

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.5 + phase: 0
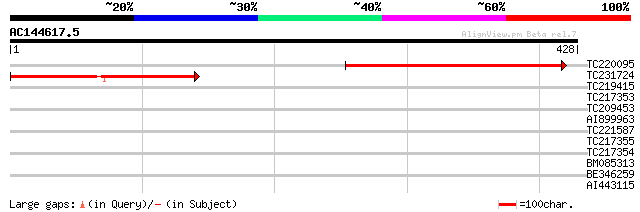
(428 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC220095 similar to GB|AAO63408.1|28950969|BT005344 At5g11330 {A... 291 5e-79
TC231724 weakly similar to GB|AAO63408.1|28950969|BT005344 At5g1... 234 7e-62
TC219415 weakly similar to UP|Q9AVE7 (Q9AVE7) Zeaxanthin epoxida... 39 0.006
TC217353 similar to UP|Q9ZSN8 (Q9ZSN8) CTF2A (Fragment), partial... 38 0.008
TC209453 similar to UP|O81335 (O81335) Geranylgeranyl hydrogenas... 34 0.14
AI899963 33 0.32
TC221587 weakly similar to UP|Q8H9C4 (Q8H9C4) Monooxygenase (Fra... 32 0.55
TC217355 similar to UP|Q9ZSN8 (Q9ZSN8) CTF2A (Fragment), partial... 29 3.6
TC217354 similar to UP|Q9SBB5 (Q9SBB5) CTF2A, partial (50%) 29 4.6
BM085313 similar to GP|10177621|db phytoene dehydrogenase-like {... 29 4.6
BE346259 29 4.6
AI443115 similar to GP|10177621|db phytoene dehydrogenase-like {... 28 7.9
>TC220095 similar to GB|AAO63408.1|28950969|BT005344 At5g11330 {Arabidopsis
thaliana;} , partial (31%)
Length = 718
Score = 291 bits (744), Expect = 5e-79
Identities = 138/167 (82%), Positives = 153/167 (90%)
Frame = +3
Query: 254 EVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIFWDNV 313
EVKGTSVT KV +DMIQKMHQEAE++WIPEL KV+KETK+PFLNFIYDSDPLEKIFWDNV
Sbjct: 3 EVKGTSVTMKVNNDMIQKMHQEAEQVWIPELVKVIKETKDPFLNFIYDSDPLEKIFWDNV 182
Query: 314 VLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPVTSKQVL 373
VLVG+AAHPTTPH LRSTNM+ILDAAVLGKCIEKWG E L SALEEYQ RLPVTSK+VL
Sbjct: 183 VLVGDAAHPTTPHCLRSTNMSILDAAVLGKCIEKWGPEKLGSALEEYQFVRLPVTSKEVL 362
Query: 374 HARRLGRIKQGLVLPDRDPFDPKSARQEECQEIILTNTPFFNDAPLS 420
++RRLGR+KQGLVLPDR+PFDPK R E+CQE++L NTPFFND PLS
Sbjct: 363 YSRRLGRLKQGLVLPDREPFDPKLIRPEDCQELLLRNTPFFNDEPLS 503
>TC231724 weakly similar to GB|AAO63408.1|28950969|BT005344 At5g11330
{Arabidopsis thaliana;} , partial (31%)
Length = 474
Score = 234 bits (596), Expect = 7e-62
Identities = 116/146 (79%), Positives = 125/146 (85%), Gaps = 3/146 (2%)
Frame = +3
Query: 1 MVGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQ 60
MVGEG KPKAVI+GGSI GISSAHAL LAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQ
Sbjct: 39 MVGEGEKPKAVIIGGSIAGISSAHALTLAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQ 218
Query: 61 IIQSWISKPP---QFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGV 117
II SW+ PP Q LHNTT LTIDQN TDSEKK + TLTRDES NFRAAHW DLHG+
Sbjct: 219 IIHSWL--PPHQKQLLHNTTLTLTIDQNHATDSEKKFSWTLTRDESFNFRAAHWADLHGL 392
Query: 118 LYESLPPQIFLWGHIFLSFHVANEKG 143
LY +LPP +FLWGH+FLS HVA++KG
Sbjct: 393 LYNALPPNVFLWGHLFLSVHVADDKG 470
>TC219415 weakly similar to UP|Q9AVE7 (Q9AVE7) Zeaxanthin epoxidase, partial
(18%)
Length = 448
Score = 38.5 bits (88), Expect = 0.006
Identities = 33/103 (32%), Positives = 50/103 (48%), Gaps = 8/103 (7%)
Frame = +3
Query: 280 WIPELAKVMKETKEPFL--NFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILD 337
W E+ ++ ET E + IYD D + V L+G+AAHP P+ + M I D
Sbjct: 147 WCDEVIALISETPEHMIIQRDIYDRDMINTWGIGRVTLLGDAAHPMQPNLGQGGCMAIED 326
Query: 338 AAVLGKCIEKW------GTEMLESALEEYQLTRLPVTSKQVLH 374
L ++K G+E++ SAL Y+ R+P +VLH
Sbjct: 327 CYQLILELDKVAKHGSDGSEVI-SALRRYEKKRIP--RVRVLH 446
>TC217353 similar to UP|Q9ZSN8 (Q9ZSN8) CTF2A (Fragment), partial (42%)
Length = 792
Score = 38.1 bits (87), Expect = 0.008
Identities = 36/156 (23%), Positives = 63/156 (40%), Gaps = 13/156 (8%)
Frame = +2
Query: 241 KLNWIWYVNQPEPEVKGTSVTTKVTSDM-IQKMHQEAEKIWIPELAKVMKETKEPFLNFI 299
K+ W N P S K+T + ++K +E K W EL ++ T + + +
Sbjct: 20 KVYWFICFNSP-------SAGPKITDSLELKKQAKELVKNWPSELLNIVDSTPD---DTV 169
Query: 300 YDSDPLEKIFWD---------NVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEK--- 347
+ +++ W VV+VG+A HP TP+ + + D+ VL K + +
Sbjct: 170 IKTPLVDRWLWPAISPSASAGRVVVVGDAWHPMTPNLGQGACCALEDSVVLAKKLARAIN 349
Query: 348 WGTEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQ 383
+E A Y R P + A +G + Q
Sbjct: 350 SEDPSVEEAFRSYGAERWPRVFPLTIRANLVGSVLQ 457
>TC209453 similar to UP|O81335 (O81335) Geranylgeranyl hydrogenase , partial
(36%)
Length = 745
Score = 33.9 bits (76), Expect = 0.14
Identities = 18/55 (32%), Positives = 27/55 (48%)
Frame = +1
Query: 5 GRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQ 59
GRK +A ++GG G S+A AL G + + E+ P P G + L L +
Sbjct: 202 GRKLRAAVIGGGPAGSSAAEALASGGVETFLFER-NPPSVAKPCGGAIPLCMLEE 363
>AI899963
Length = 345
Score = 32.7 bits (73), Expect = 0.32
Identities = 19/53 (35%), Positives = 28/53 (51%), Gaps = 1/53 (1%)
Frame = +1
Query: 372 VLHARRLGRIKQGLVLPDRDPFDPKSARQEECQE-IILTNTPFFNDAPLSFAS 423
+LH R R+ GL L DR+P+ + A E C ++ + PF ND +S S
Sbjct: 85 LLHPLRKERLHHGLFL*DRNPWGHRHAE*EYCASWTVMADRPFANDYRISSES 243
>TC221587 weakly similar to UP|Q8H9C4 (Q8H9C4) Monooxygenase (Fragment),
partial (29%)
Length = 608
Score = 32.0 bits (71), Expect = 0.55
Identities = 18/66 (27%), Positives = 32/66 (48%)
Frame = +3
Query: 312 NVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPVTSKQ 371
N+ + G+A HP TP + + D VL +C+ ++ ++ EE Q R+ + K+
Sbjct: 186 NICVGGDAFHPMTPDLGQGGCCALEDGIVLARCLAAAFSKHIKEKDEEDQFKRIEGSLKK 365
Query: 372 VLHARR 377
RR
Sbjct: 366 YAKERR 383
>TC217355 similar to UP|Q9ZSN8 (Q9ZSN8) CTF2A (Fragment), partial (29%)
Length = 631
Score = 29.3 bits (64), Expect = 3.6
Identities = 21/74 (28%), Positives = 33/74 (44%), Gaps = 3/74 (4%)
Frame = +2
Query: 313 VVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGT---EMLESALEEYQLTRLPVTS 369
VV+VG+A HP TP+ + + D+ VL K + + ++E A Y R P
Sbjct: 107 VVVVGDAWHPMTPNLGQGACCALEDSVVLAKKLARAINV**PIVEEAFRPYGTERWPRVF 286
Query: 370 KQVLHARRLGRIKQ 383
+ A +G Q
Sbjct: 287 PLTISANLVGSALQ 328
>TC217354 similar to UP|Q9SBB5 (Q9SBB5) CTF2A, partial (50%)
Length = 859
Score = 28.9 bits (63), Expect = 4.6
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = +3
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTG 45
R+ + V+VG I G+++A +L G LVLE+ S TG
Sbjct: 105 REEQVVVVGAGIAGLATALSLHRLGVRSLVLEQAQSLRTG 224
>BM085313 similar to GP|10177621|db phytoene dehydrogenase-like
{Arabidopsis thaliana}, partial (19%)
Length = 426
Score = 28.9 bits (63), Expect = 4.6
Identities = 14/35 (40%), Positives = 22/35 (62%)
Frame = +1
Query: 4 EGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEK 38
+G+K A+I+GG G+++A L G V VLE+
Sbjct: 49 KGKKWDALIIGGGHNGLTAAAYLARGGLSVAVLER 153
>BE346259
Length = 459
Score = 28.9 bits (63), Expect = 4.6
Identities = 15/51 (29%), Positives = 29/51 (56%)
Frame = -2
Query: 55 NSLSQQIIQSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLN 105
NS ++++ + W+ + P F + +D+N ++EKK +LT D SL+
Sbjct: 149 NSENERVNRVWVKETPNFYFIII--VKLDKNFRIENEKKNEESLTDDLSLS 3
>AI443115 similar to GP|10177621|db phytoene dehydrogenase-like
{Arabidopsis thaliana}, partial (20%)
Length = 421
Score = 28.1 bits (61), Expect = 7.9
Identities = 12/35 (34%), Positives = 22/35 (62%)
Frame = +2
Query: 4 EGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEK 38
+G+K A+++GG G+++A L G V +LE+
Sbjct: 8 KGKKWDALVIGGGHNGLTAAAYLARGGLSVAILER 112
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,978,269
Number of Sequences: 63676
Number of extensions: 279608
Number of successful extensions: 1399
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1391
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1396
length of query: 428
length of database: 12,639,632
effective HSP length: 100
effective length of query: 328
effective length of database: 6,272,032
effective search space: 2057226496
effective search space used: 2057226496
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144617.5


