

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146330.6 + phase: 0
(230 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
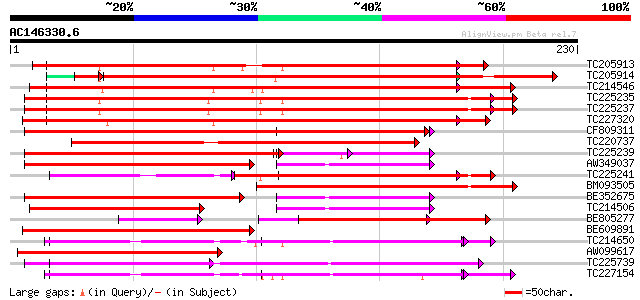
Score E
Sequences producing significant alignments: (bits) Value
TC205913 weakly similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protei... 319 8e-88
TC205914 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, ... 304 8e-85
TC214546 similar to PIR|T01932|T01932 RNA binding protein homolo... 280 4e-76
TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {A... 271 1e-73
TC225237 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 271 2e-73
TC227320 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding prot... 258 1e-69
CF809311 246 6e-66
TC220737 weakly similar to UP|Q9FFU0 (Q9FFU0) Similarity to poly... 238 2e-63
TC225239 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 156 5e-41
AW349037 145 2e-35
TC225241 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, p... 139 8e-34
BM093505 133 8e-32
BE352675 126 7e-30
TC214506 similar to PIR|T01932|T01932 RNA binding protein homolo... 113 8e-26
BE805277 109 9e-25
BE609891 106 1e-23
TC214650 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucle... 99 2e-21
AW099617 97 6e-21
TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding... 95 2e-20
TC227154 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucle... 93 9e-20
>TC205913 weakly similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45,
partial (55%)
Length = 886
Score = 319 bits (817), Expect = 8e-88
Identities = 149/185 (80%), Positives = 168/185 (90%)
Frame = +1
Query: 10 SLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAA 69
++EEVRTLWIGDLQYWVDE+YL+ CF+H GEV+SIKIIRNK+TGQPEGYGF+EFVSH++A
Sbjct: 49 AIEEVRTLWIGDLQYWVDESYLSQCFAHNGEVVSIKIIRNKLTGQPEGYGFVEFVSHASA 228
Query: 70 ERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFR 129
E L+TYNG QMPGTEQTFRLNWASFG D+GPDHSIFVGDLAPDVTD+LLQETFR
Sbjct: 229 EAFLRTYNGAQMPGTEQTFRLNWASFG------DSGPDHSIFVGDLAPDVTDFLLQETFR 390
Query: 130 THYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKK 189
HY SV+GAKVVTDP TGRSKGYGFVKF+DE++RNRAM+EMNGVYCSTRPMRISAATPKK
Sbjct: 391 AHYPSVKGAKVVTDPATGRSKGYGFVKFADEAQRNRAMTEMNGVYCSTRPMRISAATPKK 570
Query: 190 TTGYQ 194
+Q
Sbjct: 571 NASFQ 585
Score = 50.1 bits (118), Expect = 9e-07
Identities = 44/192 (22%), Positives = 83/192 (42%), Gaps = 24/192 (12%)
Frame = +1
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F +H V K++ + TG+ +GYGF++F + R +
Sbjct: 328 SIFVGDLAPDVTDFLLQETFRAHYPSVKGAKVVTDPATGRSKGYGFVKFADEAQRNRAMT 507
Query: 75 TYNGTQM-----------PGTEQTFRLNWA-----------SFGIGERRPDAGPDHS-IF 111
NG P +F+ +A S + P+ +++ +
Sbjct: 508 EMNGVYCSTRPMRISAATPKKNASFQHQYAPPKAMYQFPAYSAPVSAVAPENDVNNTTVC 687
Query: 112 VGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMN 171
+G+L +VT+ L++TF +G + K+ KGYG+V+F A+ M
Sbjct: 688 IGNLDLNVTEEELKQTFM-QFGDIVLVKIYA------GKGYGYVQFGTRVSAEDAIQRMQ 846
Query: 172 GVYCSTRPMRIS 183
G + ++IS
Sbjct: 847 GKVIGQQVIQIS 882
>TC205914 weakly similar to UP|NAM8_YEAST (Q00539) NAM8 protein, partial (9%)
Length = 1513
Score = 304 bits (778), Expect(2) = 8e-85
Identities = 150/184 (81%), Positives = 161/184 (86%)
Frame = +3
Query: 39 GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIG 98
GEVISIKIIRNK+TGQPEGYGF+EFVSH+AAERVLQTYNGTQMP T+QTFRLNWASFGIG
Sbjct: 39 GEVISIKIIRNKLTGQPEGYGFVEFVSHAAAERVLQTYNGTQMPATDQTFRLNWASFGIG 218
Query: 99 ERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFS 158
ERRPDA P+HSIFVGDLAPDVTDYLLQETFR HY SVRGAKVVTDPNT RSKGYGFVKFS
Sbjct: 219 ERRPDAAPEHSIFVGDLAPDVTDYLLQETFRAHYPSVRGAKVVTDPNTARSKGYGFVKFS 398
Query: 159 DESERNRAMSEMNGVYCSTRPMRISAATPKKTTGYQQNPYAAVVAAAPVPKGITKPYTFF 218
DE+ERNRAM+EMNGVYCSTRPMRISAATPKKTTG YAA A P P YT
Sbjct: 399 DENERNRAMTEMNGVYCSTRPMRISAATPKKTTG----AYAAPAAPVPKPVYPVPAYTSP 566
Query: 219 IIRI 222
++++
Sbjct: 567 VVQV 578
Score = 45.4 bits (106), Expect = 2e-05
Identities = 42/195 (21%), Positives = 75/195 (37%), Gaps = 27/195 (13%)
Frame = +3
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F +H V K++ + T + +GYGF++F + R +
Sbjct: 249 SIFVGDLAPDVTDYLLQETFRAHYPSVRGAKVVTDPNTARSKGYGFVKFSDENERNRAMT 428
Query: 75 TYNGTQMPGTEQTFRLNWASFGIGERRPDAGP--------------------------DH 108
NG G A P +
Sbjct: 429 EMNGVYCSTRPMRISAATPKKTTGAYAAPAAPVPKPVYPVPAYTSPVVQVQPPDYDVNNT 608
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
+IFVG+L +V++ L++ +G + K+ KG+GFV+F + A+
Sbjct: 609 TIFVGNLDLNVSEEELKQN-SLQFGEIVSVKIQP------GKGFGFVQFGTRASAEEAIQ 767
Query: 169 EMNGVYCSTRPMRIS 183
+M G + +RIS
Sbjct: 768 KMQGKMIGQQVVRIS 812
Score = 27.3 bits (59), Expect(2) = 8e-85
Identities = 9/12 (75%), Positives = 10/12 (83%)
Frame = +2
Query: 27 DENYLTHCFSHT 38
DE YL+HCF HT
Sbjct: 2 DEGYLSHCFGHT 37
>TC214546 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment) {Nicotiana tabacum;} , partial (68%)
Length = 1756
Score = 280 bits (716), Expect = 4e-76
Identities = 134/198 (67%), Positives = 158/198 (79%), Gaps = 1/198 (0%)
Frame = +3
Query: 9 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 68
AS +E+RT+W+GDL +W+DENYL +CF+HTGEV+S K+IRNK TGQ EGYGF+EF S +
Sbjct: 471 ASSDEIRTVWLGDLHHWMDENYLHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRAT 650
Query: 69 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERR-PDAGPDHSIFVGDLAPDVTDYLLQET 127
AE+VLQ YNGT MP T+Q FRLNWA+F GERR DA D SIFVGDLA DVTD +LQET
Sbjct: 651 AEKVLQNYNGTMMPNTDQAFRLNWATFSAGERRSSDATSDLSIFVGDLAIDVTDAMLQET 830
Query: 128 FRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATP 187
F Y S++GAKVV D NTGRSKGYGFV+F DE+ER RAM+EMNGVYCS+RPMRI ATP
Sbjct: 831 FAGRYSSIKGAKVVIDSNTGRSKGYGFVRFGDENERTRAMTEMNGVYCSSRPMRIGVATP 1010
Query: 188 KKTTGYQQNPYAAVVAAA 205
KKT G+QQ + V A
Sbjct: 1011KKTYGFQQQYSSQAVVLA 1064
Score = 52.0 bits (123), Expect = 2e-07
Identities = 45/194 (23%), Positives = 78/194 (40%), Gaps = 26/194 (13%)
Frame = +3
Query: 16 TLWIGDLQYWVDENYLTHCFS-HTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F+ + K++ + TG+ +GYGF+ F + R +
Sbjct: 774 SIFVGDLAIDVTDAMLQETFAGRYSSIKGAKVVIDSNTGRSKGYGFVRFGDENERTRAMT 953
Query: 75 TYNGTQM-----------PGTEQTFRLNWASFGI--------------GERRPDAGPDHS 109
NG P F+ ++S + G + +
Sbjct: 954 EMNGVYCSSRPMRIGVATPKKTYGFQQQYSSQAVVLAGGHSANGAVAQGSHSEGDINNTT 1133
Query: 110 IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSE 169
IFVG L D +D L++ F +G V K+ KG GFV+F+D A+
Sbjct: 1134IFVGGLDSDTSDEDLRQPF-LQFGEVVSVKIPV------GKGCGFVQFADRKNAEEAIQG 1292
Query: 170 MNGVYCSTRPMRIS 183
+NG + +R+S
Sbjct: 1293LNGTVIGKQTVRLS 1334
>TC225235 similar to GB|AAP37853.1|30725662|BT008494 At1g11650 {Arabidopsis
thaliana;} , partial (64%)
Length = 1154
Score = 271 bits (694), Expect = 1e-73
Identities = 136/201 (67%), Positives = 157/201 (77%), Gaps = 1/201 (0%)
Frame = +3
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QPAS +EVRTLWIGDLQYW+DENYL CF+HTGEV S+K+IRNK T Q EGYGFIEF S
Sbjct: 216 QPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVSSVKVIRNKQTSQSEGYGFIEFNSR 395
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASFGIGER-RPDAGPDHSIFVGDLAPDVTDYLLQ 125
+ AER+LQTYNG MP Q+FRLNWA+F GER R D PD++IFVGDLA DVTDYLLQ
Sbjct: 396 AGAERILQTYNGAIMPNGGQSFRLNWATFSAGERSRQDDSPDYTIFVGDLAADVTDYLLQ 575
Query: 126 ETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
ETFR Y SV+GAKVV D TGR+KGYGFV+FS+ESE+ RAM+EM GV CSTRPMRI A
Sbjct: 576 ETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSEESEQMRAMTEMQGVLCSTRPMRIGPA 755
Query: 186 TPKKTTGYQQNPYAAVVAAAP 206
+ KT Q P A+ + + P
Sbjct: 756 S-NKTPATQSQPKASYLNSQP 815
Score = 62.8 bits (151), Expect = 1e-10
Identities = 50/198 (25%), Positives = 90/198 (45%), Gaps = 16/198 (8%)
Frame = +3
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F + V K++ +++TG+ +GYGF+ F S R +
Sbjct: 525 TIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSEESEQMRAMT 704
Query: 75 TYNGT--------------QMPGTEQTFRLNWASFGIGERRPDAGPDHS-IFVGDLAPDV 119
G + P T+ + ++ + + + P+++ IFVG+L P+V
Sbjct: 705 EMQGVLCSTRPMRIGPASNKTPATQSQPKASYLNSQPQGSQNENDPNNTTIFVGNLDPNV 884
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRP 179
TD L++ F + YG + K+ K GFV+F+D S A+ +NG +
Sbjct: 885 TDDHLRQVF-SQYGELVHVKIPA------GKRCGFVQFADRSCAEEALRVLNGTLLGGQN 1043
Query: 180 MRISAATPKKTTGYQQNP 197
+R+S Q +P
Sbjct: 1044VRLSWGRSPSNKQAQADP 1097
>TC225237 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (79%)
Length = 1560
Score = 271 bits (692), Expect = 2e-73
Identities = 137/201 (68%), Positives = 156/201 (77%), Gaps = 1/201 (0%)
Frame = +3
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QPAS +EVRTLWIGDLQYW+DENYL CF+HTGEV S+K+IRNK T Q EGYGFIEF S
Sbjct: 282 QPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVTSVKVIRNKQTSQSEGYGFIEFNSR 461
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASFGIGER-RPDAGPDHSIFVGDLAPDVTDYLLQ 125
+ AER+LQTYNG MP Q+FRLNWA+F GER R D PD++IFVGDLA DVTDYLLQ
Sbjct: 462 AGAERILQTYNGAIMPNGGQSFRLNWATFSAGERSRHDDSPDYTIFVGDLAADVTDYLLQ 641
Query: 126 ETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
ETFR Y SV+GAKVV D TGR+KGYGFV+FSDESE+ RAM+EM GV CSTRPMRI A
Sbjct: 642 ETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSDESEQVRAMTEMQGVLCSTRPMRIGPA 821
Query: 186 TPKKTTGYQQNPYAAVVAAAP 206
+ KT Q P A+ + P
Sbjct: 822 S-NKTPTTQSQPKASYQNSQP 881
Score = 62.0 bits (149), Expect = 2e-10
Identities = 50/198 (25%), Positives = 90/198 (45%), Gaps = 16/198 (8%)
Frame = +3
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F + V K++ +++TG+ +GYGF+ F S R +
Sbjct: 591 TIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSDESEQVRAMT 770
Query: 75 TYNGT--------------QMPGTEQTFRLNWASFGIGERRPDAGPDHS-IFVGDLAPDV 119
G + P T+ + ++ + + + P+++ IFVG+L P+V
Sbjct: 771 EMQGVLCSTRPMRIGPASNKTPTTQSQPKASYQNSQPQGSQNENDPNNTTIFVGNLDPNV 950
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRP 179
TD L++ F + YG + K+ K GFV+F+D S A+ +NG +
Sbjct: 951 TDDHLRQVF-SQYGELVHVKIPA------GKRCGFVQFADRSCAEEALRVLNGTLLGGQN 1109
Query: 180 MRISAATPKKTTGYQQNP 197
+R+S Q +P
Sbjct: 1110VRLSWGRSPSNKQAQADP 1163
>TC227320 similar to UP|Q93YF1 (Q93YF1) Nucleic acid binding protein, partial
(70%)
Length = 1724
Score = 258 bits (660), Expect = 1e-69
Identities = 123/190 (64%), Positives = 149/190 (77%)
Frame = +1
Query: 6 HQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVS 65
HQ S E +T+WIGDL +W+DENYL CF+ TGE+ SIK+IRNK TG EGYGF+EF S
Sbjct: 397 HQNGSGGENKTIWIGDLHHWMDENYLHRCFASTGEISSIKVIRNKQTGLSEGYGFVEFYS 576
Query: 66 HSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQ 125
H+ AE+VLQ Y G MP EQ FRLNWA+F G++ D PD SIFVGDLA DVTD LL
Sbjct: 577 HATAEKVLQNYAGILMPNAEQPFRLNWATFSTGDKGSDNVPDLSIFVGDLAADVTDSLLH 756
Query: 126 ETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
ETF + Y SV+ AKVV D NTGRSKGYGFV+F D++ER +AM++MNGVYCS+RPMRI AA
Sbjct: 757 ETFASVYPSVKAAKVVFDANTGRSKGYGFVRFGDDNERTQAMTQMNGVYCSSRPMRIGAA 936
Query: 186 TPKKTTGYQQ 195
TP+K++G+QQ
Sbjct: 937 TPRKSSGHQQ 966
Score = 60.5 bits (145), Expect = 6e-10
Identities = 44/180 (24%), Positives = 82/180 (45%), Gaps = 12/180 (6%)
Frame = +1
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V ++ L F+ V + K++ + TG+ +GYGF+ F + + +
Sbjct: 706 SIFVGDLAADVTDSLLHETFASVYPSVKAAKVVFDANTGRSKGYGFVRFGDDNERTQAMT 885
Query: 75 TYNGTQM-----------PGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYL 123
NG P + S G + + +IFVG L P+V+D
Sbjct: 886 QMNGVYCSSRPMRIGAATPRKSSGHQQGGLSNGTANQSEADSTNTTIFVGGLDPNVSDED 1065
Query: 124 LQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRIS 183
L++ F + YG + K+ KG GFV+F++ + A+ ++NG + +R+S
Sbjct: 1066LRQPF-SQYGEIVSVKIPV------GKGCGFVQFANRNNAEEALQKLNGTTIGKQTVRLS 1224
>CF809311
Length = 668
Score = 246 bits (628), Expect = 6e-66
Identities = 119/164 (72%), Positives = 131/164 (79%)
Frame = +2
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP S +EVRTLWIGDLQYW+DENYL CF+HTGE+ S+K+IRNK T Q EGYGFIEF S
Sbjct: 176 QPTSADEVRTLWIGDLQYWMDENYLYTCFAHTGELASVKVIRNKQTSQSEGYGFIEFTSR 355
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQE 126
+ AERVLQTYNGT MP Q FRLNWA+F GERR D PDH+IFVGDLA DVTDYLLQE
Sbjct: 356 AGAERVLQTYNGTIMPNGGQNFRLNWATFSAGERRHDDSPDHTIFVGDLAADVTDYLLQE 535
Query: 127 TFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEM 170
TFR Y S +GAKVV D TGR KGYG V+F DESE+ RAMSEM
Sbjct: 536 TFRARYPSAKGAKVVIDRLTGRXKGYGSVRFGDESEQVRAMSEM 667
Score = 40.8 bits (94), Expect = 5e-04
Identities = 19/64 (29%), Positives = 35/64 (54%)
Frame = +2
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
++++GDL + + L F H G + KV+ + T +S+GYGF++F+ + R +
Sbjct: 203 TLWIGDLQYWMDENYLYTCF-AHTGELASVKVIRNKQTSQSEGYGFIEFTSRAGAERVLQ 379
Query: 169 EMNG 172
NG
Sbjct: 380 TYNG 391
>TC220737 weakly similar to UP|Q9FFU0 (Q9FFU0) Similarity to
polyadenylate-binding protein 5, partial (33%)
Length = 450
Score = 238 bits (606), Expect = 2e-63
Identities = 114/141 (80%), Positives = 126/141 (88%)
Frame = +1
Query: 26 VDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTYNGTQMPGTE 85
VDE YL+HCF+ GEV+SIKIIRN++TGQPEGYGF+EFVSH+ AERVLQTYN T +
Sbjct: 43 VDEAYLSHCFAPAGEVVSIKIIRNELTGQPEGYGFVEFVSHATAERVLQTYNAT-----D 207
Query: 86 QTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPN 145
QTFRLNWASFGIGERRPDA +HSIFVGDLAPD+TDYLLQE FR HY SVRGAKVV+DPN
Sbjct: 208 QTFRLNWASFGIGERRPDAALEHSIFVGDLAPDITDYLLQEMFRAHYPSVRGAKVVSDPN 387
Query: 146 TGRSKGYGFVKFSDESERNRA 166
TGRSKGYGFVKFSDE+ERNRA
Sbjct: 388 TGRSKGYGFVKFSDENERNRA 450
Score = 31.2 bits (69), Expect = 0.41
Identities = 13/39 (33%), Positives = 21/39 (53%)
Frame = +1
Query: 133 GSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMN 171
G V K++ + TG+ +GYGFV+F + R + N
Sbjct: 82 GEVVSIKIIRNELTGQPEGYGFVEFVSHATAERVLQTYN 198
>TC225239 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (23%)
Length = 854
Score = 156 bits (395), Expect(2) = 5e-41
Identities = 72/105 (68%), Positives = 81/105 (76%)
Frame = +1
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP S +EVRTLWIGDLQYW+DENYL C +HTGEV S+K+IRNK T Q EGYGFIEF S
Sbjct: 301 QPTSADEVRTLWIGDLQYWMDENYLYTCLAHTGEVASVKVIRNKQTSQSEGYGFIEFTSR 480
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIF 111
+ AERVLQTYNGT MP Q FRLNWA+ E+R D PDH+IF
Sbjct: 481 AGAERVLQTYNGTIMPNGGQNFRLNWATLSASEKRHDDSPDHTIF 615
Score = 40.8 bits (94), Expect = 5e-04
Identities = 20/64 (31%), Positives = 35/64 (54%)
Frame = +1
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
++++GDL + + L T H G V KV+ + T +S+GYGF++F+ + R +
Sbjct: 328 TLWIGDLQYWMDENYLY-TCLAHTGEVASVKVIRNKQTSQSEGYGFIEFTSRAGAERVLQ 504
Query: 169 EMNG 172
NG
Sbjct: 505 TYNG 516
Score = 28.5 bits (62), Expect(2) = 5e-41
Identities = 16/33 (48%), Positives = 19/33 (57%), Gaps = 1/33 (3%)
Frame = +3
Query: 108 HSIFVGDLAPDVTDYLLQETFRTHYG-SVRGAK 139
H I++ D TDYLLQETF +RGAK
Sbjct: 606 HHIWLETWLADXTDYLLQETFXAPIXLKLRGAK 704
>AW349037
Length = 624
Score = 145 bits (365), Expect = 2e-35
Identities = 66/93 (70%), Positives = 76/93 (80%)
Frame = -3
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QPAS +EVRTLWIGDLQYW+DENYL CF+HTGEV S+K+IRNK T Q EGYGFIEF S
Sbjct: 550 QPASADEVRTLWIGDLQYWMDENYLYTCFAHTGEVTSVKVIRNKQTSQSEGYGFIEFNSR 371
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASFGIGE 99
+ AER+LQTYNG MP Q+FRLNWA+F G+
Sbjct: 370 AGAERILQTYNGAIMPNGGQSFRLNWATFSAGQ 272
Score = 41.6 bits (96), Expect = 3e-04
Identities = 20/64 (31%), Positives = 35/64 (54%)
Frame = -3
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
++++GDL + + L F H G V KV+ + T +S+GYGF++F+ + R +
Sbjct: 523 TLWIGDLQYWMDENYLYTCF-AHTGEVTSVKVIRNKQTSQSEGYGFIEFNSRAGAERILQ 347
Query: 169 EMNG 172
NG
Sbjct: 346 TYNG 335
>TC225241 similar to UP|Q9LEB4 (Q9LEB4) RNA Binding Protein 45, partial (50%)
Length = 898
Score = 139 bits (351), Expect = 8e-34
Identities = 72/107 (67%), Positives = 82/107 (76%), Gaps = 1/107 (0%)
Frame = +3
Query: 92 WASFGIGER-RPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSK 150
WA+F GER R D PD++IFVGDLA DVTDYLLQETFR Y SV+GAKVV D TGR+K
Sbjct: 3 WATFSAGERSRQDDSPDYTIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTK 182
Query: 151 GYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKTTGYQQNP 197
GYGFV+FS+ESE+ RAM+EM GV CSTRPMRI A+ KT Q P
Sbjct: 183 GYGFVRFSEESEQMRAMTEMQGVLCSTRPMRIGPAS-NKTPATQSQP 320
Score = 45.8 bits (107), Expect = 2e-05
Identities = 26/76 (34%), Positives = 41/76 (53%)
Frame = +3
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++G+L V +++L FS GE++ +KI K G F++F S AE L+
Sbjct: 540 IFVGNLDPNVTDDHLRQVFSQYGELVHVKIPAGKRCG------FVQFADRSCAEEALRVL 701
Query: 77 NGTQMPGTEQTFRLNW 92
NGT + G Q RL+W
Sbjct: 702 NGTLLGG--QNVRLSW 743
Score = 40.8 bits (94), Expect = 5e-04
Identities = 26/74 (35%), Positives = 40/74 (53%)
Frame = +3
Query: 110 IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSE 169
IFVG+L P+VTD L++ F + YG + K+ K GFV+F+D S A+
Sbjct: 540 IFVGNLDPNVTDDHLRQVF-SQYGELVHVKIPA------GKRCGFVQFADRSCAEEALRV 698
Query: 170 MNGVYCSTRPMRIS 183
+NG + +R+S
Sbjct: 699 LNGTLLGGQNVRLS 740
Score = 37.0 bits (84), Expect = 0.007
Identities = 19/64 (29%), Positives = 33/64 (50%), Gaps = 1/64 (1%)
Frame = +3
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F + V K++ +++TG+ +GYGF+ F S R +
Sbjct: 57 TIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSEESEQMRAMT 236
Query: 75 TYNG 78
G
Sbjct: 237 EMQG 248
>BM093505
Length = 422
Score = 133 bits (334), Expect = 8e-32
Identities = 68/106 (64%), Positives = 80/106 (75%)
Frame = +3
Query: 101 RPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDE 160
R D PD++IFVGDLA DVTDYLLQETFR Y SV+GAKVV D TGR+KGYGFV+FS+E
Sbjct: 3 RQDDSPDYTIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSEE 182
Query: 161 SERNRAMSEMNGVYCSTRPMRISAATPKKTTGYQQNPYAAVVAAAP 206
SE+ RAM+EM GV CSTRPMRI A+ KT Q P A+ + + P
Sbjct: 183 SEQMRAMTEMQGVLCSTRPMRIGPAS-NKTPATQSQPKASYLNSQP 317
Score = 37.0 bits (84), Expect = 0.007
Identities = 19/64 (29%), Positives = 33/64 (50%), Gaps = 1/64 (1%)
Frame = +3
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F + V K++ +++TG+ +GYGF+ F S R +
Sbjct: 27 TIFVGDLAADVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSEESEQMRAMT 206
Query: 75 TYNG 78
G
Sbjct: 207EMQG 218
>BE352675
Length = 517
Score = 126 bits (317), Expect = 7e-30
Identities = 58/89 (65%), Positives = 68/89 (76%)
Frame = +3
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP S +EVRTLWIGDLQYW+DENYL CF+HTGE+ S+K+IRNK T Q EGYGFIEF S
Sbjct: 222 QPTSADEVRTLWIGDLQYWMDENYLYTCFAHTGELASVKVIRNKQTSQSEGYGFIEFTSR 401
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASF 95
+ AERVLQTYNGT MP ++ +F
Sbjct: 402 AGAERVLQTYNGTIMPEWRAELQIELGNF 488
Score = 40.8 bits (94), Expect = 5e-04
Identities = 19/64 (29%), Positives = 35/64 (54%)
Frame = +3
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
++++GDL + + L F H G + KV+ + T +S+GYGF++F+ + R +
Sbjct: 249 TLWIGDLQYWMDENYLYTCF-AHTGELASVKVIRNKQTSQSEGYGFIEFTSRAGAERVLQ 425
Query: 169 EMNG 172
NG
Sbjct: 426 TYNG 437
Score = 33.5 bits (75), Expect = 0.083
Identities = 14/22 (63%), Positives = 15/22 (67%)
Frame = +1
Query: 82 PGTEQTFRLNWASFGIGERRPD 103
P Q FRLNWA+F GERR D
Sbjct: 448 PNGGQNFRLNWATFSAGERRHD 513
>TC214506 similar to PIR|T01932|T01932 RNA binding protein homolog - common
tobacco (fragment) {Nicotiana tabacum;} , partial (20%)
Length = 662
Score = 113 bits (282), Expect = 8e-26
Identities = 48/71 (67%), Positives = 60/71 (83%)
Frame = +2
Query: 9 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 68
AS +E+RT+W+GDL +W+DENYL +CF+HTGEV+S K+IRNK TGQ EGYGF+EF S
Sbjct: 449 ASSDEIRTVWLGDLHHWMDENYLHNCFAHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRGT 628
Query: 69 AERVLQTYNGT 79
AE+VLQ YNGT
Sbjct: 629 AEKVLQNYNGT 661
Score = 43.9 bits (102), Expect = 6e-05
Identities = 22/64 (34%), Positives = 35/64 (54%)
Frame = +2
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMS 168
++++GDL + + L F H G V AKV+ + TG+S+GYGFV+F + +
Sbjct: 470 TVWLGDLHHWMDENYLHNCF-AHTGEVVSAKVIRNKQTGQSEGYGFVEFYSRGTAEKVLQ 646
Query: 169 EMNG 172
NG
Sbjct: 647 NYNG 658
>BE805277
Length = 407
Score = 109 bits (273), Expect = 9e-25
Identities = 55/78 (70%), Positives = 61/78 (77%)
Frame = +2
Query: 118 DVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCST 177
DVTDYLLQETFR Y SV+GAKVV D TGR+KGYGFV+FSDESE+ RAM+EM GV CST
Sbjct: 2 DVTDYLLQETFRARYNSVKGAKVVIDRLTGRTKGYGFVRFSDESEQVRAMTEMQGVLCST 181
Query: 178 RPMRISAATPKKTTGYQQ 195
RPMRI A+ K T Q
Sbjct: 182 RPMRIGPASNKTPTTQSQ 235
Score = 39.3 bits (90), Expect(2) = 8e-07
Identities = 25/70 (35%), Positives = 36/70 (50%)
Frame = +3
Query: 102 PDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDES 161
P P IFVG+L P+VTD L++ F + YG + K+ K GFV+F+D S
Sbjct: 219 PPLSPSPKIFVGNLDPNVTDDHLRQVF-SQYGELVHVKIPA------GKRCGFVQFADRS 377
Query: 162 ERNRAMSEMN 171
A+ +N
Sbjct: 378 CAEEALLVLN 407
Score = 30.4 bits (67), Expect(2) = 8e-07
Identities = 11/34 (32%), Positives = 20/34 (58%)
Frame = +2
Query: 45 KIIRNKITGQPEGYGFIEFVSHSAAERVLQTYNG 78
K++ +++TG+ +GYGF+ F S R + G
Sbjct: 65 KVVIDRLTGRTKGYGFVRFSDESEQVRAMTEMQG 166
>BE609891
Length = 295
Score = 106 bits (264), Expect = 1e-23
Identities = 51/94 (54%), Positives = 64/94 (67%)
Frame = +2
Query: 6 HQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVS 65
H PAS +EV TLWIGDLQYW+DE YL CF+ TG+V +K+I NK T + EGYGFIEF S
Sbjct: 14 HAPASADEVLTLWIGDLQYWMDETYLYTCFALTGDVTMVKVILNKQTCRSEGYGFIEFNS 193
Query: 66 HSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGE 99
+ A R+L TY G +P +R +WA+ GE
Sbjct: 194 RAGAVRILLTYYGAIVPYGGLCYRYDWATVRAGE 295
Score = 32.0 bits (71), Expect = 0.24
Identities = 17/53 (32%), Positives = 30/53 (56%)
Frame = +2
Query: 109 SIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDES 161
++++GDL + + L F G V KV+ + T RS+GYGF++F+ +
Sbjct: 44 TLWIGDLQYWMDETYLYTCFALT-GDVTMVKVILNKQTCRSEGYGFIEFNSRA 199
>TC214650 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucleolysin;
oligouridylate binding protein (AT3g14100/MAG2_5),
partial (93%)
Length = 1684
Score = 98.6 bits (244), Expect = 2e-21
Identities = 57/172 (33%), Positives = 95/172 (55%)
Frame = +2
Query: 15 RTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
R++++G++ V ++ L FS G + K+IR + YGF+++ S+A +
Sbjct: 344 RSVYVGNIHPQVTDSLLQELFSTAGALEGCKLIRK----EKSSYGFVDYFDRSSAAFAIV 511
Query: 75 TYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGS 134
T NG + G Q ++NWA +R D +IFVGDL+P+VTD L F Y S
Sbjct: 512 TLNGRNIFG--QPIKVNWAY--ASSQREDTSGHFNIFVGDLSPEVTDATLYACFSV-YPS 676
Query: 135 VRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAAT 186
A+V+ D TGRS+G+GFV F ++ + A++++ G + +R +R + AT
Sbjct: 677 CSDARVMWDQKTGRSRGFGFVSFRNQQDAQSAINDLTGKWLGSRQIRCNWAT 832
Score = 62.8 bits (151), Expect = 1e-10
Identities = 50/211 (23%), Positives = 88/211 (41%), Gaps = 30/211 (14%)
Frame = +2
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++GDL V + L CFS +++ ++ TG+ G+GF+ F + A+ +
Sbjct: 605 IFVGDLSPEVTDATLYACFSVYPSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQSAINDL 784
Query: 77 NGTQMPGTEQTFRLNWASFGIG-----------------------------ERRPDAGPD 107
G + G+ Q R NWA+ G + P+ P
Sbjct: 785 TGKWL-GSRQ-IRCNWATKGASASDEKQTSDSRSVVELTNGSSEDGQETTNDDTPEKNPQ 958
Query: 108 HS-IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRA 166
++ ++VG+LAP+VT L + F + A + D R KG+GFV++S +E A
Sbjct: 959 YTTVYVGNLAPEVTSVDLHQHFHS-----LNAGTIEDVRVQRDKGFGFVRYSTHAEAALA 1123
Query: 167 MSEMNGVYCSTRPMRISAATPKKTTGYQQNP 197
+ N +P++ S + G P
Sbjct: 1124IQMGNARILFGKPIKCSWGSKPTPPGTASTP 1216
Score = 47.8 bits (112), Expect = 4e-06
Identities = 28/83 (33%), Positives = 45/83 (53%)
Frame = +2
Query: 103 DAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESE 162
D+ S++VG++ P VTD LLQE F T G++ G K++ YGFV + D S
Sbjct: 329 DSSSCRSVYVGNIHPQVTDSLLQELFST-AGALEGCKLIRK----EKSSYGFVDYFDRSS 493
Query: 163 RNRAMSEMNGVYCSTRPMRISAA 185
A+ +NG +P++++ A
Sbjct: 494 AAFAIVTLNGRNIFGQPIKVNWA 562
>AW099617
Length = 304
Score = 97.1 bits (240), Expect = 6e-21
Identities = 47/83 (56%), Positives = 55/83 (65%)
Frame = +2
Query: 4 TYHQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEF 63
T +PAS +V+TLWI D +YW DENY+ C +HTGEV S K I NK T Q EG G IEF
Sbjct: 56 TQQEPASANKVQTLWIRDQEYWDDENYIYTCLAHTGEVTSSKEILNKRTSQSEGCG*IEF 235
Query: 64 VSHSAAERVLQTYNGTQMPGTEQ 86
H+ AER+L TYNG MP Q
Sbjct: 236 NKHAGAERILHTYNGAIMPNGRQ 304
>TC225739 homologue to UP|Q41124 (Q41124) Chloroplast RNA binding protein
precursor, partial (85%)
Length = 1436
Score = 95.1 bits (235), Expect = 2e-20
Identities = 57/176 (32%), Positives = 96/176 (54%)
Frame = +3
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
L+ G+L Y VD L G I+++ ++ TG+ G+ F+ V++
Sbjct: 504 LYFGNLPYSVDSAKLAGLIQDYGSAELIEVLYDRDTGKSRGFAFVTMSCIEDCNAVIENL 683
Query: 77 NGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVR 136
+G + G +T R+N++S + +H +FVG+L+ VT+ +L + F+ YG+V
Sbjct: 684 DGKEFLG--RTLRVNFSSKPKPKEPLYPETEHKLFVGNLSWSVTNEILTQAFQ-EYGTVV 854
Query: 137 GAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKTTG 192
GA+V+ D TGRS+GYGFV +S ++E A++ +N V R MR+S A K+ G
Sbjct: 855 GARVLYDGETGRSRGYGFVCYSTKAEMEAALAALNDVELEGRAMRVSLAQGKRA*G 1022
Score = 44.7 bits (104), Expect = 4e-05
Identities = 21/77 (27%), Positives = 38/77 (49%)
Frame = +3
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
+P E L++G+L + V LT F G V+ +++ + TG+ GYGF+ + +
Sbjct: 747 EPLYPETEHKLFVGNLSWSVTNEILTQAFQEYGTVVGARVLYDGETGRSRGYGFVCYSTK 926
Query: 67 SAAERVLQTYNGTQMPG 83
+ E L N ++ G
Sbjct: 927 AEMEAALAALNDVELEG 977
>TC227154 similar to UP|Q9LJH8 (Q9LJH8) RNA binding protein nucleolysin;
oligouridylate binding protein (AT3g14100/MAG2_5),
partial (92%)
Length = 1698
Score = 93.2 bits (230), Expect = 9e-20
Identities = 58/178 (32%), Positives = 95/178 (52%), Gaps = 6/178 (3%)
Frame = +1
Query: 15 RTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
R++++G++ V E L FS TG V K+IR YGFI + +A +
Sbjct: 307 RSVYVGNIHTQVTEPLLQEVFSGTGPVEGCKLIRK----DKSSYGFIHYFDRRSAALAIL 474
Query: 75 TYNGTQMPGTEQTFRLNWASFGIGERRPDAG------PDHSIFVGDLAPDVTDYLLQETF 128
+ NG + G Q ++NWA + G+R + ++IFVGDL+P+VTD L F
Sbjct: 475 SLNGRHLFG--QPIKVNWA-YASGQREDTSAFFF*FAGHYNIFVGDLSPEVTDATLFACF 645
Query: 129 RTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAAT 186
Y + A+V+ D TGRS+G+GFV F ++ + A++++ G + +R +R + AT
Sbjct: 646 SV-YPTCSDARVMWDQKTGRSRGFGFVSFRNQQDAQSAINDLTGKWLGSRQIRCNWAT 816
Score = 63.2 bits (152), Expect = 1e-10
Identities = 54/220 (24%), Positives = 92/220 (41%), Gaps = 31/220 (14%)
Frame = +1
Query: 17 LWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTY 76
+++GDL V + L CFS +++ ++ TG+ G+GF+ F + A+ +
Sbjct: 589 IFVGDLSPEVTDATLFACFSVYPTCSDARVMWDQKTGRSRGFGFVSFRNQQDAQSAINDL 768
Query: 77 NGTQMPGTEQTFRLNWASFGIGERR----------------------------PDAGPDH 108
G + G+ Q R NWA+ G G P+ P +
Sbjct: 769 TGKWL-GSRQ-IRCNWATKGAGGTEEKQNSDAKSVVELTYGSSDGKETSNSDAPENNPQY 942
Query: 109 S-IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRA- 166
+ ++VG+LAP+ T L F + GA V+ + R KG+GFV++S +E A
Sbjct: 943 TTVYVGNLAPEATQLDLHHHFHS-----LGAGVIEEVRVQRDKGFGFVRYSTHAEAALAI 1107
Query: 167 -MSEMNGVYCSTRPMRISAATPKKTTGYQQNPYAAVVAAA 205
M + C + ++ S + G NP AA+
Sbjct: 1108QMGNAQSLLCG-KQIKCSWGSKPTPAGTASNPLPPPAAAS 1224
Score = 40.8 bits (94), Expect = 5e-04
Identities = 24/83 (28%), Positives = 43/83 (50%)
Frame = +1
Query: 103 DAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESE 162
D S++VG++ VT+ LLQE F + G V G K++ + YGF+ + D
Sbjct: 292 DPSTCRSVYVGNIHTQVTEPLLQEVF-SGTGPVEGCKLIRKDKS----SYGFIHYFDRRS 456
Query: 163 RNRAMSEMNGVYCSTRPMRISAA 185
A+ +NG + +P++++ A
Sbjct: 457 AALAILSLNGRHLFGQPIKVNWA 525
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,992,212
Number of Sequences: 63676
Number of extensions: 123719
Number of successful extensions: 878
Number of sequences better than 10.0: 205
Number of HSP's better than 10.0 without gapping: 719
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 821
length of query: 230
length of database: 12,639,632
effective HSP length: 94
effective length of query: 136
effective length of database: 6,654,088
effective search space: 904955968
effective search space used: 904955968
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146330.6


