

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140104.12 - phase: 0
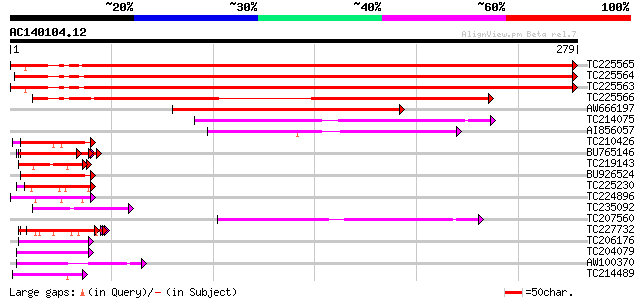
(279 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC225565 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ... 518 e-148
TC225564 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ... 511 e-145
TC225563 similar to UP|RS2_ARATH (P49688) 40S ribosomal protein ... 508 e-144
TC225566 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 ... 290 6e-79
AW666197 similar to GP|18146720|dbj ribosomal protein S2 {Arabid... 153 7e-38
TC214075 similar to UP|P93014 (P93014) 30S ribosomal protein S5,... 80 1e-15
AI856057 similar to PIR|G84749|G847 30S ribosomal protein S5 [im... 54 1e-07
TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protei... 49 3e-06
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 48 4e-06
TC219143 similar to UP|Q9FJJ8 (Q9FJJ8) Arabidopsis thaliana geno... 48 6e-06
BU926524 similar to GP|9758900|dbj| contains similarity to GAR1 ... 46 2e-05
TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein ... 45 4e-05
TC224896 similar to UP|Q94EH8 (Q94EH8) At1g66260/T6J19_1, partia... 45 5e-05
TC235092 similar to UP|O91332 (O91332) EBNA-1, partial (5%) 44 8e-05
TC207560 similar to UP|Q6GKU7 (Q6GKU7) At1g64880, partial (29%) 44 1e-04
TC227732 similar to UP|Q6P623 (Q6P623) Fibrillarin, partial (16%) 44 1e-04
TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein... 43 1e-04
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 41 5e-04
AW100370 similar to GP|16265863|gb| suppressor of K+ transport g... 41 7e-04
TC214489 similar to UP|O65450 (O65450) Glycine-rich protein, par... 41 7e-04
>TC225565 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (92%)
Length = 1164
Score = 518 bits (1335), Expect = e-148
Identities = 266/280 (95%), Positives = 268/280 (95%), Gaps = 1/280 (0%)
Frame = +1
Query: 1 MAERGG-DRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLV 59
MAERGG DR GFG GFGGRG RGG DRGRG RRRAGGRR+EEEKWVPVTKLGRLV
Sbjct: 82 MAERGGGDRAGFGRGFGGRG-----RGG--DRGRG-RRRAGGRRDEEEKWVPVTKLGRLV 237
Query: 60 KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV 119
KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV
Sbjct: 238 KEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVV 417
Query: 120 GDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS 179
GDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS
Sbjct: 418 GDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGS 597
Query: 180 VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGF 239
VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCL+KTYGF
Sbjct: 598 VTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLLKTYGF 777
Query: 240 LTPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
LTPEFWKETRFSKSPFQEYTDLLAKPT KALILEEERVEA
Sbjct: 778 LTPEFWKETRFSKSPFQEYTDLLAKPTGKALILEEERVEA 897
>TC225564 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (93%)
Length = 1047
Score = 511 bits (1315), Expect = e-145
Identities = 260/277 (93%), Positives = 262/277 (93%)
Frame = +1
Query: 3 ERGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEG 62
ERGGDRGGFG GFGGRG RGG DRGRG RR G RREEEEKWVPVTKLGRLVKEG
Sbjct: 88 ERGGDRGGFGRGFGGRG-----RGG--DRGRG--RRRGPRREEEEKWVPVTKLGRLVKEG 240
Query: 63 KIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDN 122
+IRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKI PVQKQTRAGQRTRFKAFVVVGDN
Sbjct: 241 RIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKITPVQKQTRAGQRTRFKAFVVVGDN 420
Query: 123 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 182
NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV
Sbjct: 421 NGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTV 600
Query: 183 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 242
RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP
Sbjct: 601 RMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYGFLTP 780
Query: 243 EFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
EFWKETRFSKSPFQEYTDLLAKPT K LILEEERV+A
Sbjct: 781 EFWKETRFSKSPFQEYTDLLAKPTGKTLILEEERVDA 891
>TC225563 similar to UP|RS2_ARATH (P49688) 40S ribosomal protein S2, partial
(90%)
Length = 1117
Score = 508 bits (1308), Expect = e-144
Identities = 262/284 (92%), Positives = 264/284 (92%), Gaps = 5/284 (1%)
Frame = +2
Query: 1 MAERGG-----DRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKL 55
MAERGG DRGGFG GFGGRG RGG DRGRG RR G RREEEEKWVPVTKL
Sbjct: 77 MAERGGGDRGSDRGGFGRGFGGRG-----RGG--DRGRG--RRRGPRREEEEKWVPVTKL 229
Query: 56 GRLVKEGKIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKA 115
GRLVKEG+IRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKI PVQKQTRAGQRTRFKA
Sbjct: 230 GRLVKEGRIRSLEQIYLHSLPIKEHQIIDTLVGPTLKDEVMKITPVQKQTRAGQRTRFKA 409
Query: 116 FVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTG 175
FVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTG
Sbjct: 410 FVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTG 589
Query: 176 KCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMK 235
KCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMK
Sbjct: 590 KCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMK 769
Query: 236 TYGFLTPEFWKETRFSKSPFQEYTDLLAKPTAKALILEEERVEA 279
TYGFLTPEFWKETRFSKSPFQEYTDLLAKPT K LILEEERV+A
Sbjct: 770 TYGFLTPEFWKETRFSKSPFQEYTDLLAKPTGKTLILEEERVDA 901
>TC225566 similar to UP|Q9SCM3 (Q9SCM3) 40S ribosomal protein S2 homolog
(At3g57490), partial (63%)
Length = 631
Score = 290 bits (741), Expect = 6e-79
Identities = 161/227 (70%), Positives = 165/227 (71%)
Frame = +1
Query: 12 GSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGKIRSLEQIY 71
G GFGGRG RGG DRGRG RR AG RR EEE+WVPVTKLGRLVKEGKIRSLEQIY
Sbjct: 109 GCGFGGRG-----RGG--DRGRGWRRTAG-RRGEEEEWVPVTKLGRLVKEGKIRSLEQIY 264
Query: 72 LHSLPIKEHQIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVK 131
LHSLPIKE+QIIDT VGPTLKDEVMKIMPVQK
Sbjct: 265 LHSLPIKEYQIIDTFVGPTLKDEVMKIMPVQK---------------------------- 360
Query: 132 CSKEVATAIRGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGS 191
LSVIPVRRGYWGNKIGKPH VP KVTGKCGSVTVRMVPAPRGS
Sbjct: 361 -----------------LSVIPVRRGYWGNKIGKPHNVPFKVTGKCGSVTVRMVPAPRGS 489
Query: 192 GIVAARVPKKVLQFAGIDDVFTSSRGSTKTLGNFVKATFDCLMKTYG 238
GIVA RVPKKVLQFAGID VFTSSRGSTKTLGNFVKATFDCL+K+YG
Sbjct: 490 GIVAIRVPKKVLQFAGIDGVFTSSRGSTKTLGNFVKATFDCLLKSYG 630
>AW666197 similar to GP|18146720|dbj ribosomal protein S2 {Arabidopsis
thaliana}, partial (32%)
Length = 354
Score = 153 bits (387), Expect = 7e-38
Identities = 75/114 (65%), Positives = 89/114 (77%)
Frame = +2
Query: 81 QIIDTLVGPTLKDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAI 140
+IIDTLVGPTLKDEVMKI PVQKQTRAGQRTRFKAF V+ +NN H+GL CSKE AT I
Sbjct: 5 EIIDTLVGPTLKDEVMKITPVQKQTRAGQRTRFKAFXVISNNNSHMGLCXXCSKEXATTI 184
Query: 141 RGAIILAKLSVIPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIV 194
G IIL KLSVIP+++GY NKI PHT+P KVT S+T++++P P S I+
Sbjct: 185 HGTIILTKLSVIPIKKGY*RNKIRNPHTIPYKVTKXYKSITIKIIPTP*NSKII 346
>TC214075 similar to UP|P93014 (P93014) 30S ribosomal protein S5, partial
(76%)
Length = 1140
Score = 79.7 bits (195), Expect = 1e-15
Identities = 48/148 (32%), Positives = 79/148 (52%)
Frame = +2
Query: 92 KDEVMKIMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSV 151
++ V+++ V K + G++ RF+A VVVGD G VG+GV +KEV A++ + + A+ ++
Sbjct: 479 EERVVQVRRVTKVVKGGKQLRFRAVVVVGDKKGQVGVGVGKAKEVIAAVQKSAVNARRNI 658
Query: 152 IPVRRGYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDV 211
I V + K T P + G G+ V + PA G+G++A + VL+ AG+D+
Sbjct: 659 IKV-------PMTKYSTFPHRADGDYGAAKVMLRPASPGTGVIAGGSVRIVLEMAGVDNA 817
Query: 212 FTSSRGSTKTLGNFVKATFDCLMKTYGF 239
GS L N +AT + K F
Sbjct: 818 LGKQLGSNNALNN-ARATVAAVQKMKQF 898
>AI856057 similar to PIR|G84749|G847 30S ribosomal protein S5 [imported] -
Arabidopsis thaliana, partial (19%)
Length = 376
Score = 53.5 bits (127), Expect = 1e-07
Identities = 35/126 (27%), Positives = 59/126 (46%), Gaps = 1/126 (0%)
Frame = -1
Query: 98 IMPVQKQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAI-RGAIILAKLSVIPVRR 156
+ V K + + RF+A +V+ D HVG+GV K+V A+ + A+ + S+
Sbjct: 373 VRSVSKXVKXEHQLRFRAMIVLDDKKDHVGVGVDKGKQVIAAV*KSAVNYTRTSIKVA-- 200
Query: 157 GYWGNKIGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSR 216
I K + P K G G+ V + PA G+G++A + VL+ AG+++
Sbjct: 199 ------IAKYSSFPHKDDGDYGASKVMLRPASPGTGVIAGGSVRIVLEMAGVNNALGKQL 38
Query: 217 GSTKTL 222
S L
Sbjct: 37 RSNNAL 20
>TC210426 similar to UP|Q9FK53 (Q9FK53) Similarity to GAR1 protein, partial
(50%)
Length = 434
Score = 48.9 bits (115), Expect = 3e-06
Identities = 25/37 (67%), Positives = 25/37 (67%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
G G G G GGRGG RGGRGG G RGRGG R GGR
Sbjct: 219 GQSAGRGGGGGGRGGGRGGRGGGGFRGRGGPR--GGR 323
Score = 43.5 bits (101), Expect = 1e-04
Identities = 27/46 (58%), Positives = 27/46 (58%), Gaps = 5/46 (10%)
Frame = +3
Query: 2 AERGGDRGGFGSGFGGRGG----DRGG-RGGRGDRGRGGRRRAGGR 42
A RGG GG G G GGRGG RGG RGGRG RGG R GR
Sbjct: 228 AGRGGGGGGRGGGRGGRGGGGFRGRGGPRGGRGGPPRGGGFRGRGR 365
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 48.1 bits (113), Expect = 4e-06
Identities = 24/41 (58%), Positives = 25/41 (60%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREE 45
GG GG G G GG GG+ GG GG G G GG GGRREE
Sbjct: 272 GGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREE 394
Score = 48.1 bits (113), Expect = 4e-06
Identities = 23/37 (62%), Positives = 23/37 (62%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GG GG GG GG G GRGGRR GG
Sbjct: 290 GGGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREEGG 400
Score = 43.9 bits (102), Expect = 8e-05
Identities = 21/37 (56%), Positives = 22/37 (58%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GGRGG GG GG G G GG+ GG
Sbjct: 62 GGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGG 172
Score = 43.5 bits (101), Expect = 1e-04
Identities = 21/37 (56%), Positives = 22/37 (58%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG+RGG G G GG GG G GG G GRGG GG
Sbjct: 244 GGERGGGGGGGGGGGGGGGXGGGGGGGGRGGGGGGGG 354
Score = 43.5 bits (101), Expect = 1e-04
Identities = 22/38 (57%), Positives = 22/38 (57%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
GG GG G G GGRGG GG GG G G GG GGR
Sbjct: 117 GGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGR 230
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/37 (56%), Positives = 22/37 (58%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G G GGRGG+ GG GG G G GG GG
Sbjct: 52 GGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGG 162
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/38 (55%), Positives = 22/38 (57%)
Frame = +2
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
RGG GG G G GG GG +GG GG G G GG GG
Sbjct: 101 RGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGG 214
Score = 42.7 bits (99), Expect = 2e-04
Identities = 19/31 (61%), Positives = 21/31 (67%)
Frame = +2
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
GG+ GG G G GG GG GGRGGR + G GG
Sbjct: 314 GGEGGGGGGGGGGGGGGGGGRGGRREEGGGG 406
Score = 41.2 bits (95), Expect = 5e-04
Identities = 20/36 (55%), Positives = 21/36 (57%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
G GG G G GG GG GG GGRG+ G GG GG
Sbjct: 255 GGGGGGGGGGGGGGGXGGGGGGRGEGGGGGGGGGGG 362
Score = 38.5 bits (88), Expect = 0.003
Identities = 22/46 (47%), Positives = 23/46 (49%), Gaps = 8/46 (17%)
Frame = +2
Query: 4 RGGDRGGFGSGFGGRGGDRG--------GRGGRGDRGRGGRRRAGG 41
+GG GG G G GG GG G GRGGRG G GG GG
Sbjct: 155 KGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGG 292
Score = 37.7 bits (86), Expect = 0.006
Identities = 21/39 (53%), Positives = 21/39 (53%), Gaps = 2/39 (5%)
Frame = +2
Query: 5 GGDRGGFGSGF--GGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG GG G GF G R RGGRGG G G GG GG
Sbjct: 185 GGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGG 301
>TC219143 similar to UP|Q9FJJ8 (Q9FJJ8) Arabidopsis thaliana genomic DNA,
chromosome 5, TAC clone:K19B1, partial (88%)
Length = 689
Score = 47.8 bits (112), Expect = 6e-06
Identities = 22/36 (61%), Positives = 23/36 (63%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAG 40
GG GG G +GG G RGGRGGRG GRGG R G
Sbjct: 447 GGGGGGGGKHYGGGKGGRGGRGGRGGGGRGGHGRGG 554
Score = 41.2 bits (95), Expect = 5e-04
Identities = 24/38 (63%), Positives = 26/38 (68%), Gaps = 4/38 (10%)
Frame = +3
Query: 5 GGDRGG--FGSGFGGRGGDRGGRGG--RGDRGRGGRRR 38
GG GG +G G GGRGG RGGRGG RG GRGG+ R
Sbjct: 453 GGGGGGKHYGGGKGGRGG-RGGRGGGGRGGHGRGGKWR 563
Score = 31.2 bits (69), Expect = 0.54
Identities = 15/29 (51%), Positives = 17/29 (57%)
Frame = +3
Query: 14 GFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
G GG GG + GG+G RG G R GGR
Sbjct: 447 GGGGGGGGKHYGGGKGGRGGRGGRGGGGR 533
Score = 27.3 bits (59), Expect = 7.8
Identities = 14/25 (56%), Positives = 16/25 (64%)
Frame = +3
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGR 28
+GG G G G GGRGG GRGG+
Sbjct: 489 KGGRGGRGGRGGGGRGGH--GRGGK 557
>BU926524 similar to GP|9758900|dbj| contains similarity to GAR1
protein~gene_id:MRG7.14 {Arabidopsis thaliana}, partial
(23%)
Length = 341
Score = 45.8 bits (107), Expect = 2e-05
Identities = 24/37 (64%), Positives = 24/37 (64%)
Frame = +2
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
G G G G GGRGG RGGRGG G RGRG R GGR
Sbjct: 65 GQSAGRGGGGGGRGGFRGGRGGGGFRGRGAPR--GGR 169
Score = 40.0 bits (92), Expect = 0.001
Identities = 24/43 (55%), Positives = 24/43 (55%), Gaps = 5/43 (11%)
Frame = +2
Query: 5 GGDRGGF-----GSGFGGRGGDRGGRGGRGDRGRGGRRRAGGR 42
GG RGGF G GF GRG RGGRGG RGG R G R
Sbjct: 92 GGGRGGFRGGRGGGGFRGRGAPRGGRGG---PPRGGGRGGGFR 211
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/39 (61%), Positives = 24/39 (61%), Gaps = 4/39 (10%)
Frame = +2
Query: 4 RGGDRGG-FGSGFGGRGGDRGGRGG---RGDRGRGGRRR 38
RGG RGG G GF GRG RGGRGG G RG G R R
Sbjct: 101 RGGFRGGRGGGGFRGRGAPRGGRGGPPRGGGRGGGFRGR 217
>TC225230 similar to UP|Q9SZZ1 (Q9SZZ1) Fibrillarin-like protein (Fibrillarin
2) (AtFib2), partial (66%)
Length = 773
Score = 45.1 bits (105), Expect = 4e-05
Identities = 29/49 (59%), Positives = 29/49 (59%), Gaps = 10/49 (20%)
Frame = +1
Query: 4 RGGDRG--GFGSGFGGRGGDRG-----GRGGRGDRGRGGRR---RAGGR 42
RGG RG G G G GGRGGDRG GGRG GRGG R R GGR
Sbjct: 73 RGGGRGDRGRGRGGGGRGGDRGTPFKARGGGRGGGGRGGGRGGGRGGGR 219
Score = 45.1 bits (105), Expect = 4e-05
Identities = 26/42 (61%), Positives = 26/42 (61%), Gaps = 7/42 (16%)
Frame = +1
Query: 8 RGGFGSGFGGRGGDRGGRG-GRGDRGRGGRR------RAGGR 42
RGGFG G G RGG RG RG GRG GRGG R R GGR
Sbjct: 43 RGGFGGGGGFRGGGRGDRGRGRGGGGRGGDRGTPFKARGGGR 168
Score = 37.4 bits (85), Expect = 0.008
Identities = 25/47 (53%), Positives = 25/47 (53%), Gaps = 8/47 (17%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGD---RGGRGGRGDRG-----RGGRRRAGGR 42
RGG GG G GGRG RGG G GDRG RGG R GGR
Sbjct: 43 RGGFGGGGGFRGGGRGDRGRGRGGGGRGGDRGTPFKARGGGRGGGGR 183
Score = 36.6 bits (83), Expect = 0.013
Identities = 22/40 (55%), Positives = 23/40 (57%), Gaps = 6/40 (15%)
Frame = +1
Query: 4 RGGDRG------GFGSGFGGRGGDRGGRGGRGDRGRGGRR 37
RGGDRG G G G GGRG GGRGG GRGG +
Sbjct: 121 RGGDRGTPFKARGGGRGGGGRG---GGRGGGRGGGRGGMK 231
Score = 35.4 bits (80), Expect = 0.029
Identities = 20/33 (60%), Positives = 21/33 (63%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGR 36
RGG RGG G G GGRGG RG GGRG G +
Sbjct: 154 RGGGRGGGGRG-GGRGGGRG--GGRGGMKGGSK 243
>TC224896 similar to UP|Q94EH8 (Q94EH8) At1g66260/T6J19_1, partial (28%)
Length = 1102
Score = 44.7 bits (104), Expect = 5e-05
Identities = 28/51 (54%), Positives = 30/51 (57%), Gaps = 9/51 (17%)
Frame = +2
Query: 1 MAERGGDRGGF------GSGFGGRGGDRGG--RGGRGDRGRG-GRRRAGGR 42
M RGG GG G+G+G RGG R G RGG G RGRG GR AGGR
Sbjct: 200 MTSRGGQAGGGPAMPTRGAGWGRRGGPRSGSGRGGAGGRGRGRGRGGAGGR 352
Score = 39.3 bits (90), Expect = 0.002
Identities = 24/46 (52%), Positives = 26/46 (56%), Gaps = 1/46 (2%)
Frame = +2
Query: 4 RGGDRGGFG-SGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEK 48
RGG R G G G GGRG RG RGG G RGRG + EE +K
Sbjct: 269 RGGPRSGSGRGGAGGRGRGRG-RGGAGGRGRGRKDAVEKSAEELDK 403
>TC235092 similar to UP|O91332 (O91332) EBNA-1, partial (5%)
Length = 641
Score = 43.9 bits (102), Expect = 8e-05
Identities = 25/50 (50%), Positives = 28/50 (56%)
Frame = -2
Query: 12 GSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKE 61
G+ G RG RG RGGRG+ GR GRRR G R KW L RLV +
Sbjct: 160 GARRGARGRSRGRRGGRGE-GRSGRRRNGQGRRSSGKWRVFRVLERLVAQ 14
>TC207560 similar to UP|Q6GKU7 (Q6GKU7) At1g64880, partial (29%)
Length = 743
Score = 43.5 bits (101), Expect = 1e-04
Identities = 37/131 (28%), Positives = 57/131 (43%)
Frame = +2
Query: 103 KQTRAGQRTRFKAFVVVGDNNGHVGLGVKCSKEVATAIRGAIILAKLSVIPVRRGYWGNK 162
K T+ GQ ++ A V G+ NG +G V A++ A ++ V R
Sbjct: 92 KVTKGGQVVKYTAMVACGNYNGVIGFAKAKGPAVPVALQKAYEKCFQNLHYVERH----- 256
Query: 163 IGKPHTVPCKVTGKCGSVTVRMVPAPRGSGIVAARVPKKVLQFAGIDDVFTSSRGSTKTL 222
+ HT+ V V + PAP +G+ A R + +L AG+ +V + GS +
Sbjct: 257 --EEHTIAHAVQTSYKKTKVYLWPAPTTTGMKAGRSVEAILHLAGLKNVKSKVIGS-RNP 427
Query: 223 GNFVKATFDCL 233
N VKA F L
Sbjct: 428 HNTVKAVFKAL 460
>TC227732 similar to UP|Q6P623 (Q6P623) Fibrillarin, partial (16%)
Length = 1693
Score = 43.5 bits (101), Expect = 1e-04
Identities = 26/49 (53%), Positives = 30/49 (61%), Gaps = 5/49 (10%)
Frame = +3
Query: 5 GGDRGGF-GSGFGGRGGDRGGRGGRGDRG----RGGRRRAGGRREEEEK 48
G DRGGF G GF GRG +RGG GGRG+ G RG R GGR + +
Sbjct: 681 GSDRGGFRGRGFRGRG-ERGGFGGRGEHGGFGGRGERGGFGGRGRSDRE 824
Score = 43.1 bits (100), Expect = 1e-04
Identities = 26/52 (50%), Positives = 30/52 (57%), Gaps = 10/52 (19%)
Frame = +3
Query: 6 GDRGGFGS-------GFGGRGGDRGGRGGRGDRGRGGRRRAGG---RREEEE 47
G+RGGFG GFGGR G GGRGG RGRG ++GG RR+ E
Sbjct: 780 GERGGFGGRGRSDREGFGGRWGSEGGRGG---RGRGRNDQSGGWNNRRDSGE 926
Score = 42.7 bits (99), Expect = 2e-04
Identities = 28/62 (45%), Positives = 33/62 (53%), Gaps = 18/62 (29%)
Frame = +3
Query: 6 GDRGGFGS-----GFGGRGGDRGGRGGRG-------------DRGRGGRRRAGGRREEEE 47
G+RGGFG GFGGRG +RGG GGRG + GRGGR R GR ++
Sbjct: 726 GERGGFGGRGEHGGFGGRG-ERGGFGGRGRSDREGFGGRWGSEGGRGGRGR--GRNDQSG 896
Query: 48 KW 49
W
Sbjct: 897 GW 902
Score = 40.8 bits (94), Expect = 7e-04
Identities = 22/37 (59%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Frame = +3
Query: 9 GGFGSGFGGRGG-DRGGRGGRGDRGRGGRRRAGGRRE 44
G + G+ GRGG DRGG GRG RGRG R GGR E
Sbjct: 648 GNWRGGYRGRGGSDRGGFRGRGFRGRGERGGFGGRGE 758
Score = 28.9 bits (63), Expect = 2.7
Identities = 22/50 (44%), Positives = 22/50 (44%), Gaps = 5/50 (10%)
Frame = +3
Query: 2 AERGGDRGGFGS-----GFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEE 46
A D G GS G GG G RGG GRG RGG R G R E
Sbjct: 585 ANASSDDGNEGSNENSDGVGG-GNWRGGYRGRGGSDRGGFRGRGFRGRGE 731
>TC206176 weakly similar to UP|Q8LPA7 (Q8LPA7) Cold shock protein-1, partial
(77%)
Length = 883
Score = 43.1 bits (100), Expect = 1e-04
Identities = 21/37 (56%), Positives = 22/37 (58%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
GG RGG G G+GG GG GG G G G GG R GG
Sbjct: 315 GGGRGGGGGGYGGGGGYGGGGGYGGGGGYGGGGRGGG 425
Score = 39.7 bits (91), Expect = 0.002
Identities = 20/33 (60%), Positives = 21/33 (63%), Gaps = 1/33 (3%)
Frame = +3
Query: 4 RGGDRGGFGSGFG-GRGGDRGGRGGRGDRGRGG 35
RGG GG+G G G G GG GG GG G GRGG
Sbjct: 324 RGGGGGGYGGGGGYGGGGGYGGGGGYGGGGRGG 422
Score = 33.9 bits (76), Expect = 0.084
Identities = 19/33 (57%), Positives = 19/33 (57%), Gaps = 2/33 (6%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRGGR--GGRGDRGRGG 35
GGDR G G G G GG GGR GG G G GG
Sbjct: 489 GGDRYGGGGGGGRYGGGGGGRYVGGGGGGGGGG 587
Score = 32.3 bits (72), Expect = 0.24
Identities = 18/40 (45%), Positives = 21/40 (52%), Gaps = 4/40 (10%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDRGGRGGR----GDRGRGGRRRAGG 41
G GG+G G G GG RGG GG G+ G R +GG
Sbjct: 366 GGGGGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGG 485
Score = 32.3 bits (72), Expect = 0.24
Identities = 23/54 (42%), Positives = 23/54 (42%), Gaps = 17/54 (31%)
Frame = +3
Query: 5 GGDRGGFGSGF--------------GGRGGDRGGRGGRGDR---GRGGRRRAGG 41
GG RGG G GG GGDR G GG G R G GGR GG
Sbjct: 405 GGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGGGGGGRYVGGG 566
Score = 31.2 bits (69), Expect = 0.54
Identities = 23/55 (41%), Positives = 23/55 (41%), Gaps = 18/55 (32%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGGDRG---------------GRGGRGDR---GRGGRRRAGG 41
GG GG G G GGRGG G GG GDR G GG R GG
Sbjct: 375 GGYGGGGGYGGGGRGGGGGACYNCGESGHLARDCSGGGGGDRYGGGGGGGRYGGG 539
Score = 29.6 bits (65), Expect = 1.6
Identities = 16/30 (53%), Positives = 17/30 (56%)
Frame = +3
Query: 6 GDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
G GG+G G G GG GG GG G G GG
Sbjct: 348 GGGGGYGGGGGYGGG--GGYGGGGRGGGGG 431
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 41.2 bits (95), Expect = 5e-04
Identities = 21/38 (55%), Positives = 22/38 (57%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGG 41
RGG GG G G G GG GG GG G G GG R+GG
Sbjct: 325 RGGGGGGGGGGGGYGGGRGGGYGGGGRGGGGGYNRSGG 438
Score = 38.1 bits (87), Expect = 0.004
Identities = 18/32 (56%), Positives = 19/32 (59%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGG 35
R G GG+G G G GG GG GG DRG GG
Sbjct: 427 RSGGGGGYGGGGGYGGGGGGGYGGGRDRGYGG 522
Score = 35.8 bits (81), Expect = 0.022
Identities = 24/41 (58%), Positives = 25/41 (60%), Gaps = 2/41 (4%)
Frame = +1
Query: 5 GGDRGGFGSGFGGRGGDRGGRGGRGDRG--RGGRRRAGGRR 43
GG GG G G+GG G DR G GG GDRG RGG GG R
Sbjct: 460 GGYGGGGGGGYGG-GRDR-GYGGGGDRGYSRGGDGGDGGWR 576
>AW100370 similar to GP|16265863|gb| suppressor of K+ transport growth
defect-like protein {Musa acuminata}, partial (19%)
Length = 411
Score = 40.8 bits (94), Expect = 7e-04
Identities = 26/64 (40%), Positives = 33/64 (50%)
Frame = +1
Query: 4 RGGDRGGFGSGFGGRGGDRGGRGGRGDRGRGGRRRAGGRREEEEKWVPVTKLGRLVKEGK 63
+GG R G S G GG RGG GGRG GG R E+ W PV + RL ++ +
Sbjct: 76 QGGGRRGASSSAGRGGGRRGGGGGRG---------GGGGRGEQRWWDPVWRAERL-RQQQ 225
Query: 64 IRSL 67
IR +
Sbjct: 226 IRPM 237
>TC214489 similar to UP|O65450 (O65450) Glycine-rich protein, partial (9%)
Length = 443
Score = 40.8 bits (94), Expect = 7e-04
Identities = 20/45 (44%), Positives = 26/45 (57%), Gaps = 8/45 (17%)
Frame = +3
Query: 2 AERGGDRGGFGSGFGGRGGDRGGRGG--------RGDRGRGGRRR 38
+E GG GG G G+G GGD+GG G G+RG+GG+ R
Sbjct: 69 SEGGGSSGGEGGGYGAGGGDKGGGIGGDEKGDDHHGERGKGGKDR 203
Score = 33.5 bits (75), Expect = 0.11
Identities = 19/35 (54%), Positives = 21/35 (59%), Gaps = 4/35 (11%)
Frame = +3
Query: 4 RGG---DRGGFGSGFGGRGGDRGGRG-GRGDRGRG 34
RGG DRGG G G GG+ GG G G GD+G G
Sbjct: 36 RGGSRRDRGGGSEGGGSSGGEGGGYGAGGGDKGGG 140
Score = 28.9 bits (63), Expect = 2.7
Identities = 18/39 (46%), Positives = 18/39 (46%), Gaps = 5/39 (12%)
Frame = +3
Query: 8 RGGFGSGFGGRGGDR-----GGRGGRGDRGRGGRRRAGG 41
R G G G GRGG R G GG G GG AGG
Sbjct: 6 RPGRG*GRTGRGGSRRDRGGGSEGGGSSGGEGGGYGAGG 122
Score = 27.7 bits (60), Expect = 6.0
Identities = 15/45 (33%), Positives = 20/45 (44%), Gaps = 20/45 (44%)
Frame = +3
Query: 5 GGDRGGFGSGFGGRGG--------------------DRGGRGGRG 29
GG+ GG+G+G G +GG DRG +GG G
Sbjct: 90 GGEGGGYGAGGGDKGGGIGGDEKGDDHHGERGKGGKDRGHKGGEG 224
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.319 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,086,939
Number of Sequences: 63676
Number of extensions: 156242
Number of successful extensions: 4414
Number of sequences better than 10.0: 512
Number of HSP's better than 10.0 without gapping: 2763
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3806
length of query: 279
length of database: 12,639,632
effective HSP length: 96
effective length of query: 183
effective length of database: 6,526,736
effective search space: 1194392688
effective search space used: 1194392688
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140104.12


