPVPKPPYYYHSPPPPSPS--PPPPYYYKSPPPPS-------PVP 326
Query: 356 HPVNYRHS---DGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQH 412
P Y HS + + PP PS + + + PPP Y +
Sbjct: 327 KPPYYYHSPPPPSPSPPPPYYYKSPPPPSPSPPPPYYY-KSPPPPSPSPPPPYYYHSPPP 503
Query: 413 FVENMEREHFYHN--NHERLKPPPYDYRERWDVPPPYPGP----RYHDEDMPS-----PY 461
E++ ++YH+ PPPY Y+ PPP P P YH PS PY
Sbjct: 504 PKEHVHPPYYYHSPPPPSPSPPPPYYYK---SPPPPSPSPPPPYYYHSPPPPSPSPPPPY 674
Query: 462 GCH---PCEPPRIPDHGWRFPP 480
H P P P + ++ PP
Sbjct: 675 YYHSPPPPSPSPPPPYYYKSPP 740
Score = 45.8 bits (107), Expect = 5e-05
Identities = 63/244 (25%), Positives = 77/244 (30%), Gaps = 10/244 (4%)
Frame = +3
Query: 238 ENNIHLP----SAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPL----PMPHLVSSTSDP 289
+ ++H P S PP P S PPP PPPP P PPPP P P S S P
Sbjct: 507 KEHVHPPYYYHSPPP----PSPSPPPPYYYKSPPPPSPSPPPPYYYHSPPP---PSPSPP 665
Query: 290 CRTVFNSRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSF 349
++S P P P P + P Y ++ S
Sbjct: 666 PPYYYHS------------PPPPSPSPPPPYYYKSPPPPSPSPPPPYYYQSPPPPS---- 797
Query: 350 NSFPVPH-PVNYRHSDGVTMHDRG-HSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPY 407
P PH P Y+ T H H + PP PS PPPPY
Sbjct: 798 ---PTPHPPYYYKSPPPPTSHPPPYHYVSPPPPSPS-------------------PPPPY 911
Query: 408 SNRQHFVENMEREHFYHNNHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCE 467
H+ PPP P P P P+Y + P P +
Sbjct: 912 ----HYT----------------SPPP---------PSPAPAPKYIYKSPPPPVYIYASP 1004
Query: 468 PPRI 471
PP I
Sbjct: 1005PPPI 1016
>BE657727 similar to SP|P10495|GRP1_ Glycine-rich cell wall structural
protein 1.0 precursor (GRP 1.0). [Kidney bean French
bean], partial (22%)
Length = 338
Score = 57.0 bits (136), Expect = 2e-08
Identities = 23/37 (62%), Positives = 24/37 (64%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP L P + PPP APPP PPPPPPP PP P P
Sbjct: 174 P*PPPSPLKPPAPYPPPRAPPPDPPPPPPPEPPPPYP 284
Score = 45.4 bits (106), Expect = 6e-05
Identities = 19/38 (50%), Positives = 20/38 (52%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPH 281
P PP P PPP PPPP PPP P P P P P+
Sbjct: 210 PYPPPRAPPPDPPPPPPPEPPPPYPPPAPCPAPSPPPY 323
Score = 43.5 bits (101), Expect = 2e-04
Identities = 20/38 (52%), Positives = 20/38 (52%), Gaps = 1/38 (2%)
Frame = +3
Query: 244 PSAP-PHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P AP P P PPP P PPPP PPP P P P P
Sbjct: 201 PPAPYPPPRAPPPDPPPPPPPEPPPPYPPPAPCPAPSP 314
Score = 42.0 bits (97), Expect = 7e-04
Identities = 20/44 (45%), Positives = 20/44 (45%)
Frame = +3
Query: 247 PPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDPC 290
PP P PP PPP PPP PPPPP P P PC
Sbjct: 168 PPP*PPPSPLKPPAPYPPPRAPPPDPPPPPPPEPPPPYPPPAPC 299
>TC226468 weakly similar to UP|O65450 (O65450) Glycine-rich protein, partial
(20%)
Length = 1041
Score = 57.0 bits (136), Expect = 2e-08
Identities = 48/178 (26%), Positives = 68/178 (37%), Gaps = 10/178 (5%)
Frame = -2
Query: 244 PSAPPHQL-VPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHTES 302
P APP Q+ +PQ P P++PPP PPPP PPPLP P + + + + ++
Sbjct: 752 PKAPPPQIPIPQPPKPSPVSPPPLPPPPVASPPPLPPPKIAPTPAKTPPSPAPAKATPAP 573
Query: 303 QCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHPVNYRH 362
P P P+ P ++ P V P A + E P P P ++RH
Sbjct: 572 APAPTKPA-PTPSPVPPPPAATPAPAPV-IEVPAPAPAPVIE-------VPAPAPAHHRH 420
Query: 363 SDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHR---------REVPPPPPYSNRQ 411
R H H +H RH+ R+ PP PP S +
Sbjct: 419 --------RRHR---------------HRHRHRRHQAPAPAPTIIRKSPPAPPDSTAE 315
Score = 45.1 bits (105), Expect = 8e-05
Identities = 43/164 (26%), Positives = 53/164 (32%), Gaps = 1/164 (0%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAP-PPPPPPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHTES 302
PSAP PPP P P PP P P PPPLP P + S P + + T
Sbjct: 785 PSAPKVAPASSPKAPPPQIPIPQPPKPSPVSPPPLPPPPVASPPPLPPPKIAPTPAKTPP 606
Query: 303 QCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHPVNYRH 362
P P AP P + V P A + + P P PV
Sbjct: 605 ---SPAPAKATPAPAPAPTKPAPTPSPV----PPPPAATPAPAPVIEVPAPAPAPV---- 459
Query: 363 SDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPP 406
I P P++ H +H R + P P P
Sbjct: 458 ------------IEVPAPAPAHHRHRRHRHRHRHRRHQAPAPAP 363
Score = 32.7 bits (73), Expect = 0.40
Identities = 48/217 (22%), Positives = 64/217 (29%), Gaps = 1/217 (0%)
Frame = -2
Query: 245 SAPPHQLVPQSSVPPPLAPPPPPPPPP-PPPPPLPMPHLVSSTSDPCRTVFNSRGHTESQ 303
S PP + P+ + P AP P P PPP +P+P Q
Sbjct: 830 SQPPANVAPKPAPVKPSAPKVAPASSPKAPPPQIPIP----------------------Q 717
Query: 304 CVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHPVNYRHS 363
K +P+ P PL P + P AP + S + P P P +
Sbjct: 716 PPKPSPVSP--PPLPPPPVASPPPLPPPKIAPTPAKTPPSPAPAKATPAPAPAPTKPAPT 543
Query: 364 DGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMEREHFY 423
PP P+ EVP P P H R
Sbjct: 542 PSPVP-------PPPAATPAPAPVIEVPAPAPAPVIEVPAPAP----AHHRHRRHRHRHR 396
Query: 424 HNNHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSP 460
H H+ P P R+ PP P + P+P
Sbjct: 395 HRRHQAPAPAPTIIRKS---PPAPPDSTAESDTAPAP 294
>TC225027 similar to UP|Q39835 (Q39835) Extensin, partial (49%)
Length = 747
Score = 56.6 bits (135), Expect = 3e-08
Identities = 64/253 (25%), Positives = 78/253 (30%), Gaps = 14/253 (5%)
Frame = +2
Query: 242 HLPSAPPHQLVPQSSVPPPLAPPPPPP-----PPPP---------PPPPLPMPHLVSSTS 287
H PS PP S+PPP PP P PPPP PPPP P+ S
Sbjct: 140 HYPSPPPPPKYYYLSLPPPSPSPPKKPYYYHSPPPPYKKPYYYHSPPPPPKKPYYYHSPP 319
Query: 288 DPCRTVFNSRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDR 347
P S P + P P P ++H+P
Sbjct: 320 PP------------SPPPPKKPYYYHSPP--PPSPPPPYKKPYYYHSPP----------- 424
Query: 348 SFNSFPVPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPY 407
P P Y HS PP P + + H PPPPP
Sbjct: 425 --PP*PPPPKPYYHHS-------------PPPPPPKKKPYYYHS----------PPPPPP 529
Query: 408 SNRQHFVENMEREHFYHNNHERLKPPPYDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCE 467
+ ++ + H Y + H PP Y PPP P Y PSP P
Sbjct: 530 KKKPYYYHSPPPPHPYPHPHPHPHPPFVHYS-----PPPSPKKPY---KYPSP--PPPVH 679
Query: 468 PPRIPDHGWRFPP 480
PP P + PP
Sbjct: 680 PPYTPHPVYHSPP 718
Score = 45.4 bits (106), Expect = 6e-05
Identities = 50/195 (25%), Positives = 59/195 (29%), Gaps = 30/195 (15%)
Frame = +2
Query: 242 HLPSAPPHQLVPQSSVPPPLAPPP--------PPPPPPPPP--------------PPLPM 279
H P PP + S PPP PPP PPPP PPPP PP P
Sbjct: 269 HSPPPPPKKPYYYHSPPPPSPPPPKKPYYYHSPPPPSPPPPYKKPYYYHSPPPP*PPPPK 448
Query: 280 PHLVSSTSDPCRTVFNSRGHTESQCVKDNPLHPMDRP--LAAPRSSQPISNAVHHHAP-- 335
P+ S P P +P +P P ++H+P
Sbjct: 449 PYYHHSPPPP---------------------PPKKKPYYYHSPPPPPPKKKPYYYHSPPP 565
Query: 336 --EYREAHISESDRSFNSFPVPHP--VNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHG 391
Y H P PHP V+Y + S PP H P H
Sbjct: 566 PHPYPHPH-----------PHPHPPFVHYSPPPSPKKPYKYPSPPPPVHPPYTPHPVYHS 712
Query: 392 EQHARHRREVPPPPP 406
PPPPP
Sbjct: 713 ----------PPPPP 727
Score = 45.1 bits (105), Expect = 8e-05
Identities = 64/266 (24%), Positives = 84/266 (31%), Gaps = 9/266 (3%)
Frame = +2
Query: 238 ENNIHLPSAPPHQLVPQSSVPPPLAPPPPPPP-------PPPPPPPLPMPHLVSSTSDPC 290
+N ++ PP + P S PP P PPPPP PPP P P P+ S P
Sbjct: 77 DNYLYSSPPPPKKYPPVS--PPYHYPSPPPPPKYYYLSLPPPSPSPPKKPYYYHSPPPPY 250
Query: 291 RTVFNSRGHTESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFN 350
+ + P P P S P ++H+P
Sbjct: 251 KKPYYYHSPPPP------PKKPYYYHSPPPPSPPPPKKPYYYHSPPPP------------ 376
Query: 351 SFPVPHPVNYRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNR 410
S P P+ Y + HS PP P + + PPPPP
Sbjct: 377 SPPPPYKKPYYY----------HSPPPP*PPPPKPY----------YHHSPPPPPP---- 484
Query: 411 QHFVENMEREHFYHNNHERLKPPPYDYRERWDVPPPYPGP--RYHDEDMPSPYGCHPCEP 468
++ ++YH+ PP PPP YH P PY HP
Sbjct: 485 ------KKKPYYYHS-------PP---------PPPPKKKPYYYHSPPPPHPYP-HPHPH 595
Query: 469 PRIPDHGWRFPPRSMNHRNSMPFRPP 494
P P + PP S P PP
Sbjct: 596 PHPPFVHYS-PPPSPKKPYKYPSPPP 670
Score = 31.6 bits (70), Expect = 0.90
Identities = 18/40 (45%), Positives = 19/40 (47%), Gaps = 6/40 (15%)
Frame = +2
Query: 245 SAPPHQLVPQS--SVPPPLAPPPPPPP----PPPPPPPLP 278
S PP P S PPP+ PP P P PPPPP P
Sbjct: 620 SPPPSPKKPYKYPSPPPPVHPPYTPHPVYHSPPPPPTNKP 739
>CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein DZ-HRGP
{Volvox carteri f. nagariensis}, partial (23%)
Length = 159
Score = 56.2 bits (134), Expect = 3e-08
Identities = 26/47 (55%), Positives = 26/47 (55%)
Frame = -1
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDP 289
LP PP P PPP PPPPPP PPPPPPP P L S S P
Sbjct: 150 LPPPPPPPPSPLXXPPPPPPPPPPPPSPPPPPPPPPPGLLFSGGSLP 10
Score = 50.4 bits (119), Expect = 2e-06
Identities = 21/37 (56%), Positives = 22/37 (58%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
P PP P S + P PPPPPPPPP PPPP P P
Sbjct: 156 PPLPPPPPPPPSPLXXPPPPPPPPPPPPSPPPPPPPP 46
Score = 48.5 bits (114), Expect = 7e-06
Identities = 20/34 (58%), Positives = 20/34 (58%)
Frame = -1
Query: 247 PPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMP 280
PP P P PL PPPPPPPPPPPP P P
Sbjct: 159 PPPLPPPPPPPPSPLXXPPPPPPPPPPPPSPPPP 58
Score = 47.0 bits (110), Expect = 2e-05
Identities = 23/48 (47%), Positives = 25/48 (51%), Gaps = 2/48 (4%)
Frame = -3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPP--PPPPPPPPPLPMPHLVSSTSDP 289
P PP + PPP PPPPP PPPP PPPP P S+S P
Sbjct: 145 PPPPPPSFSXLXAPPPPPPPPPPPLPPPPPXPPPPRPAFFRWFSSSPP 2
Score = 32.3 bits (72), Expect = 0.53
Identities = 17/42 (40%), Positives = 17/42 (40%), Gaps = 9/42 (21%)
Frame = -1
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPP---------PPPP 276
P PP PPPPPPPPPP PPPP
Sbjct: 87 PPPPPS-------------PPPPPPPPPPGLLFSGGSLPPPP 1
>TC204047 hydroxyproline-rich glycoprotein
Length = 887
Score = 55.8 bits (133), Expect = 4e-08
Identities = 55/226 (24%), Positives = 76/226 (33%), Gaps = 6/226 (2%)
Frame = +1
Query: 242 HLPSAPPHQLVPQSSVPPPLAPPPPPPPPP--PPPPPLPMPHLVSSTSDPCRTVFNSRGH 299
H P PP + S PPP+ P P P P PPPP P+ S P + + +
Sbjct: 1 HSPPPPPKKPYKYPSPPPPVKKPYPHPHPVYHSPPPPPKKPYKYPSPPPPVK-----KPY 165
Query: 300 TESQCVKDNPLHPMDRPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHPVN 359
V +P P +P P P+ +P PHPV
Sbjct: 166 PHPHPVYHSPPPPPKKPYKYPSPPPPVKKP----------------------YPHPHPVY 279
Query: 360 YRHSDGVTMHDRGHSIRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMER 419
HS PP P ++ PPPP ++ H +
Sbjct: 280 -------------HSPPPPPKKP--------------YKYPSPPPPVHTYPPHVPTPV-- 372
Query: 420 EHFYHNNHERLKPPPYDYRERWDVPPP----YPGPRYHDEDMPSPY 461
YH+ PPPY+ ++ PPP P P Y+ + P PY
Sbjct: 373 ---YHSP----PPPPYEKPYKYKSPPPPVHSPPPPHYYYKSPPPPY 489
Score = 40.8 bits (94), Expect = 0.001
Identities = 49/226 (21%), Positives = 63/226 (27%)
Frame = +1
Query: 255 SSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDPCRTVFNSRGHTESQCVKDNPLHPMD 314
S PPP P P PPPP P P PH V + P P
Sbjct: 4 SPPPPPKKPYKYPSPPPPVKKPYPHPHPVYHSPPP----------------------PPK 117
Query: 315 RPLAAPRSSQPISNAVHHHAPEYREAHISESDRSFNSFPVPHPVNYRHSDGVTMHDRGHS 374
+P P P+ +P PHPV HS
Sbjct: 118 KPYKYPSPPPPVKKP----------------------YPHPHPVY-------------HS 192
Query: 375 IRPPRHVPSNQFSFVHGEQHARHRREVPPPPPYSNRQHFVENMEREHFYHNNHERLKPPP 434
PP P S PPPP Y + H PP
Sbjct: 193 PPPPPKKPYKYPS---------------PPPPVKKP------------YPHPHPVYHSPP 291
Query: 435 YDYRERWDVPPPYPGPRYHDEDMPSPYGCHPCEPPRIPDHGWRFPP 480
++ + P P P + +P+P P PP + ++ PP
Sbjct: 292 PPPKKPYKYPSPPPPVHTYPPHVPTPVYHSPPPPPYEKPYKYKSPP 429
>TC225231 homologue to UP|PGKH_TOBAC (Q42961) Phosphoglycerate kinase,
chloroplast precursor , partial (86%)
Length = 1778
Score = 55.5 bits (132), Expect = 6e-08
Identities = 28/54 (51%), Positives = 31/54 (56%), Gaps = 5/54 (9%)
Frame = +2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPP-----PPLPMPHLVSSTSDPCRT 292
P PP L SS PPPL PPPPPPPPPPPP PP P ++ + P RT
Sbjct: 29 PHPPPLSL---SSTPPPLPPPPPPPPPPPPPRACSSPPPSAPARCAAWASPRRT 181
Score = 40.0 bits (92), Expect = 0.003
Identities = 18/37 (48%), Positives = 20/37 (53%)
Frame = +2
Query: 253 PQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDP 289
PQ PP PPP PPPPPPP P P + +S P
Sbjct: 23 PQPHPPPLSLSSTPPPLPPPPPPPPPPPPPRACSSPP 133
Score = 30.8 bits (68), Expect = 1.5
Identities = 18/41 (43%), Positives = 20/41 (47%), Gaps = 4/41 (9%)
Frame = -2
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPP----PPPPPPPLPMP 280
PS PP P+SS P + PPP P PPP PL P
Sbjct: 304 PSGPPRSGPPRSSSP---STPPPSRPFLLLSPPPRAPLHSP 191
Score = 28.5 bits (62), Expect = 7.6
Identities = 15/36 (41%), Positives = 16/36 (43%)
Frame = +2
Query: 241 IHLPSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPP 276
+ L S PP P PPP PPP PPP P
Sbjct: 44 LSLSSTPPPLPPPPPPPPPP--PPPRACSSPPPSAP 145
Score = 28.1 bits (61), Expect = 9.9
Identities = 14/28 (50%), Positives = 14/28 (50%), Gaps = 6/28 (21%)
Frame = +2
Query: 259 PPLAPPPPPPPP------PPPPPPLPMP 280
P L P P PPP PPP PP P P
Sbjct: 8 PSLQWPQPHPPPLSLSSTPPPLPPPPPP 91
>TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (11%)
Length = 678
Score = 54.7 bits (130), Expect = 1e-07
Identities = 31/61 (50%), Positives = 34/61 (54%), Gaps = 14/61 (22%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVP-----PPLAPPPPPP---PPPPPPPPLPMP------HLVSSTSD 288
LP +PP L P S P PPLAPPPP P PPPPP PPLP+P +LV D
Sbjct: 318 LPPSPPPPLPPSPSPPNLPSPPPLAPPPPLPPLAPPPPPLPPLPIPPNGPGEYLVPPPRD 497
Query: 289 P 289
P
Sbjct: 498 P 500
Score = 50.8 bits (120), Expect = 1e-06
Identities = 24/41 (58%), Positives = 27/41 (65%), Gaps = 4/41 (9%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPP----PPPPPPPPLPM 279
+P APP L P S PPPL P PPPP PPPP PPP+P+
Sbjct: 186 IPLAPPPPLPP--SPPPPLPPSPPPPLPPSPPPPLPPPIPL 302
Score = 49.7 bits (117), Expect = 3e-06
Identities = 26/48 (54%), Positives = 27/48 (56%), Gaps = 1/48 (2%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPP-PPPPPPPPPPLPMPHLVSSTSDP 289
LP PP + P P PLAPPPP PP PPPP PP P P L S P
Sbjct: 147 LPPPPPKECPPP---PIPLAPPPPLPPSPPPPLPPSPPPPLPPSPPPP 281
Score = 48.1 bits (113), Expect = 9e-06
Identities = 24/41 (58%), Positives = 25/41 (60%), Gaps = 3/41 (7%)
Frame = +3
Query: 243 LPSAPPHQLVPQS--SVPPPLAPPPPPP-PPPPPPPPLPMP 280
LP +PP L P S PPPL P PPPP PP P PP LP P
Sbjct: 258 LPPSPPPPLPPPIPLSTPPPLPPSPPPPLPPSPSPPNLPSP 380
Score = 47.8 bits (112), Expect = 1e-05
Identities = 27/54 (50%), Positives = 28/54 (51%), Gaps = 7/54 (12%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPP-------PPPPPPPPLPMPHLVSSTSDP 289
LP +PP L P S PPPL P PPPP PPP PP P P L S S P
Sbjct: 210 LPPSPPPPLPP--SPPPPLPPSPPPPLPPPIPLSTPPPLPPSPPPPLPPSPSPP 365
Score = 47.0 bits (110), Expect = 2e-05
Identities = 26/52 (50%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVPPPLAPP-----PPPPPPPPPPPPLPMPHLVSSTSDP 289
LP +PP L P S PPPL PP PPP PP PPPP P P + S P
Sbjct: 234 LPPSPPPPLPP--SPPPPLPPPIPLSTPPPLPPSPPPPLPPSPSPPNLPSPP 383
Score = 37.7 bits (86), Expect = 0.013
Identities = 22/60 (36%), Positives = 26/60 (42%), Gaps = 4/60 (6%)
Frame = +3
Query: 234 ATQFENNIHLPSAPPHQLVPQSSVPPPLAPPPPPPPPP----PPPPPLPMPHLVSSTSDP 289
ATQ + PH + + +PPP PPPP P PP PP P P L S P
Sbjct: 78 ATQTTLTVGKLDQVPHIMCKECELPPPPPKECPPPPIPLAPPPPLPPSPPPPLPPSPPPP 257
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 54.7 bits (130), Expect = 1e-07
Identities = 25/46 (54%), Positives = 28/46 (60%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHLVSSTSDP 289
P +PP +PPPL+PPPP PPPPPPPPP P P L S P
Sbjct: 288 PLSPP--------IPPPLSPPPP*PPPPPPPPPPPPPPLEPPPSLP 401
Score = 49.7 bits (117), Expect = 3e-06
Identities = 22/39 (56%), Positives = 22/39 (56%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLPMPHL 282
P PP P PPP PP PPPP PPPP PLP P L
Sbjct: 342 PPPPPPPPPPPPLEPPPSLPPLPPPP*PPPPDPLPPPWL 458
Score = 48.9 bits (115), Expect = 5e-06
Identities = 23/40 (57%), Positives = 24/40 (59%), Gaps = 2/40 (5%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPP--PPPPPLPMP 280
LP PP P + PPP PPPP PPPP PPPP LP P
Sbjct: 513 LPPLPP*LPPPPALPPPPWLPPPP*PPPPLLPPPPALPPP 632
Score = 48.5 bits (114), Expect = 7e-06
Identities = 26/51 (50%), Positives = 28/51 (53%), Gaps = 5/51 (9%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPP---PPP--PPPPLPMPHLVSSTSDP 289
P PP L P S+PP PP PPPP PPP PPPPLP P L + P
Sbjct: 357 PPPPPPPLEPPPSLPPLPPPP*PPPPDPLPPPWLPPPPLPPPWLPLPSPPP 509
Score = 46.6 bits (109), Expect = 3e-05
Identities = 22/39 (56%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPP--PPPPPPPLPMP 280
P PP P PPP PPPPPPP PPP PPLP P
Sbjct: 300 PIPPPLSPPPP*PPPPPPPPPPPPPPLEPPPSLPPLPPP 416
Score = 44.3 bits (103), Expect = 1e-04
Identities = 23/45 (51%), Positives = 23/45 (51%), Gaps = 8/45 (17%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPP-------PP-PPPPLPMP 280
P PP P PPP PPPPPPPP PP PPPP P P
Sbjct: 297 PPIPPPLSPPPP*PPPPPPPPPPPPPPLEPPPSLPPLPPPP*PPP 431
Score = 43.9 bits (102), Expect = 2e-04
Identities = 21/40 (52%), Positives = 22/40 (54%), Gaps = 3/40 (7%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPP---PPPPLPMP 280
P PP P PPPL PPP PP PP PPPP P+P
Sbjct: 327 PP*PPPPPPPPPPPPPPLEPPPSLPPLPPPP*PPPPDPLP 446
Score = 42.0 bits (97), Expect = 7e-04
Identities = 19/32 (59%), Positives = 20/32 (62%), Gaps = 2/32 (6%)
Frame = +3
Query: 247 PPHQLVPQSSVPPPLAPPPP--PPPPPPPPPP 276
PP L P +PPP PPPP PPPP PPPP
Sbjct: 540 PPPALPPPPWLPPPP*PPPPLLPPPPALPPPP 635
Score = 41.6 bits (96), Expect = 9e-04
Identities = 16/26 (61%), Positives = 18/26 (68%)
Frame = +3
Query: 255 SSVPPPLAPPPPPPPPPPPPPPLPMP 280
S + PP+ PP PPPP PPPPP P P
Sbjct: 285 SPLSPPIPPPLSPPPP*PPPPPPPPP 362
Score = 41.2 bits (95), Expect = 0.001
Identities = 20/41 (48%), Positives = 22/41 (52%), Gaps = 2/41 (4%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPP--PPPPPPLPMPHL 282
P PP P +PPP PPPP PPP P P PP +P L
Sbjct: 402 PLPPPP*PPPPDPLPPPWLPPPPLPPPWLPLPSPPPALPPL 524
Score = 40.4 bits (93), Expect = 0.002
Identities = 24/42 (57%), Positives = 26/42 (61%), Gaps = 1/42 (2%)
Frame = +3
Query: 243 LPSAPPHQLVPQSSVPPPLAPPPPPPPPP-PPPPPLPMPHLV 283
LPS PP L P +PP L PPP PPPP PPPP P P L+
Sbjct: 492 LPSPPP-ALPP---LPP*LPPPPALPPPPWLPPPP*PPPPLL 605
Score = 40.0 bits (92), Expect = 0.003
Identities = 22/45 (48%), Positives = 22/45 (48%), Gaps = 11/45 (24%)
Frame = +3
Query: 247 PPHQLVPQSSVPPPLAPP------PPPPPPP-----PPPPPLPMP 280
PP L P PPPL PP PPP PP PPPP LP P
Sbjct: 429 PPDPLPPPWLPPPPLPPPWLPLPSPPPALPPLPP*LPPPPALPPP 563
Score = 40.0 bits (92), Expect = 0.003
Identities = 19/37 (51%), Positives = 21/37 (56%), Gaps = 2/37 (5%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPP--PPPPPPPPPPPLP 278
P PP +P +PPP PPP P P PPP PPLP
Sbjct: 417 P*PPPPDPLPPPWLPPPPLPPPWLPLPSPPPALPPLP 527
Score = 37.0 bits (84), Expect = 0.021
Identities = 21/41 (51%), Positives = 22/41 (53%), Gaps = 4/41 (9%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPP--PPPPPPPPP--LPMP 280
P PP +P PPP PP PP PPPP PPP LP P
Sbjct: 468 PLPPPWLPLPS---PPPALPPLPP*LPPPPALPPPPWLPPP 581
Score = 36.6 bits (83), Expect = 0.028
Identities = 15/30 (50%), Positives = 17/30 (56%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPP 273
P+ PP +P PPP PPPP PPPP
Sbjct: 546 PALPPPPWLPPPP*PPPPLLPPPPALPPPP 635
Score = 36.2 bits (82), Expect = 0.036
Identities = 19/35 (54%), Positives = 19/35 (54%)
Frame = +3
Query: 244 PSAPPHQLVPQSSVPPPLAPPPPPPPPPPPPPPLP 278
P PP L P S PP L P PP PPPP PP P
Sbjct: 465 PPLPPPWL-PLPSPPPALPPLPP*LPPPPALPPPP 566
Score = 30.8 bits (68), Expect = 1.5
Identities = 17/33 (51%), Positives = 18/33 (54%), Gaps = 10/33 (30%)
Frame = +3
Query: 258 PPPLAPP---PPPPPP-------PPPPPPLPMP 280
PPPL+PP P PP P PP PPPL P
Sbjct: 228 PPPLSPPSLSPFPPFP***SPLSPPIPPPLSPP 326
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.137 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,353,339
Number of Sequences: 63676
Number of extensions: 1000727
Number of successful extensions: 77709
Number of sequences better than 10.0: 2307
Number of HSP's better than 10.0 without gapping: 11588
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33060
length of query: 516
length of database: 12,639,632
effective HSP length: 101
effective length of query: 415
effective length of database: 6,208,356
effective search space: 2576467740
effective search space used: 2576467740
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC146865.12


