

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146330.2 + phase: 0
(223 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
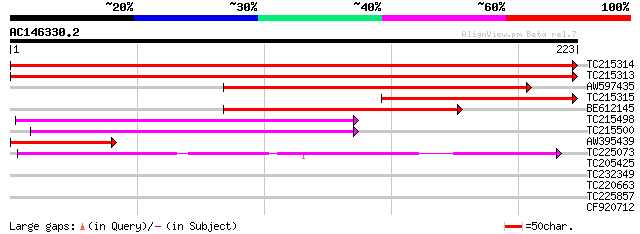
Score E
Sequences producing significant alignments: (bits) Value
TC215314 homologue to UP|PSB1_PETHY (O82531) Proteasome subunit ... 439 e-124
TC215313 homologue to UP|PSB1_PETHY (O82531) Proteasome subunit ... 433 e-122
AW597435 similar to SP|O82531|PSB1 Proteasome subunit beta type ... 204 3e-53
TC215315 homologue to UP|PSB1_PETHY (O82531) Proteasome subunit ... 148 2e-36
BE612145 similar to SP|O82531|PSB1_ Proteasome subunit beta type... 144 3e-35
TC215498 homologue to UP|Q8VZC4 (Q8VZC4) Proteasome subunit, com... 81 4e-16
TC215500 homologue to UP|Q8VZC4 (Q8VZC4) Proteasome subunit, com... 80 7e-16
AW395439 similar to SP|O82531|PSB1_ Proteasome subunit beta type... 77 5e-15
TC225073 homologue to UP|O23716 (O23716) Multicatalytic endopept... 72 2e-13
TC205425 similar to UP|O23718 (O23718) 20S proteasome beta subun... 34 0.060
TC232349 similar to UP|O81241 (O81241) Dc3 promoter-binding fact... 29 1.5
TC220663 similar to UP|Q9LG20 (Q9LG20) F14J16.17, partial (9%) 27 5.6
TC225857 similar to UP|Q6T284 (Q6T284) Predicted protein, partia... 27 7.4
CF920712 27 9.6
>TC215314 homologue to UP|PSB1_PETHY (O82531) Proteasome subunit beta type 1
(20S proteasome alpha subunit F) (20S proteasome subunit
beta-6) , complete
Length = 947
Score = 439 bits (1130), Expect = e-124
Identities = 216/223 (96%), Positives = 220/223 (97%)
Frame = +2
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 60
MTKQHANWSPYDNNGGSCVAIAG+DYCVIAADTRMSTGYNILTRDYSKIS LAEKCVMAS
Sbjct: 149 MTKQHANWSPYDNNGGSCVAIAGADYCVIAADTRMSTGYNILTRDYSKISQLAEKCVMAS 328
Query: 61 SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD 120
SGFQADVKALQKVLSARHL YQHQHNKQMSCPAMAQLLSNTLYYKRFFPYY+FNVLGGLD
Sbjct: 329 SGFQADVKALQKVLSARHLIYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYAFNVLGGLD 508
Query: 121 SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 180
+EGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE
Sbjct: 509 NEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 688
Query: 181 AVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD 223
AVDLVKTVFASATERDIYTGDKVEIVILNA GIHRE+MDLRKD
Sbjct: 689 AVDLVKTVFASATERDIYTGDKVEIVILNASGIHREYMDLRKD 817
>TC215313 homologue to UP|PSB1_PETHY (O82531) Proteasome subunit beta type 1
(20S proteasome alpha subunit F) (20S proteasome subunit
beta-6) , complete
Length = 1152
Score = 433 bits (1113), Expect = e-122
Identities = 213/223 (95%), Positives = 218/223 (97%)
Frame = +3
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMAS 60
MTKQ ANWSPYDNNGGSCVAIAG+DYCVIAADTRMSTGYNILTRDYSKIS LAEKCVMAS
Sbjct: 144 MTKQQANWSPYDNNGGSCVAIAGADYCVIAADTRMSTGYNILTRDYSKISQLAEKCVMAS 323
Query: 61 SGFQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLD 120
SGFQADVKALQKVLSARHL YQHQHNKQMSCPAMAQLLSNTLYYKRFFPYY+FNVLGGLD
Sbjct: 324 SGFQADVKALQKVLSARHLIYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYAFNVLGGLD 503
Query: 121 SEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 180
+EGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE
Sbjct: 504 NEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAE 683
Query: 181 AVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREFMDLRKD 223
AVDLVKTVFASATERDIYTGDKVEIV+LNA GI RE+MDLRKD
Sbjct: 684 AVDLVKTVFASATERDIYTGDKVEIVVLNASGIRREYMDLRKD 812
>AW597435 similar to SP|O82531|PSB1 Proteasome subunit beta type 1 (EC
3.4.25.1) (20S proteasome alpha subunit F), partial
(51%)
Length = 364
Score = 204 bits (519), Expect = 3e-53
Identities = 100/121 (82%), Positives = 110/121 (90%)
Frame = +2
Query: 85 HNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSEGKGCVFTYDAVGSYERVGYSSQ 144
HNKQMSCPAMAQLLSNTLYYKRFFPYY+FNVLGGLD+EGKGCVFTYDAVGSY++V YSSQ
Sbjct: 2 HNKQMSCPAMAQLLSNTLYYKRFFPYYAFNVLGGLDNEGKGCVFTYDAVGSYKKVKYSSQ 181
Query: 145 GSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAEAVDLVKTVFASATERDIYTGDKVE 204
GSGSTLIMPFL+N LKS SPLLLP Q+AVT LSEA A DLVKT+F SA ER+IYT + +E
Sbjct: 182 GSGSTLIMPFLNNXLKSASPLLLPTQNAVTALSEAXACDLVKTIFTSAPERNIYTRE*IE 361
Query: 205 I 205
I
Sbjct: 362 I 364
>TC215315 homologue to UP|PSB1_PETHY (O82531) Proteasome subunit beta type 1
(20S proteasome alpha subunit F) (20S proteasome subunit
beta-6) , partial (35%)
Length = 445
Score = 148 bits (374), Expect = 2e-36
Identities = 75/77 (97%), Positives = 76/77 (98%)
Frame = +2
Query: 147 GSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAEAVDLVKTVFASATERDIYTGDKVEIV 206
GSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAEAVDLVKTVFASATERDIYTGDKVEIV
Sbjct: 2 GSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSEAEAVDLVKTVFASATERDIYTGDKVEIV 181
Query: 207 ILNAGGIHREFMDLRKD 223
ILNA GIHRE+MDLRKD
Sbjct: 182 ILNASGIHREYMDLRKD 232
>BE612145 similar to SP|O82531|PSB1_ Proteasome subunit beta type 1 (EC
3.4.25.1) (20S proteasome alpha subunit F), partial
(42%)
Length = 283
Score = 144 bits (363), Expect = 3e-35
Identities = 73/94 (77%), Positives = 78/94 (82%)
Frame = +2
Query: 85 HNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSEGKGCVFTYDAVGSYERVGYSSQ 144
HNKQMSCPAMAQLLSNTLYYKR FPYY+ NV+GGLD+EG GCVFTYDA GSY R GYSSQ
Sbjct: 2 HNKQMSCPAMAQLLSNTLYYKRCFPYYAINVIGGLDNEG*GCVFTYDA*GSY*RDGYSSQ 181
Query: 145 GSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSE 178
GSGS LIM +DN+LK PS LL AQDA T L E
Sbjct: 182 GSGSKLIMACIDNELKYPSALL*TAQDAETALVE 283
>TC215498 homologue to UP|Q8VZC4 (Q8VZC4) Proteasome subunit, complete
Length = 967
Score = 80.9 bits (198), Expect = 4e-16
Identities = 45/135 (33%), Positives = 69/135 (50%)
Frame = +1
Query: 3 KQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSG 62
+Q S ++ NG + VA+ G + IA+D R+ + D+ +IS + +K +A SG
Sbjct: 67 QQIERMSIFEYNGSAVVAMVGKNCFAIASDRRLGVQLQTIATDFQRISKIHDKLFIALSG 246
Query: 63 FQADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSE 122
D + L + L RH YQ + + M A L+S LY KRF PY+ V+ GL E
Sbjct: 247 LATDAQTLYQRLVFRHKLYQLREERDMKPETFASLVSALLYEKRFGPYFCQPVIAGLGDE 426
Query: 123 GKGCVFTYDAVGSYE 137
K + T D +G+ E
Sbjct: 427 DKPFICTMDCLGAKE 471
>TC215500 homologue to UP|Q8VZC4 (Q8VZC4) Proteasome subunit, complete
Length = 975
Score = 80.1 bits (196), Expect = 7e-16
Identities = 43/129 (33%), Positives = 67/129 (51%)
Frame = +1
Query: 9 SPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGFQADVK 68
S ++ NG + VA+ G + IA+D R+ + D+ +IS + +K + SG D +
Sbjct: 97 SIFEYNGSALVAMVGKNCFAIASDRRLGVQLQTIATDFQRISKIHDKLFIGLSGLATDAQ 276
Query: 69 ALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFNVLGGLDSEGKGCVF 128
L + L RH YQ + + M A L+S LY KRF PY+ V+ GL + K +
Sbjct: 277 TLYQRLLFRHKLYQLREERDMKPQTFASLVSAILYEKRFGPYFCQPVIAGLGDDDKPFIC 456
Query: 129 TYDAVGSYE 137
T DA+G+ E
Sbjct: 457 TMDAIGAKE 483
>AW395439 similar to SP|O82531|PSB1_ Proteasome subunit beta type 1 (EC
3.4.25.1) (20S proteasome alpha subunit F), partial
(17%)
Length = 127
Score = 77.4 bits (189), Expect = 5e-15
Identities = 33/42 (78%), Positives = 39/42 (92%)
Frame = +2
Query: 1 MTKQHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNIL 42
M +Q A+W+PYDN+GGSCVAIAG++YCVIAADTRMS GYNIL
Sbjct: 2 MRRQRASWAPYDNDGGSCVAIAGAEYCVIAADTRMSAGYNIL 127
>TC225073 homologue to UP|O23716 (O23716) Multicatalytic endopeptidase
complex, proteasome, beta subunit (AT4g31300/F8F16_120)
(20S proteasome subunit PBA1) , partial (93%)
Length = 1080
Score = 72.0 bits (175), Expect = 2e-13
Identities = 58/220 (26%), Positives = 99/220 (44%), Gaps = 6/220 (2%)
Frame = +1
Query: 4 QHANWSPYDNNGGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGF 63
Q ++S + G + + + + V+ AD+R STG + R KI+ L + + SG
Sbjct: 136 QKVDFSAPHSMGTTIIGVTYNGGVVLGADSRTSTGVYVANRASDKITQLTDNVYVCRSGS 315
Query: 64 QADVKALQKVLSARHLTYQHQHNKQMSCPAMAQLLSNTLYYKRFFPYYSFN------VLG 117
AD +++S + HQH Q+ PA ++ +N + R Y + N ++G
Sbjct: 316 AAD----SQIVSDYVRYFLHQHTIQLGQPATVKVAANLV---RLLAYNNKNFLQTGLIVG 474
Query: 118 GLDSEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLS 177
G D G ++ G+ + ++ GSGS+ + F D K ++
Sbjct: 475 GWDKYEGGQIYGVPLGGTIVQQPFAIGGSGSSYLYGFFDQAWKE-------------GMT 615
Query: 178 EAEAVDLVKTVFASATERDIYTGDKVEIVILNAGGIHREF 217
+ EA DLVK + A RD +G V VI+N+ G+ R F
Sbjct: 616 KDEAEDLVKKAVSLAIARDGASGGVVRTVIINSEGVTRNF 735
>TC205425 similar to UP|O23718 (O23718) 20S proteasome beta subunit PBG1 ,
partial (92%)
Length = 1076
Score = 33.9 bits (76), Expect = 0.060
Identities = 33/147 (22%), Positives = 64/147 (43%), Gaps = 3/147 (2%)
Frame = +1
Query: 15 GGSCVAIAGSDYCVIAADTRMSTGYNILTRDYSKISHLAEKCVMASSGFQADVKALQKVL 74
G S VAI D ++AAD S G + + +I + + ++ +SG +D + + + L
Sbjct: 187 GSSVVAIKYKDGILMAADMGGSYGSTLRYKSVERIKPIGKHSLLGASGEISDFQEILRYL 366
Query: 75 SARHL-TYQHQHNKQMSCPAMAQLLSNTLYYKR--FFPYYSFNVLGGLDSEGKGCVFTYD 131
L + + L+ +Y +R F P ++ VLGG+ G+ + +
Sbjct: 367 DELILYDNMWDDGNSLGPKEVHNYLTRVMYNRRNKFNPLWNALVLGGV-KNGQKYLGMVN 543
Query: 132 AVGSYERVGYSSQGSGSTLIMPFLDNQ 158
+G + + G G+ L P L ++
Sbjct: 544 MIGINFEDNHVATGLGNHLARPILRDE 624
>TC232349 similar to UP|O81241 (O81241) Dc3 promoter-binding factor-3
(Fragment), partial (50%)
Length = 834
Score = 29.3 bits (64), Expect = 1.5
Identities = 13/31 (41%), Positives = 19/31 (60%)
Frame = +3
Query: 145 GSGSTLIMPFLDNQLKSPSPLLLPAQDAVTP 175
G+G++L + F DNQ+ PS L+ D TP
Sbjct: 270 GAGASLDVSFADNQMAMPSSLMGTMSDTQTP 362
>TC220663 similar to UP|Q9LG20 (Q9LG20) F14J16.17, partial (9%)
Length = 749
Score = 27.3 bits (59), Expect = 5.6
Identities = 25/84 (29%), Positives = 34/84 (39%)
Frame = +3
Query: 119 LDSEGKGCVFTYDAVGSYERVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLSE 178
LD E + V E VG S Q STL L + + LPA+ + S
Sbjct: 48 LDHEDVDYEVAENIVHKGENVGSSRQTDNSTLPSNLLSRKRSQDTNSSLPAKSS---SST 218
Query: 179 AEAVDLVKTVFASATERDIYTGDK 202
+E VDL S +++ TG K
Sbjct: 219 SEFVDLTMDTPKSRPDKEHQTGPK 290
>TC225857 similar to UP|Q6T284 (Q6T284) Predicted protein, partial (9%)
Length = 1483
Score = 26.9 bits (58), Expect = 7.4
Identities = 11/32 (34%), Positives = 20/32 (62%)
Frame = -1
Query: 134 GSYERVGYSSQGSGSTLIMPFLDNQLKSPSPL 165
GS+ GY + +G+T+IM + ++ +P PL
Sbjct: 709 GSFSYHGYEKKENGTTIIMLTKEGRMPTPLPL 614
>CF920712
Length = 365
Score = 26.6 bits (57), Expect = 9.6
Identities = 23/76 (30%), Positives = 33/76 (43%), Gaps = 5/76 (6%)
Frame = -1
Query: 138 RVGYSSQGSGSTLIMPFLDNQLKSPSPLLLPAQDAVTPLS-----EAEAVDLVKTVFASA 192
R+ YSS G + MP L+ SP+ PA + PLS E ++ L+ V
Sbjct: 287 RLSYSSTCFGCST-MPILNPSR*MNSPIFTPASSIIIPLS*TTQYE*KSFKLIAVVIVLM 111
Query: 193 TERDIYTGDKVEIVIL 208
RD+ V I I+
Sbjct: 110 YVRDLQLRQNVLI*II 63
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.133 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,277,875
Number of Sequences: 63676
Number of extensions: 120812
Number of successful extensions: 624
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 621
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 624
length of query: 223
length of database: 12,639,632
effective HSP length: 94
effective length of query: 129
effective length of database: 6,654,088
effective search space: 858377352
effective search space used: 858377352
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC146330.2


