

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140027.12 + phase: 0 /pseudo
(526 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
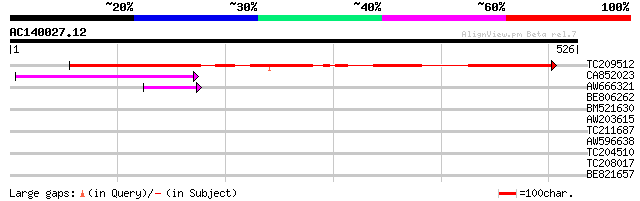
Score E
Sequences producing significant alignments: (bits) Value
TC209512 similar to UP|Q6SF22 (Q6SF22) Tetracycline resistance p... 384 e-107
CA852023 120 1e-27
AW666321 44 2e-04
BE806262 40 0.003
BM521630 30 3.4
AW203615 similar to GP|15983410|gb| AT4g16570/dl4310w {Arabidops... 30 3.4
TC211687 similar to UP|Q7VNN2 (Q7VNN2) Conserved probable membra... 29 5.9
AW596638 28 7.7
TC204510 similar to UP|RR1_SPIOL (P29344) 30S ribosomal protein ... 28 7.7
TC208017 28 7.7
BE821657 23 8.6
>TC209512 similar to UP|Q6SF22 (Q6SF22) Tetracycline resistance protein,
partial (5%)
Length = 1375
Score = 384 bits (986), Expect = e-107
Identities = 237/455 (52%), Positives = 277/455 (60%), Gaps = 3/455 (0%)
Frame = +3
Query: 56 NGLQQTITGIFKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYV 115
NG+QQTI GIFKM VLPLLGQLSDE+GRKPLLL+TIST+I PF LL W+QS+E+VYAYYV
Sbjct: 18 NGVQQTIVGIFKMVVLPLLGQLSDEYGRKPLLLITISTAIFPFVLLVWHQSEEYVYAYYV 197
Query: 116 LRTFSHIISQGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYI 175
LRT S+IISQGSIFCISVAY ADVV+ESKR AVF WITGL SA+HV+ +V A LP+ YI
Sbjct: 198 LRTISNIISQGSIFCISVAYAADVVNESKRAAVFGWITGLLSASHVLGDVLAWSLPEKYI 377
Query: 176 FVILNYGLLLSGFNNTIDLLSTLYAFLPS*NCETGSRKEPRVRLLY*I*GMPQK**SSGC 235
F + +I LL++ ++ ET + P+ SGC
Sbjct: 378 FAV------------SIVLLTSCPVYMKFFLVET-------------VIPAPKNDRESGC 482
Query: 236 LSHV--N*HQSFFELTCIHES*IF-PTLRGVALVSFFYKLGMTGIHSVLLQIFSLLCHYY 292
+ + Q + + E IF PTLRG+ALVSFFY+LGM+GI SVLL YY
Sbjct: 483 WAKIVDVPRQRYISMRRAAEIVIFSPTLRGMALVSFFYELGMSGISSVLL--------YY 638
Query: 293 LTNFF**IP*YNRQSHNSYAVLFESCFWFQQKSVLRTSDDGRNWFHLFSARFQIVLLPIL 352
L F +N+ + ++ + S Q++LLPIL
Sbjct: 639 LKAVF----GFNKNQFSELLMM----------------------VGIGSIFSQMLLLPIL 740
Query: 353 NPLVGEKVILCSALLASIAYAWLSGLAWAPWVSKIVTFYINLIQIYMFIWLAENFITYIS 412
NPLVGEKVILCSALLASIAYAWL GLAWAPW
Sbjct: 741 NPLVGEKVILCSALLASIAYAWLYGLAWAPW----------------------------- 833
Query: 413 IMKVIEFNPLWQVPYLGGSFGIIYILEKPATYGIISKASSSTNQGKAQTFIAGANSISGL 472
VPYL SFGIIY+L KPATY IIS ASSSTNQGKAQTFIAG SIS L
Sbjct: 834 ------------VPYLSASFGIIYVLVKPATYAIISNASSSTNQGKAQTFIAGTQSISDL 977
Query: 473 LSPIVMSPLTSLFLSSDAPFECKGFSIICASVCMV 507
LSPI MSPLTS FLSS+APFECKGFSII AS+CM+
Sbjct: 978 LSPIAMSPLTSWFLSSNAPFECKGFSIISASICMI 1082
>CA852023
Length = 827
Score = 120 bits (301), Expect = 1e-27
Identities = 64/170 (37%), Positives = 97/170 (56%)
Frame = +3
Query: 6 GFFELKPLFHLLLPLSIHWIAEAMTVSVLVDVTTTALCPQQSSCSKAIYINGLQQTITGI 65
G L L HL + + + + + + DVT ALCP Q CS AIY++G QQ + G+
Sbjct: 105 GMEGLGGLGHLFVTMFVTGFGGVIVLPAITDVTMAALCPGQDQCSLAIYLSGFQQAMAGV 284
Query: 66 FKMAVLPLLGQLSDEHGRKPLLLLTISTSIIPFALLAWNQSKEFVYAYYVLRTFSHIISQ 125
+ + PL+G LSD +GRK LL L ++ S+IP +LA+++ +F YAYYV +T + + +
Sbjct: 285 GSVVMTPLIGNLSDRYGRKALLTLPLTVSVIPQVILAYSRDTQFFYAYYVGKTLAAMAGE 464
Query: 126 GSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYI 175
GS C+++AYVAD V + KR + G + + ARFL YI
Sbjct: 465 GSFHCLALAYVADKVPDGKRHQHSEYFAGGWISIVRWWTLAARFLSTAYI 614
>AW666321
Length = 422
Score = 43.5 bits (101), Expect = 2e-04
Identities = 21/54 (38%), Positives = 32/54 (58%)
Frame = +1
Query: 125 QGSIFCISVAYVADVVHESKRVAVFSWITGLSSAAHVIANVFARFLPQNYIFVI 178
+GS C+++AYVAD V + KR + F + G+ SA+ V + ARFL F +
Sbjct: 13 EGSFHCLALAYVADKVPDGKRTSAFGILAGVGSASFVGGTLAARFLSTALTFQV 174
>BE806262
Length = 417
Score = 40.0 bits (92), Expect = 0.003
Identities = 36/127 (28%), Positives = 53/127 (41%)
Frame = +3
Query: 258 PTLRGVALVSFFYKLGMTGIHSVLLQIFSLLCHYYLTNFF**IP*YNRQSHNSYAVLFES 317
PT A+VSFF L G+ +VLL YYL F + + N +A
Sbjct: 69 PTFSQAAMVSFFNSLADGGLMAVLL--------YYLKARF-------QFNKNQFA----- 188
Query: 318 CFWFQQKSVLRTSDDGRNWFHLFSARFQIVLLPILNPLVGEKVILCSALLASIAYAWLSG 377
+L + G LF +PIL P++GE+ +L + LL S ++
Sbjct: 189 -------DLLMITGIGATLAQLF-------FMPILVPVIGEEKLLSTGLLISCINVFVYS 326
Query: 378 LAWAPWV 384
+AW WV
Sbjct: 327 IAWTAWV 347
>BM521630
Length = 445
Score = 29.6 bits (65), Expect = 3.4
Identities = 20/81 (24%), Positives = 39/81 (47%), Gaps = 7/81 (8%)
Frame = +2
Query: 337 FHLFSARFQIVLLPILNPLV----GEKVILCSALLA-SIAYAWLSGLAWAP--WVSKIVT 389
F + R +++P+ +P+ G + A+LA ++ WL AP W + +
Sbjct: 161 FSIRGTRETGIIVPMEDPMTILQKGYSFTIVLAVLAFGLSTRWLLYTEQAPSAWFNFALC 340
Query: 390 FYINLIQIYMFIWLAENFITY 410
I +I Y+F+W+A+ + Y
Sbjct: 341 GLIGIITAYIFVWIAKYYTDY 403
>AW203615 similar to GP|15983410|gb| AT4g16570/dl4310w {Arabidopsis
thaliana}, partial (14%)
Length = 429
Score = 29.6 bits (65), Expect = 3.4
Identities = 14/48 (29%), Positives = 27/48 (56%), Gaps = 5/48 (10%)
Frame = +2
Query: 81 HGRKPLLLLTISTS-----IIPFALLAWNQSKEFVYAYYVLRTFSHII 123
H + +LL+ ++T + P + +++ K+F+Y +LR SHII
Sbjct: 266 HQQLQVLLVGVTTGNSVFGLFPAVVFPYSKGKKFIYMLLILRQVSHII 409
>TC211687 similar to UP|Q7VNN2 (Q7VNN2) Conserved probable membrane protein,
partial (5%)
Length = 632
Score = 28.9 bits (63), Expect = 5.9
Identities = 11/26 (42%), Positives = 16/26 (61%)
Frame = -1
Query: 499 IICASVCMVSYLFLSPIHHLVVFFFF 524
I+CAS+C++S+ S H FF F
Sbjct: 386 ILCASLCLLSFSLYSVFLHCACFFLF 309
>AW596638
Length = 324
Score = 28.5 bits (62), Expect = 7.7
Identities = 10/20 (50%), Positives = 13/20 (65%)
Frame = +1
Query: 505 CMVSYLFLSPIHHLVVFFFF 524
C+ S+ F SP H +FFFF
Sbjct: 85 CLFSFFFFSPSHMFFLFFFF 144
>TC204510 similar to UP|RR1_SPIOL (P29344) 30S ribosomal protein S1,
chloroplast precursor (CS1), partial (89%)
Length = 1475
Score = 28.5 bits (62), Expect = 7.7
Identities = 17/70 (24%), Positives = 37/70 (52%), Gaps = 2/70 (2%)
Frame = -1
Query: 444 YGIISKASSSTNQGKAQTFIAGANSISG--LLSPIVMSPLTSLFLSSDAPFECKGFSIIC 501
+ I++ SS+ + +G + ++G + SP++++PL+ LS A ++I+C
Sbjct: 1334 HAILNHDSSAGGTSLKSSPSSGKSEVNGPRIPSPLIVNPLSGWNLSMSARAIASAWAILC 1155
Query: 502 ASVCMVSYLF 511
+V +S F
Sbjct: 1154 LNV*AISSAF 1125
>TC208017
Length = 626
Score = 28.5 bits (62), Expect = 7.7
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 1/71 (1%)
Frame = +1
Query: 455 NQGKAQTFIAGANSISGLLSPIVMSPLTSLFLSSDAPFECKGFSIIC-ASVCMVSYLFLS 513
NQG I + G+++ + ++ + +S F+CK SI C A+ Y+ LS
Sbjct: 400 NQGSRVNIICNRS*PIGIVNFLEQ*LISGVMMSGSMYFDCKNGSIFCFATTPGRLYM*LS 579
Query: 514 PIHHLVVFFFF 524
P +LV F F
Sbjct: 580 PPIYLVYF*VF 612
>BE821657
Length = 677
Score = 23.5 bits (49), Expect(2) = 8.6
Identities = 12/52 (23%), Positives = 26/52 (49%), Gaps = 9/52 (17%)
Frame = -2
Query: 333 GRNWFHLFSARFQ---------IVLLPILNPLVGEKVILCSALLASIAYAWL 375
G++W S+ F+ + + +NPL V+LCS ++ +A+ ++
Sbjct: 532 GQHWLLR*SSNFEYQNHLKNCYLSFIHTVNPLWVNNVMLCSVIMRVVAFEYV 377
Score = 23.1 bits (48), Expect(2) = 8.6
Identities = 7/14 (50%), Positives = 8/14 (57%)
Frame = -1
Query: 370 IAYAWLSGLAWAPW 383
I + WLS L W W
Sbjct: 278 IQFGWLSNLRWGGW 237
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.339 0.147 0.482
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,696,985
Number of Sequences: 63676
Number of extensions: 466106
Number of successful extensions: 4256
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 4195
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4253
length of query: 526
length of database: 12,639,632
effective HSP length: 102
effective length of query: 424
effective length of database: 6,144,680
effective search space: 2605344320
effective search space used: 2605344320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140027.12


