

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144388.8 - phase: 0
(160 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
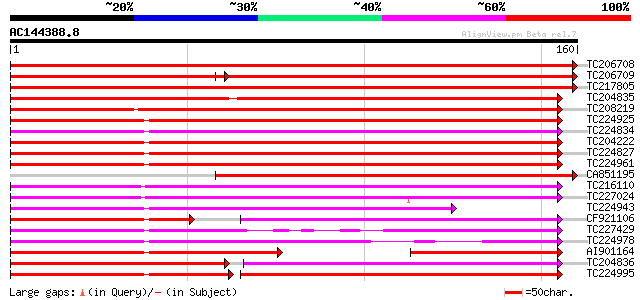
Score E
Sequences producing significant alignments: (bits) Value
TC206708 weakly similar to UP|Q6QHU3 (Q6QHU3) Major cherry aller... 281 1e-76
TC206709 weakly similar to UP|PRS1_SOLTU (P17641) Pathogenesis-r... 177 4e-72
TC217805 weakly similar to UP|BV1L_BETVE (P43185) Major pollen a... 254 1e-68
TC204835 weakly similar to UP|O23747 (O23747) Pollen allergen, B... 133 4e-32
TC208219 similar to UP|DRR4_PEA (P27047) Disease resistance resp... 122 6e-29
TC224925 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-... 117 3e-27
TC224834 homologue to UP|SAM2_SOYBN (P26987) Stress-induced prot... 113 3e-26
TC204222 UP|SAM2_SOYBN (P26987) Stress-induced protein SAM22 (St... 113 4e-26
TC224827 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-... 112 8e-26
TC224961 UP|Q43453 (Q43453) G.max mRNA from stress-induced gene ... 112 1e-25
CA851195 weakly similar to GP|1545897|emb| pollen allergen Car b... 111 2e-25
TC216110 weakly similar to UP|P93333 (P93333) PR10-1 protein, pa... 105 1e-23
TC227024 similar to UP|Q6T6J0 (Q6T6J0) Pathogenesis-related clas... 101 1e-22
TC224943 homologue to UP|SAM2_SOYBN (P26987) Stress-induced prot... 94 3e-20
CF921106 65 1e-19
TC227429 UP|Q8LJU1 (Q8LJU1) PR10-like protein, partial (98%) 83 5e-17
TC224978 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-... 83 5e-17
AI901164 SP|P26987|SAM Stress-induced protein SAM22. [Soybean] {... 66 2e-16
TC204836 weakly similar to UP|Q9ZRU8 (Q9ZRU8) Stress and pathoge... 81 2e-16
TC224995 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-... 68 2e-12
>TC206708 weakly similar to UP|Q6QHU3 (Q6QHU3) Major cherry allergen Pru av
1.0201, partial (78%)
Length = 857
Score = 281 bits (718), Expect = 1e-76
Identities = 134/160 (83%), Positives = 148/160 (91%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGVTTFT EYSS+VAPSRMF ALI+DSRNL+PKLLPQFVKDVN+I+GDG AGSIEQVNFN
Sbjct: 99 MGVTTFTQEYSSSVAPSRMFKALIVDSRNLLPKLLPQFVKDVNVIQGDGEAGSIEQVNFN 278
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E SPFKYLK +IDVLDK+NL+CKYTMIEGDPLGDKL+SI YEVKFEAT+DGGCLCKM S+
Sbjct: 279 EASPFKYLKHRIDVLDKDNLVCKYTMIEGDPLGDKLDSIGYEVKFEATSDGGCLCKMTSN 458
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENPQVYA 160
Y TIG+ DVKEE+VKEGRES I +Y VVESYLLENPQVYA
Sbjct: 459 YNTIGEIDVKEEEVKEGRESGIAVYRVVESYLLENPQVYA 578
>TC206709 weakly similar to UP|PRS1_SOLTU (P17641) Pathogenesis-related
protein STH-21, partial (34%)
Length = 951
Score = 177 bits (450), Expect(2) = 4e-72
Identities = 83/102 (81%), Positives = 92/102 (89%)
Frame = +1
Query: 59 FNEGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMA 118
F +PFKYLK +IDVLDK+NL+CKYTMIEGDPLGDKLESI YEVKFEAT+DGGCLCKM
Sbjct: 364 FYADNPFKYLKHRIDVLDKDNLVCKYTMIEGDPLGDKLESIGYEVKFEATSDGGCLCKMT 543
Query: 119 SSYKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENPQVYA 160
S+Y TIG+FDVKEE+VKEGRES I +Y VVESYLLENPQVYA
Sbjct: 544 SNYNTIGEFDVKEEEVKEGRESGIAVYRVVESYLLENPQVYA 669
Score = 110 bits (275), Expect(2) = 4e-72
Identities = 52/62 (83%), Positives = 58/62 (92%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MG+TTFT EYSS+VAPSRMF ALI+DSRN +PKLLPQFVKDVN+I+GDG AGSIEQVNFN
Sbjct: 96 MGITTFTQEYSSSVAPSRMFKALIVDSRNSLPKLLPQFVKDVNVIQGDGEAGSIEQVNFN 275
Query: 61 EG 62
EG
Sbjct: 276 EG 281
>TC217805 weakly similar to UP|BV1L_BETVE (P43185) Major pollen allergen Bet
v 1-L (Bet v I-L), partial (72%)
Length = 824
Score = 254 bits (649), Expect = 1e-68
Identities = 119/160 (74%), Positives = 140/160 (87%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MG TTFTH+YSS VAPSRMF ALI DSR L+PKLLPQ +K+VN+I+GDG AGSIEQVNF
Sbjct: 94 MGATTFTHDYSSPVAPSRMFKALITDSRTLLPKLLPQIIKEVNLIQGDGEAGSIEQVNFA 273
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E PFKY+K +ID +DK+N +CKYTMIEGDPL DKLESIAYEVKFEAT+DGGCLCKM +
Sbjct: 274 EAFPFKYVKHRIDEVDKDNFVCKYTMIEGDPLEDKLESIAYEVKFEATSDGGCLCKMTTK 453
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENPQVYA 160
Y IG F+VKEE++KEGRES++G+ +VVE+YLLENPQVYA
Sbjct: 454 YNVIGGFEVKEEEIKEGRESSLGVCKVVEAYLLENPQVYA 573
>TC204835 weakly similar to UP|O23747 (O23747) Pollen allergen, Betv1
(Fragment), partial (76%)
Length = 831
Score = 133 bits (334), Expect = 4e-32
Identities = 65/156 (41%), Positives = 96/156 (60%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV T E+ S V+ ++++ A+++D+ N+ PK LP F+K V IEGDGG G+I+++
Sbjct: 121 MGVFTSESEHVSPVSAAKLYKAIVLDASNVFPKALPNFIKSVETIEGDGGPGTIKKLTLA 300
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
EG Y+K +D +D EN + Y++IEG L + LE I YE K AT DGG + K S
Sbjct: 301 EG--LGYVKHHVDAIDTENYVYNYSVIEGSALSEPLEKICYEYKLVATPDGGSIVKSTSK 474
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y T GD + EE VK G+E + G + +E ++ NP
Sbjct: 475 YYTKGDEQLAEEYVKTGKERSAGFTKAIEDFIQANP 582
>TC208219 similar to UP|DRR4_PEA (P27047) Disease resistance response protein
DRRG49-C, complete
Length = 748
Score = 122 bits (307), Expect = 6e-29
Identities = 60/156 (38%), Positives = 100/156 (63%)
Frame = +2
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +STVAP+R++ AL+ D+ NL+PK + + +K V I+EG+GG G+I+++ F
Sbjct: 68 MGVFTFEDETTSTVAPARLYKALVKDADNLVPKAV-EAIKSVEIVEGNGGPGTIKKLTFV 244
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E KY+ K++ +D+ N Y+++ G L D +E I++E K A +GG + K+
Sbjct: 245 EDGQTKYVLHKVEAIDEANWGYNYSVVGGVGLPDTVEKISFEAKLVADPNGGSIAKITVK 424
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD + EE++K G+ +++ +E Y+L NP
Sbjct: 425 YQTKGDANPSEEELKSGKAKGDALFKALEGYVLANP 532
>TC224925 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-induced gene
(H4), complete
Length = 1041
Score = 117 bits (292), Expect = 3e-27
Identities = 59/156 (37%), Positives = 97/156 (61%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK + F + V +EG+GG G+I+++ F
Sbjct: 318 MGVFTFEDETTSPVAPATLYKALVTDADNVIPKAVDAF-RSVENVEGNGGPGTIKKITFL 494
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ GD L D +E I +E K A +GG K+
Sbjct: 495 EDGETKFVLHKIEAIDEANLGYSYSVVGGDGLPDTVEKITFECKLAAGANGGSAGKLTVK 674
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD ++D+K G+ + +++ VE+YLL +P
Sbjct: 675 YQTKGDAQPNQDDLKIGKAKSDALFKAVEAYLLAHP 782
>TC224834 homologue to UP|SAM2_SOYBN (P26987) Stress-induced protein SAM22
(Starvation-associated message 22) (Allergen Gly m 4),
complete
Length = 776
Score = 113 bits (283), Expect = 3e-26
Identities = 57/156 (36%), Positives = 92/156 (58%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MG+ TF E +S VAP+ ++ AL+ D+ N+IPK L F K V +EG+GG G+I+++ F
Sbjct: 162 MGIFTFEDEITSPVAPATLYKALVTDADNIIPKALDSF-KSVENVEGNGGPGTIKKITFV 338
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ G L D E I + K A +GG K+
Sbjct: 339 EDGETKFVLHKIEAVDEANLGYSYSVVGGAALPDTAEKITFHSKLAAGPNGGSAGKLTVE 518
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD ++ +K G+ +++ +E+YLL NP
Sbjct: 519 YQTKGDAQPNQDQLKTGKAKADALFKAIEAYLLANP 626
>TC204222 UP|SAM2_SOYBN (P26987) Stress-induced protein SAM22
(Starvation-associated message 22) (Allergen Gly m 4),
complete
Length = 896
Score = 113 bits (282), Expect = 4e-26
Identities = 57/156 (36%), Positives = 95/156 (60%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK L F K V +EG+GG G+I+++ F
Sbjct: 172 MGVFTFEDEINSPVAPATLYKALVTDADNVIPKALDSF-KSVENVEGNGGPGTIKKITFL 348
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ G L D E I ++ K A +GG K+
Sbjct: 349 EDGETKFVLHKIESIDEANLGYSYSVVGGAALPDTAEKITFDSKLVAGPNGGSAGKLTVK 528
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD + ++++K G+ +++ +E+YLL +P
Sbjct: 529 YETKGDAEPNQDELKTGKAKADALFKAIEAYLLAHP 636
>TC224827 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-induced gene
(H4), complete
Length = 776
Score = 112 bits (280), Expect = 8e-26
Identities = 59/156 (37%), Positives = 95/156 (60%)
Frame = +2
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK + F + V +EG+GG G+I+++ F
Sbjct: 89 MGVFTFEDETTSPVAPATLYKALVTDADNVIPKAVDAF-RSVENLEGNGGPGTIKKITFV 265
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ G L D +E I +E K A +GG K+
Sbjct: 266 EDGESKFVLHKIESVDEANLGYSYSVVGGVGLPDTVEKITFECKLAAGANGGSAGKLTVK 445
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD +D+K G+ + +++ VE+YLL NP
Sbjct: 446 YQTKGDAQPNPDDLKIGKVKSDALFKAVEAYLLANP 553
>TC224961 UP|Q43453 (Q43453) G.max mRNA from stress-induced gene (H4),
complete
Length = 1048
Score = 112 bits (279), Expect = 1e-25
Identities = 58/156 (37%), Positives = 95/156 (60%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MG+ TF E +S VAP+ ++ AL+ D+ N+IPK + F + V +EG+GG G+I+++ F
Sbjct: 166 MGIFTFEDETTSPVAPATLYKALVTDADNVIPKAVEAF-RSVENLEGNGGPGTIKKITFV 342
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ G L D +E I +E K A +GG K+
Sbjct: 343 EDGESKFVLHKIESVDEANLGYSYSVVGGVGLPDTVEKITFECKLAAGANGGSAGKLTVK 522
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD +D+K G+ + +++ VE+YLL NP
Sbjct: 523 YQTKGDAQPNPDDLKIGKVKSDALFKAVEAYLLANP 630
>CA851195 weakly similar to GP|1545897|emb| pollen allergen Car b 1 {Carpinus
betulus}, partial (44%)
Length = 706
Score = 111 bits (277), Expect = 2e-25
Identities = 52/102 (50%), Positives = 71/102 (68%)
Frame = -3
Query: 59 FNEGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMA 118
F G+ K +K +ID +D+E L YT+IEG+ L DK SIA+E+KFEA DGG + K+
Sbjct: 518 FPAGNQVKNIKNRIDEIDEETLTYNYTLIEGEALKDKFASIAHEIKFEAAPDGGSISKVT 339
Query: 119 SSYKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENPQVYA 160
S Y GD ++ EED+K +E +GIY+VVE+YLL+NP VYA
Sbjct: 338 SKYYLKGDVEINEEDIKASKEIVLGIYKVVEAYLLQNPDVYA 213
>TC216110 weakly similar to UP|P93333 (P93333) PR10-1 protein, partial (92%)
Length = 772
Score = 105 bits (261), Expect = 1e-23
Identities = 50/156 (32%), Positives = 91/156 (58%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV T ++ + V P+R+F A+ +D NL PKL+ + + +G+GG G+I+++
Sbjct: 136 MGVVTQIYDTPAAVPPTRLFKAMTLDFHNLFPKLVDS-IHSIVFTQGNGGPGTIKKITTI 312
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
EG KY+ ++D +D+ N + +++ EG L D LE +++E + +GG + K++
Sbjct: 313 EGDKTKYVLHRVDAIDEANFVYNFSITEGTALADTLEKVSFESQLVEAPNGGSIRKVSVQ 492
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
+ T GD + EE++ + G+ ++VE YLL NP
Sbjct: 493 FFTKGDATLSEEELTANKAKIQGLVKLVEGYLLANP 600
>TC227024 similar to UP|Q6T6J0 (Q6T6J0) Pathogenesis-related class 10 protein
SPE-16 (Fragment), complete
Length = 914
Score = 101 bits (252), Expect = 1e-22
Identities = 50/157 (31%), Positives = 91/157 (57%), Gaps = 1/157 (0%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MG F E SSTVAP+ ++ AL D+ +IPK++ ++ + I+EG+GG G+++++ +
Sbjct: 151 MGAFAFDEENSSTVAPATLYKALTKDADTIIPKIIGA-IQTIEIVEGNGGPGTVKKITAS 327
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDG-GCLCKMAS 119
EG ++ QK+D +D+ NL+ Y+++ G L + LE + ++ K DG G + K
Sbjct: 328 EGDQTSFVLQKVDAIDEANLVYDYSIVGGTGLHESLEKVTFQTKVVPGTDGNGSIAKATL 507
Query: 120 SYKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
++ T D + + E + GI++ +E Y+L NP
Sbjct: 508 TFHTKDDAPLSDAVRDETKARGAGIFKAIEGYVLANP 618
>TC224943 homologue to UP|SAM2_SOYBN (P26987) Stress-induced protein SAM22
(Starvation-associated message 22) (Allergen Gly m 4),
complete
Length = 696
Score = 94.0 bits (232), Expect = 3e-20
Identities = 50/126 (39%), Positives = 76/126 (59%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK L F K V +EG+GG G+I+++ F
Sbjct: 88 MGVFTFEDEINSPVAPATLYKALVTDADNVIPKALDSF-KSVENVEGNGGPGTIKKITFL 264
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ G L D E I ++ K A +GG K+
Sbjct: 265 EDGETKFVLHKIESIDEANLGYSYSVVGGAALPDTAEKITFDSKLVAGPNGGSAGKLTVK 444
Query: 121 YKTIGD 126
Y+T GD
Sbjct: 445 YETKGD 462
>CF921106
Length = 611
Score = 64.7 bits (156), Expect(2) = 1e-19
Identities = 32/91 (35%), Positives = 53/91 (58%)
Frame = +1
Query: 66 KYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASSYKTIG 125
K++ KI+ +D+ NL Y+++ G L D +E I +E K A +GG K+ Y+T G
Sbjct: 226 KFVLHKIESVDEANLGYSYSVVGGVGLPDTVEKITFECKLAAGTNGGSAGKLTVKYQTKG 405
Query: 126 DFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
D +D+K G+ + +++ VE+YLL NP
Sbjct: 406 DAQPNPDDLKIGKVKSDALFKAVEAYLLANP 498
Score = 47.4 bits (111), Expect(2) = 1e-19
Identities = 23/52 (44%), Positives = 35/52 (67%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAG 52
MG+ TF E +S VAP+ ++ AL+ D+ N+IPK + F + V +EG+GG G
Sbjct: 69 MGIFTFEDETTSPVAPATLYKALVTDADNVIPKAVDAF-RSVENLEGNGGPG 221
>TC227429 UP|Q8LJU1 (Q8LJU1) PR10-like protein, partial (98%)
Length = 699
Score = 83.2 bits (204), Expect = 5e-17
Identities = 52/156 (33%), Positives = 86/156 (54%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK L F K V +EG+GG G+I+++ F
Sbjct: 64 MGVFTFEDEINSPVAPATLYKALVTDADNVIPKALDSF-KSVENVEGNGGPGTIKKITFL 240
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E ++ +D E K+ + K+ESI + TN GG K+
Sbjct: 241 EDDVNEW-------IDGET---KFVL-------HKIESI------DETNGGGSAGKLTVK 351
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
Y+T GD + ++++K G+ +++ +E+YLL +P
Sbjct: 352 YETKGDAEPNQDELKTGKAKADALFKAIEAYLLAHP 459
>TC224978 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-induced gene
(H4), partial (84%)
Length = 727
Score = 83.2 bits (204), Expect = 5e-17
Identities = 50/156 (32%), Positives = 84/156 (53%)
Frame = +1
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MG+ TF E +S VAP+ ++ AL+ D+ N+IPK + F + V + G+GG G+I+++ F
Sbjct: 73 MGIFTFEDETTSPVAPATLYKALVTDADNVIPKAVEAF-RSVENLAGNGGPGTIKKITFV 249
Query: 61 EGSPFKYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASS 120
E K++ KI+ +D+ NL Y+++ G L D +E I +E CK+A+
Sbjct: 250 EDGESKFVLHKIESVDEANLGYSYSVLGGVGLPDTVEKITFE------------CKLAAG 393
Query: 121 YKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
K G+ + +++ VE+YLL NP
Sbjct: 394 -------------AKIGKVKSDALFKAVEAYLLANP 462
>AI901164 SP|P26987|SAM Stress-induced protein SAM22. [Soybean] {Glycine
max}, partial (76%)
Length = 418
Score = 66.2 bits (160), Expect(2) = 2e-16
Identities = 33/77 (42%), Positives = 51/77 (65%)
Frame = +2
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK L F K V +EG+GG G+I+++ F
Sbjct: 38 MGVFTFEDEINSPVAPATLYKALVTDADNVIPKALDSF-KSVENVEGNGGPGTIKKITFL 214
Query: 61 EGSPFKYLKQKIDVLDK 77
E K++ KI+ +D+
Sbjct: 215 EDGETKFVLHKIESIDE 265
Score = 35.4 bits (80), Expect(2) = 2e-16
Identities = 13/43 (30%), Positives = 27/43 (62%)
Frame = +3
Query: 114 LCKMASSYKTIGDFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
L K+ Y+T GD + ++++K G+ +++ +E+YLL +P
Sbjct: 258 LMKLTVKYETKGDAEPNQDELKTGKAKADALFKAIEAYLLAHP 386
>TC204836 weakly similar to UP|Q9ZRU8 (Q9ZRU8) Stress and
pathogenesis-related protein, partial (86%)
Length = 1022
Score = 80.9 bits (198), Expect = 2e-16
Identities = 38/90 (42%), Positives = 53/90 (58%)
Frame = +2
Query: 67 YLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASSYKTIGD 126
Y+K +D +D EN + Y++IEG L + LE I YE K AT DGG + K S Y T GD
Sbjct: 638 YVKHHVDAIDTENYVYNYSVIEGSALSEPLEKICYEYKLVATPDGGSIVKSTSKYYTKGD 817
Query: 127 FDVKEEDVKEGRESTIGIYEVVESYLLENP 156
+ EE VK G+E + G + +E ++ NP
Sbjct: 818 EQLAEEYVKTGKERSAGFTKAIEDFIQANP 907
Score = 62.8 bits (151), Expect = 7e-11
Identities = 27/62 (43%), Positives = 43/62 (68%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV T E+ S V+ ++++ A+++D+ N+ PK LP F+K V IEGDGG G+I+++
Sbjct: 105 MGVFTSESEHVSPVSAAKLYKAIVLDASNVFPKALPNFIKSVETIEGDGGPGTIKKLTLA 284
Query: 61 EG 62
EG
Sbjct: 285 EG 290
>TC224995 homologue to UP|Q43453 (Q43453) G.max mRNA from stress-induced gene
(H4), complete
Length = 894
Score = 68.2 bits (165), Expect = 2e-12
Identities = 32/91 (35%), Positives = 55/91 (60%)
Frame = +1
Query: 66 KYLKQKIDVLDKENLICKYTMIEGDPLGDKLESIAYEVKFEATNDGGCLCKMASSYKTIG 125
K++ KI+ +D+ NL Y+++ GD L D +E I +E K A +GG K+ Y+T G
Sbjct: 370 KFVLHKIEAIDEANLGYSYSVVGGDGLPDTVEKITFECKLAAGANGGSAGKLTVKYQTKG 549
Query: 126 DFDVKEEDVKEGRESTIGIYEVVESYLLENP 156
D ++D+K G+ + +++ VE+YLL +P
Sbjct: 550 DAQPNQDDLKIGKAKSDALFKAVEAYLLAHP 642
Score = 57.8 bits (138), Expect = 2e-09
Identities = 28/63 (44%), Positives = 44/63 (69%)
Frame = +3
Query: 1 MGVTTFTHEYSSTVAPSRMFTALIIDSRNLIPKLLPQFVKDVNIIEGDGGAGSIEQVNFN 60
MGV TF E +S VAP+ ++ AL+ D+ N+IPK + F + V +EG+GG G+I+++ F
Sbjct: 63 MGVFTFEDETTSPVAPATLYKALVTDADNVIPKAVDAF-RSVENVEGNGGPGTIKKITFL 239
Query: 61 EGS 63
EG+
Sbjct: 240 EGT 248
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.315 0.137 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,573,726
Number of Sequences: 63676
Number of extensions: 61845
Number of successful extensions: 258
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 239
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 241
length of query: 160
length of database: 12,639,632
effective HSP length: 90
effective length of query: 70
effective length of database: 6,908,792
effective search space: 483615440
effective search space used: 483615440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC144388.8


