

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.2 - phase: 0
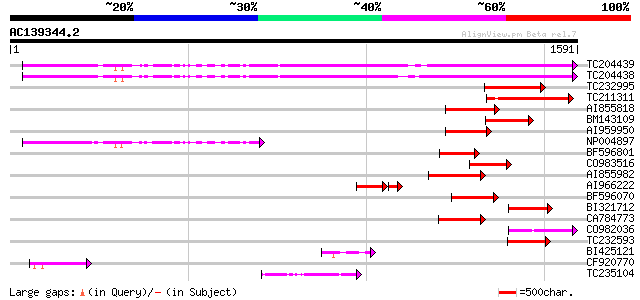
(1591 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete 971 0.0
TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, co... 965 0.0
TC232995 228 2e-59
TC211311 weakly similar to UP|O24587 (O24587) Pol protein, parti... 227 4e-59
AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza ... 221 2e-57
BM143109 169 7e-42
AI959950 160 5e-39
NP004897 gag-protease polyprotein 159 1e-38
BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Gl... 153 7e-37
CO983516 152 1e-36
AI855982 143 5e-34
AI966222 117 5e-32
BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vi... 127 4e-29
BI321712 122 1e-27
CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {V... 121 3e-27
CO982036 121 3e-27
TC232593 weakly similar to UP|Q9XG91 (Q9XG91) Tpv2-1c protein (F... 120 6e-27
BI425121 114 4e-25
CF920770 109 8e-24
TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotei... 107 3e-23
>TC204439 UP|Q84VI4 (Q84VI4) Gag-pol polyprotein, complete
Length = 4731
Score = 971 bits (2510), Expect = 0.0
Identities = 597/1582 (37%), Positives = 856/1582 (53%), Gaps = 27/1582 (1%)
Frame = +1
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG D T + +L
Sbjct: 49 DGSNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTDELKPEEDWTKEEDEL 228
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ VK ++ +L ++E
Sbjct: 229 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKITHEGTSKVKMSRLQLLATKFEN 408
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 409 LKMKEEECIHDFHMNILEIANACTALGERITDEKLVRKILRSLPKRFDMKVTAIEEAQDI 588
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ KKSK++A S E EE+ D D+
Sbjct: 589 CNMRVDELIGSLQTFELGLSDR-AEKKSKNLAFVSN------------DEGEEDEYDLDT 729
Query: 272 DEDQSVKMAMLSNKLEYLARKQKK----------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + + + K F ++GS K K+ D K C
Sbjct: 730 DEGLTNAVVLLGKQFNKVLNRMDKRQKPHVQNIPFDIRKGS-KYQKRSDVKPSHSKGIQC 906
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH IA+CP K+ KG S S D +SE SD
Sbjct: 907 HGCEGYGHIIAECPTHLKKHRKGLSVCQS-------------------DTESEQESD--- 1020
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G+ T +ED ++ S+I EL S ++L E +
Sbjct: 1021 SDRDVNALTGIFET------------AEDSSDTDSEITFDELAASYRKLCIKSEKILQQE 1164
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGN 493
LK+ DL ++++ E+ ELKG +++ E+ KS+ + +KGS
Sbjct: 1165 AQLKKVIADLEAEKEAHKEEIS----ELKGEVGFLNSKLENMTKSI-----KMLNKGSDT 1317
Query: 494 KHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLK 551
LD+ ++ G KN G +G+G++ + + ++
Sbjct: 1318 -----LDEVLLLG--------------KNAGNQRGLGFNPKSAGRTTM------------ 1404
Query: 552 STFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVH 610
+ FVP + T+ Q + G Q K KS K + +
Sbjct: 1405 TEFVPAKNRTGATMSQHRSRHHGMQQK--------------KSKRKKWRCHYCGK----- 1527
Query: 611 QNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSW 670
IKP ++ S K + K K ++ L +H + A+E W
Sbjct: 1528 YGHIKPFCYHLHGHPHHGTQSSNSRKKMMWVPKHKAVS----LVVHTSLRASAKE---DW 1686
Query: 671 YLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDG 729
YLDSGCSRHMTG K L + V FG GKIIG G + + + S+N V LV G
Sbjct: 1687 YLDSGCSRHMTGVKEFLLNIEPCSTSYVTFGDGSKGKIIGMGKLVHDGLPSLNKVLLVKG 1866
Query: 730 LKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVC 789
L NL+SISQ CD ++V F+K+ C LV + + KG R ++ + C
Sbjct: 1867 LTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTC 2043
Query: 790 LLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFK 849
L S D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S +
Sbjct: 2044 LSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQ 2223
Query: 850 SKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFS 909
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 2224 KLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSETFEVFK 2403
Query: 910 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 969
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNR
Sbjct: 2404 ELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTSEGITHEFSAAITPQQNGIVERKNR 2583
Query: 970 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 1029
TLQE AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P++ +FH
Sbjct: 2584 TLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPSVKHFHI 2763
Query: 1030 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPG 1089
FG CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P
Sbjct: 2764 FGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLSPA 2943
Query: 1090 SKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQN 1149
K + + + DA++S + +++ S+S + S I Q
Sbjct: 2944 RKKDVEEDVRTSGDNVADAAKSGENAEN-------------------SDSATDESNINQP 3066
Query: 1150 ESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPK 1209
+ S + + HP+ELIIG + TRS + S +S IEPK
Sbjct: 3067 DKRSST-------------RIQKMHPKELIIGDPNRGVTTRSREVEIVSNSCFVSKIEPK 3207
Query: 1210 TVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNK 1269
V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNK
Sbjct: 3208 NVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNK 3387
Query: 1270 ARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEE 1329
ARLVAQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + EE
Sbjct: 3388 ARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEE 3567
Query: 1330 VYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRR 1389
VYV+QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF +
Sbjct: 3568 VYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVK 3747
Query: 1390 TLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGV 1449
++++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +
Sbjct: 3748 QDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSI 3927
Query: 1450 YVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTAS 1509
++ Q++Y K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTAS
Sbjct: 3928 FLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTAS 4107
Query: 1510 RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYA 1569
RPDI ++V +CAR+Q++P+ SHLT VKRI +Y+ GT++ G++Y + L+G+CDAD+A
Sbjct: 4108 RPDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDADWA 4287
Query: 1570 GDRIERKSTSGNCQFLGENLIS 1591
G +RKSTSG C +LG NLIS
Sbjct: 4288 GSADDRKSTSGGCFYLGNNLIS 4353
>TC204438 homologue to UP|Q84VH6 (Q84VH6) Gag-pol polyprotein, complete
Length = 4734
Score = 965 bits (2495), Expect = 0.0
Identities = 595/1582 (37%), Positives = 857/1582 (53%), Gaps = 27/1582 (1%)
Frame = +1
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWDILEDGVDDLD-LDEEGAAIDR----RIHTPAQKKL 91
D + +WK M +F+ LD W + G + LD EG + T + +L
Sbjct: 49 DGTNYEYWKARMVAFLKSLDSRTWKAVIKGWEHPKMLDTEGKPTNELKPEEDWTKEEDEL 228
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ VK ++ +L ++E
Sbjct: 229 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 408
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 409 LKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 588
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
+ V++L+ SL+ E+ L++ T KKSK++A S E EE+ D D+
Sbjct: 589 CNMRVDELIGSLQTFELGLSDR-TEKKSKNLAFVSN------------DEGEEDEYDLDT 729
Query: 272 DEDQSVKMAMLSNK----LEYLARKQKK------FLSKRGSYKNSKKEDQKG-------C 314
DE + + +L + L + R+QK F ++GS + KK D+K C
Sbjct: 730 DEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGS-EYQKKSDEKPSHSKGIQC 906
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH A+CP K++ KG S S +D +SE SD
Sbjct: 907 HGCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESD--- 1023
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G + +ED ++ S+I EL S +EL E +
Sbjct: 1024 SDRDVNALTGRFES------------AEDSSDTDSEITFDELAISYRELCIKSEKILQQE 1167
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGN 493
LK+ +L ++++ E+ ELKG +++ E+ KS+ + +KGS
Sbjct: 1168 AQLKKVIANLEAEKEAHEEEIS----ELKGEVGFLNSKLENMTKSI-----KMLNKGSD- 1317
Query: 494 KHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLK 551
LD+ + G KN G +G+G++ + + ++
Sbjct: 1318 ----MLDEVLQLG--------------KNVGNQRGLGFNHKSAGRTTM------------ 1407
Query: 552 STFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVH 610
+ FVP + + T+ Q + G+Q K KS K + +
Sbjct: 1408 TEFVPAKNSTGATMSQHRSRHHGTQQK--------------KSKRKKWRCHYCGK----- 1530
Query: 611 QNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSW 670
IKP +++S + + K K+++ L +H + A+E W
Sbjct: 1531 YGHIKPFCYHLHGHPHHGTQSSSSGRKMMWVPKHKIVS----LVVHTSLRASAKE---DW 1689
Query: 671 YLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSI-SINNVWLVDG 729
YLDSGCSRHMTG K + + V FG GKI G G + + + S+N V LV G
Sbjct: 1690 YLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKLVHDGLPSLNKVLLVKG 1869
Query: 730 LKHNLLSISQFCDNEYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKINFSDLADQKVVC 789
L NL+SISQ CD ++V F+K+ C LV + + KG R ++ + C
Sbjct: 1870 LTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSKDNCYLWTPQETSYSSTC 2046
Query: 790 LLSMNDKKWVWHKRLGHANWRLISKISKLQLVKGLPNIDYHSDALCGACQKGKIVKSSFK 849
L S D+ +WH+R GH + R + KI V+G+PN+ +CG CQ GK VK S +
Sbjct: 2047 LFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEGRICGECQIGKQVKMSHQ 2226
Query: 850 SKDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFIKSKDYACEVFS 909
+TSR LELLH+DL GP+ SL G +Y V+VDD+SR+TWV FI+ K EVF
Sbjct: 2227 KLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRFTWVNFIREKSDTFEVFK 2406
Query: 910 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 969
++Q EK+ I ++RSDHG EFEN F FC GI HEFS+ TPQQNG+VERKNR
Sbjct: 2407 ELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEFSAAITPQQNGIVERKNR 2586
Query: 970 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 1029
TLQE AR M+H L + WAEA+NT+CYI NR+ +R T YE++KGR+P + +FH
Sbjct: 2587 TLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTTLYEIWKGRKPTVKHFHI 2766
Query: 1030 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPG 1089
FG CYIL ++ +K D K+ GIFLGYS S+AYRV+NS T+ V ES++V DD P
Sbjct: 2767 FGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRTRTVMESINVVVDDLTPA 2946
Query: 1090 SKTSEQSESNAGTTDSEDASESDQPSDSEKYTKVESSPEAEITPEAESNSEAESSPIVQN 1149
K D E+ + A+ AE+ +N
Sbjct: 2947 RK-----------------------KDVEEDVRTSGDNVADTAKSAEN---------AEN 3030
Query: 1150 ESASEDFQDNTQQVIQPKFKHKSSHPEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPK 1209
++ D + Q +P + + HP+ELIIG + TRS + S +S IEPK
Sbjct: 3031 SDSATDEPNINQPDKRPSIRIQKMHPKELIIGDPNRGVTTRSREIEIVSNSCFVSKIEPK 3210
Query: 1210 TVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNK 1269
V+EAL+D+ WI AMQEEL QF+RN+VW+LVP+P N+I TKW+F+NK NE+G +TRNK
Sbjct: 3211 NVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNK 3390
Query: 1270 ARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEE 1329
ARLVAQGY+Q EG+++ ETFAPVARLE+IRLLL A LYQMDVKS FLNG + EE
Sbjct: 3391 ARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEE 3570
Query: 1330 VYVKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRR 1389
YV+QP GF D HPDHVY+LKK+LYGLKQAPRAWY+RL+ FL + + +G +D TLF +
Sbjct: 3571 AYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVK 3750
Query: 1390 TLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGV 1449
++++I QIYVDDI+FG + + + F + MQ EFEMS++GEL +FLG+Q+ Q ++ +
Sbjct: 3751 QDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSI 3930
Query: 1450 YVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTAS 1509
++ Q+KY K ++KKF +E+ TP LSK++ GT VDQ LYR MIGSLLYLTAS
Sbjct: 3931 FLSQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTAS 4110
Query: 1510 RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYA 1569
RPDI ++V +CAR+Q++P+ SHL VKRI +Y+ GT++ G++Y D L+G+CDAD+A
Sbjct: 4111 RPDITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWA 4290
Query: 1570 GDRIERKSTSGNCQFLGENLIS 1591
G +RKSTSG C +LG NLIS
Sbjct: 4291 GSADDRKSTSGGCFYLGTNLIS 4356
>TC232995
Length = 1009
Score = 228 bits (581), Expect = 2e-59
Identities = 108/173 (62%), Positives = 139/173 (79%)
Frame = +2
Query: 1332 VKQPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTL 1391
V+QPPGFE P+HVYKL+K+LYGLKQAPRAWY+RLSNFL++ +F RG+VDTTLF +
Sbjct: 2 VEQPPGFEISDKPNHVYKLQKALYGLKQAPRAWYERLSNFLLEKEFSRGKVDTTLFIKRK 181
Query: 1392 KKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYV 1451
DIL+VQIYVDDIIFGSTN SLCKEFS MQ EFEMSMMGELK+FLG+QI Q++ G+++
Sbjct: 182 HNDILLVQIYVDDIIFGSTNDSLCKEFSLDMQSEFEMSMMGELKYFLGLQIKQTQ*GIFI 361
Query: 1452 HQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLL 1504
+Q+KY KEL+K+F ++ K M+TPM C L K+++G +D K YR IG ++
Sbjct: 362 NQSKYCKELIKRFGMDSAKHMSTPMSTNCYLDKDESGQSIDIKQYRDAIGEVV 520
>TC211311 weakly similar to UP|O24587 (O24587) Pol protein, partial (15%)
Length = 1213
Score = 227 bits (578), Expect = 4e-59
Identities = 121/245 (49%), Positives = 159/245 (64%)
Frame = +3
Query: 1337 GFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDIL 1396
GFED + P HV+ + L L +F + D R K+ L
Sbjct: 330 GFEDKERPCHVFMV*NKL*ELGMKG*V------HF*FQMDSPEE*RTPHYSERLKKETFL 491
Query: 1397 IVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKY 1456
I+ IYVDDIIFG+T+ +CKEF +LM+D FE SM GELKF LG+QI Q G+++HQ KY
Sbjct: 492 IIHIYVDDIIFGATSKRMCKEFFELMKDGFETSMKGELKFLLGLQIIQKVYGIFIHQEKY 671
Query: 1457 TKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFS 1516
TK LK+F++++ K M TPMH + + K++ G K Y GMI SL YLT+SRPDI+F
Sbjct: 672 TKSHLKRFRMDEAKPMATPMHRSTIIDKDEKGNHTS*KEYSGMIDSLSYLTSSRPDIVFV 851
Query: 1517 VCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERK 1576
VCLCARFQS P+ SH+TAVKRI RYL GTTN L ++K ++ L+G+CD +AGD++ERK
Sbjct: 852 VCLCARFQSYPKISHVTAVKRILRYLVGTTNHCLWFKKRSEFDLLGYCDVYFAGDKVERK 1031
Query: 1577 STSGN 1581
STS N
Sbjct: 1032 STSRN 1046
>AI855818 weakly similar to GP|21741393|e OSJNBb0051N19.6 {Oryza sativa
(japonica cultivar-group)}, partial (10%)
Length = 463
Score = 221 bits (563), Expect = 2e-57
Identities = 106/153 (69%), Positives = 128/153 (83%)
Frame = -3
Query: 1222 LAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQE 1281
+AMQEELNQF+RN+VW LV KP +I TKWVFRNKL+E G + RNKARLVA+GY+Q+E
Sbjct: 461 IAMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIIIRNKARLVAKGYNQEE 282
Query: 1282 GINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDL 1341
GI+Y ET+APVARLE IR+LL+Y LYQMDVKS FLNG+I+EEVYV+QPPGFE
Sbjct: 281 GIDYEETYAPVARLEVIRMLLAYVSIMNFKLYQMDVKSAFLNGLIQEEVYVEQPPGFEIP 102
Query: 1342 KHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIK 1374
P HVYKL+K+LYGLKQAPRAWY+R+SNFL++
Sbjct: 101 DKPTHVYKLQKALYGLKQAPRAWYERISNFLLE 3
>BM143109
Length = 415
Score = 169 bits (429), Expect = 7e-42
Identities = 83/137 (60%), Positives = 107/137 (77%)
Frame = +1
Query: 1334 QPPGFEDLKHPDHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKK 1393
QPP ++ + P+HV+KLKK LYGLKQA RAWY+ LS FL+ F +G+VDT LF
Sbjct: 4 QPPVRKNSEKPNHVFKLKKVLYGLKQALRAWYELLSKFLLDKGFSKGKVDTNLFI*KKLN 183
Query: 1394 DILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQ 1453
DIL+VQIYVDDIIFGSTN SLCK+FS+ MQ+EFEMSMM EL FFLG+QI Q+K G+++ Q
Sbjct: 184 DILLVQIYVDDIIFGSTNDSLCKKFSQDMQNEFEMSMMRELNFFLGLQIKQTKNGIFISQ 363
Query: 1454 TKYTKELLKKFKLEDCK 1470
+KY K+L+ +F +E+ K
Sbjct: 364 SKYCKDLIHRFGMENDK 414
>AI959950
Length = 466
Score = 160 bits (404), Expect = 5e-39
Identities = 81/130 (62%), Positives = 102/130 (78%)
Frame = -1
Query: 1223 AMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEG 1282
AMQEEL+QFQ+N+V LV P +K ++ KW+F NKL+E G+V R KARLVA+GYSQQEG
Sbjct: 391 AMQEELDQFQKNNV*KLVKLPKRKKVVGVKWIFCNKLDEDGKVVRYKARLVAKGYSQQEG 212
Query: 1283 INYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLK 1342
I+Y +TFA VARLE I +LLS+A + LYQMDVKS FLNG+I++EVYV+QPPGFE+
Sbjct: 211 IDYPKTFALVARLEVICILLSFATYSNMKLYQMDVKSAFLNGLIQKEVYVEQPPGFENET 32
Query: 1343 HPDHVYKLKK 1352
HV+KL K
Sbjct: 31 LHQHVFKLNK 2
>NP004897 gag-protease polyprotein
Length = 1923
Score = 159 bits (401), Expect = 1e-38
Identities = 177/704 (25%), Positives = 293/704 (41%), Gaps = 26/704 (3%)
Frame = +1
Query: 37 DPEEFSWWKTNMYSFIMGLDEELWD-ILEDGVDDLDLDEEGAAID----RRIHTPAQKKL 91
D + +WK M +F+ LD W +++D LD EG D T + +L
Sbjct: 49 DGTNYEYWKARMVAFLKSLDSRTWKAVIKDWEHPKMLDTEGKPTDGLKPEEDWTKEEDEL 228
Query: 92 YKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYEL 151
+ K + + + + ++ + AK + L EG+ VK ++ +L ++E
Sbjct: 229 ALGNSKALNALFNGVDKNIFRLINTCTVAKDAWEILKTTHEGTSKVKMSRLQLLATKFEN 408
Query: 152 FRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL 211
+MK++E I + + + + L + V KILRSLP R+ KVTAIEEA+D+
Sbjct: 409 LKMKEEECIHDFHMNILEIANACTALGERMTDEKLVRKILRSLPKRFDMKVTAIEEAQDI 588
Query: 212 NTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDS 271
L V++L+ SL+ E+ L++ T KKSK++A S E EE+ D D+
Sbjct: 589 CNLRVDELIGSLQTFELGLSD-RTEKKSKNLAFVSN------------DEGEEDEYDLDT 729
Query: 272 DEDQSVKMAMLSNK----LEYLARKQK------KFLSKRGSYKNSKKEDQK-------GC 314
DE + + +L + L + R+QK F ++GS + K+ D+K C
Sbjct: 730 DEGLTNAVVLLGKQFNKVLNRMDRRQKPHVRNIPFDIRKGS-EYQKRSDEKPSHSKGFQC 906
Query: 315 FNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C+ GH A+CP K++ KG S S +D +SE SD
Sbjct: 907 HGCEGYGHIKAECPTHLKKQRKGLSVCRS------------------DDTESEQESD--- 1023
Query: 375 ADDDAKAAMGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNEL 434
+D D A G +ED ++ S+I EL S +EL E +
Sbjct: 1024SDRDVNALTGRF------------ESAEDSSDTDSEITFDELATSYRELCIKSEKILQQE 1167
Query: 435 TDLKEKYVDLMKQQKSTLLELKASEEELKG-FNLISTTYEDRLKSLCQKLQEKCDKGSGN 493
LK+ +L ++++ E+ ELKG +++ E+ KS+ + +KGS
Sbjct: 1168AQLKKVIANLEAEKEAHEEEI----SELKGEVGFLNSKLENMTKSI-----KMLNKGSD- 1317
Query: 494 KHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKSYCDCIKDGLK 551
LD+ + G KN G +G+G++ + + ++
Sbjct: 1318----MLDEVLQLG--------------KNVGNQRGLGFNHKSAGRITM------------ 1407
Query: 552 STFVP-EGTNAVTVVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVH 610
+ FVP + + T+ Q + G+Q K KS K + +
Sbjct: 1408TEFVPAKISTGATMSQHRSRHHGTQQK--------------KSKRKKWRCHYCGK----- 1530
Query: 611 QNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVCLRAREKQRSW 670
IKP +++S + + K K+++ L +H + A+E W
Sbjct: 1531YGHIKPFCYHLHGHPHHGTQSSSSRRKMMWVPKHKIVS----LVVHTSLRASAKE---DW 1689
Query: 671 YLDSGCSRHMTGEKALFLTLTMKDGGEVKFGGNQTGKIIGTGTI 714
YLDSGCSRHMTG K + + V FG GKI G G +
Sbjct: 1690YLDSGCSRHMTGVKEFLVNIEPCSTSYVTFGDGSKGKITGMGKL 1821
>BF596801 weakly similar to GP|29423270|g gag-pol polyprotein {Glycine max},
partial (7%)
Length = 336
Score = 153 bits (386), Expect = 7e-37
Identities = 73/111 (65%), Positives = 89/111 (79%)
Frame = +3
Query: 1207 EPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNEQGEVT 1266
EPK ++EA+ DD WI+ MQEELNQF+RN+VW LV KP +I TKWVFRNKL+E G +
Sbjct: 3 EPKNIKEAIVDDNWIIVMQEELNQFERNNVWKLVEKPENYPVIGTKWVFRNKLDEHGIII 182
Query: 1267 RNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDV 1317
RNKARLVA+GY+Q+EGI+Y ET+APVARLE IR+LL+YA LYQMDV
Sbjct: 183 RNKARLVAKGYNQEEGIDYEETYAPVARLEAIRMLLAYASIMNFKLYQMDV 335
>CO983516
Length = 724
Score = 152 bits (384), Expect = 1e-36
Identities = 74/120 (61%), Positives = 91/120 (75%)
Frame = +2
Query: 1289 FAPVARLETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVY 1348
F PVARLE+IRLLL A LYQMDVKS FLNG + EEVYV+QP GF D HPDHVY
Sbjct: 365 FHPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEVYVEQPKGFIDPTHPDHVY 544
Query: 1349 KLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFG 1408
+LKK+LYGLKQAPRAWY+RL+ L + + +G +D TLF + ++++I QIYVDDI+FG
Sbjct: 545 RLKKALYGLKQAPRAWYERLTELLTQQGYRKGGIDKTLFVKQDAENLMIAQIYVDDIVFG 724
>AI855982
Length = 484
Score = 143 bits (361), Expect = 5e-34
Identities = 76/161 (47%), Positives = 103/161 (63%)
Frame = +2
Query: 1175 PEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRN 1234
P + IIG TR + + + +S+IEPK ++EA+ DD WI+AMQEELNQF+RN
Sbjct: 2 PLDNIIGDISKGVTTRHSLKDLCNNMAFVSMIEPKNIKEAIVDDNWIIAMQEELNQFERN 181
Query: 1235 DVWDLVPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVAR 1294
+VW LV KP +I TKWVFRNKL+E + +KARLVA+GY+Q +G++Y T+A +AR
Sbjct: 182 NVWKLVEKPDNYPVI*TKWVFRNKLDEHRIIIIHKARLVAEGYNQVDGLDYEHTYASIAR 361
Query: 1295 LETIRLLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQP 1335
L I + LSY LY S L+G++ EVYV QP
Sbjct: 362 L*VIIMPLSYVYIMNSTLYHYACVSALLHGLLLHEVYVDQP 484
>AI966222
Length = 430
Score = 117 bits (294), Expect(2) = 5e-32
Identities = 52/88 (59%), Positives = 70/88 (79%)
Frame = +1
Query: 973 EMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQFGC 1032
EMART +++N KHF AE +N CY+QN+IYIRP+L++T YEL+KGR+PNISYF+ F C
Sbjct: 1 EMARTTLNDNLTPKHF*AEVMNIVCYLQNKIYIRPILKRTPYELWKGRKPNISYFYPFRC 180
Query: 1033 TCYILNTKDYLKKFDAKAQRGIFLGYSE 1060
C+I+NTKD L K D+K+ GIF+ YS+
Sbjct: 181 KCFIINTKDNLGKIDSKSDCGIFIAYSK 264
Score = 40.4 bits (93), Expect(2) = 5e-32
Identities = 18/39 (46%), Positives = 26/39 (66%)
Frame = +2
Query: 1062 SKAYRVYNSETQCVEESMHVKFDDREPGSKTSEQSESNA 1100
SKA+RVYNS T +EE++H++F +P + E ES A
Sbjct: 269 SKAFRVYNSGTLVIEEAIHIRFGKNKPNKELLELDESFA 385
>BF596070 similar to GP|27901698|gb gag-pol polyprotein {Vitis vinifera},
partial (34%)
Length = 407
Score = 127 bits (319), Expect = 4e-29
Identities = 64/131 (48%), Positives = 91/131 (68%)
Frame = -2
Query: 1240 VPKPFQKNIIETKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIR 1299
VP P K + +WV+ K+ GEV R KARLVA+GY+Q GI+Y +TF+PVA+L T+R
Sbjct: 406 VPLPPGKTPVGCRWVYTVKVGPTGEVDRLKARLVAKGYTQVYGIDYCDTFSPVAKLTTVR 227
Query: 1300 LLLSYAINHGIILYQMDVKSVFLNGVIEEEVYVKQPPGFEDLKHPDHVYKLKKSLYGLKQ 1359
L L+ A L+Q+D+K+ FL+G +EE++Y++QPPGF V KL +SLYGLKQ
Sbjct: 226 LFLAMAAICHWPLHQLDIKNAFLHGDLEEDIYMEQPPGFVAQGEYGLVCKLHRSLYGLKQ 47
Query: 1360 APRAWYDRLSN 1370
+PRAW+ + S+
Sbjct: 46 SPRAWFGKFSH 14
>BI321712
Length = 399
Score = 122 bits (307), Expect = 1e-27
Identities = 58/124 (46%), Positives = 85/124 (67%)
Frame = -3
Query: 1400 IYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKE 1459
+YVDD+IF N S+ +EF K M +EFEM+ MG + ++LGI++ Q +G+++ Q Y KE
Sbjct: 379 LYVDDLIFTGNNPSMFEEFKKDMSNEFEMTDMGLMAYYLGIEVKQEDKGIFITQEGYAKE 200
Query: 1460 LLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCL 1519
+LKKFK++D + TPM LSK + G VD LY+ +IGSL YLT +RPDIL+ V +
Sbjct: 199 VLKKFKMDDANPVGTPMECGSKLSKHEKGENVDPTLYKSLIGSLRYLTCTRPDILYVVGV 20
Query: 1520 CARF 1523
+R+
Sbjct: 19 VSRY 8
>CA784773 weakly similar to GP|27901698|gb gag-pol polyprotein {Vitis
vinifera}, partial (34%)
Length = 409
Score = 121 bits (303), Expect = 3e-27
Identities = 59/134 (44%), Positives = 86/134 (64%)
Frame = +3
Query: 1202 LLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIETKWVFRNKLNE 1261
L S+ P T+ EAL GW AM +E+ + N W+LVP P K + +WV+ K+
Sbjct: 3 LSSLTVPSTIREALDHPGWRQAMVDEMQALENNGTWELVPLPPGKTTVGCRWVYTVKVGP 182
Query: 1262 QGEVTRNKARLVAQGYSQQEGINYTETFAPVARLETIRLLLSYAINHGIILYQMDVKSVF 1321
G+V R KARLVA+GY+Q GI Y +TF+PV L T+RL L+ A L+Q+D+K+ F
Sbjct: 183 NGKVDRLKARLVAKGYTQVYGIEYCDTFSPVFFLTTVRLFLAMAAIRHWPLHQLDIKNAF 362
Query: 1322 LNGVIEEEVYVKQP 1335
L+G +EE++Y++QP
Sbjct: 363 LHGDLEEDIYMEQP 404
>CO982036
Length = 674
Score = 121 bits (303), Expect = 3e-27
Identities = 69/195 (35%), Positives = 115/195 (58%), Gaps = 3/195 (1%)
Frame = -2
Query: 1400 IYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKE 1459
+YVD II GS+ +L + + + F + ++G+L +F+ I++ + ++ +T +
Sbjct: 640 VYVDIIITGSS-CTLIQNLTSKLNSSFPLKLLGKLDYFVEIEVKSMPDLLFSLRTSIFEI 464
Query: 1460 LLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCL 1519
+K + + + +++PM TC LSK D+ YR ++G+L Y T RP+I F+V
Sbjct: 463 FCRKPR*Q-AQPISSPMTTTCKLSKSDSDLFSGPTFYRSVVGALQYTTVIRPEISFAVNK 287
Query: 1520 CARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYK---LIGFCDADYAGDRIERK 1576
+F S+P +SH T VKRI RYLKG+ + GL + ++ + + GFCDAD+A +++
Sbjct: 286 VCQFMSNPLDSHWTEVKRILRYLKGSLSYGL*LKPAISSQPLPIRGFCDADWASAVDDKR 107
Query: 1577 STSGNCQFLGENLIS 1591
STSG FLG NLIS
Sbjct: 106 STSGAAVFLGPNLIS 62
>TC232593 weakly similar to UP|Q9XG91 (Q9XG91) Tpv2-1c protein (Fragment),
partial (16%)
Length = 562
Score = 120 bits (300), Expect = 6e-27
Identities = 57/122 (46%), Positives = 85/122 (68%)
Frame = +1
Query: 1396 LIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTK 1455
LIV +YVDD++ +A L +EF + M FEM+ +G + +FLGI+I QS+ V + Q K
Sbjct: 193 LIVSLYVDDLLVTRDDARLVEEFKQEMMQAFEMTNLGLMTYFLGIEIKQSQNKVLICQRK 372
Query: 1456 YTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILF 1515
Y KE+LKKF++E+CK ++TPM+ +K D +D+ YR +IG L+YLTA+RPDILF
Sbjct: 373 YAKEILKKFQMEECKSVSTPMNQKEKFNKVDGADKIDEGYYRSLIGCLMYLTATRPDILF 552
Query: 1516 SV 1517
++
Sbjct: 553 AI 558
>BI425121
Length = 412
Score = 114 bits (284), Expect = 4e-25
Identities = 69/162 (42%), Positives = 89/162 (54%), Gaps = 10/162 (6%)
Frame = +3
Query: 875 SLYGSKYGLVIVDDYSRWTWVKFIKSKDYAC----------EVFSSFCTQIQSEKELKIL 924
SL KY + VDDYSR+TWV F+ K + +VF ++ E L+IL
Sbjct: 6 SLGCKKYEFLTVDDYSRYTWVYFLAHKHESLRYFIRGFKMKKVFVFLLLEVTMELSLRIL 185
Query: 925 KVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNL 984
FCE++GI H S PRTPQ+N VVERKNRTLQE ART++
Sbjct: 186 S*NH-------------FCERNGIFHNLS*PRTPQENRVVERKNRTLQEKARTILF---- 314
Query: 985 AKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISY 1026
T+C++QN+I IRPM++KT YEL+KGRR ISY
Sbjct: 315 ----------TTCFVQNKILIRPMIKKTPYELWKGRRHIISY 410
>CF920770
Length = 581
Score = 109 bits (273), Expect = 8e-24
Identities = 62/187 (33%), Positives = 104/187 (55%), Gaps = 12/187 (6%)
Frame = -2
Query: 56 DEELWDILEDG------VDDLDLDEEGAAIDRRIHTPAQK------KLYKKHHKIRGIIV 103
D +W+ +E G V+ + +D ++ I P + K + + K + II
Sbjct: 574 DLNIWEAIEIGPYIPTTVERVSIDGSSSSESITIEKPRDRWSEEDRKRVQYNLKAKNIIT 395
Query: 104 ASIPRTEYMKMSDKSTAKAMFASLCANFEGSKNVKEAKALMLVHQYELFRMKDDESIEEM 163
+++ EY ++S+ +AK M+ +L EG+ +VK ++ L H+YELFRM +E+I+ M
Sbjct: 394 SALGMDEYFRVSNCKSAKEMWDTLRLTHEGTTDVKRSRINALTHEYELFRMNTNENIQSM 215
Query: 164 YSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSL 223
RF +V+ L L K + D ++K+LR L W+PKVTAI E++DL+ +S+ L L
Sbjct: 214 QKRFTHIVNHLAALGKEFQNEDLINKVLRCLSREWQPKVTAISESRDLSNMSLATLFGKL 35
Query: 224 KVHEMSL 230
+ HEM L
Sbjct: 34 QEHEMEL 14
>TC235104 weakly similar to UP|Q850H8 (Q850H8) Gag-pol polyprotein
(Fragment), partial (28%)
Length = 865
Score = 107 bits (268), Expect = 3e-23
Identities = 89/287 (31%), Positives = 142/287 (49%), Gaps = 6/287 (2%)
Frame = +2
Query: 707 KIIGTGTIGN----SSISINNVWLVDGLKHNLLSISQFCD-NEYDVTFSKTNCTLVNKDD 761
+++ TG IG+ SS+S+N+V + G N+ S+SQ VTF + +
Sbjct: 41 RVVATG-IGHVSPTSSLSLNSVVFILGCPFNITSLSQLTRFRNCSVTFDANSFVIQECGT 217
Query: 762 KSITFKGKRVENVYKINFSDLADQKVVCLLSMNDKKWVWHKRLGHANWRLISKISKLQLV 821
G +Y + + VC + K + H+RLGH + +SKL+++
Sbjct: 218 GWTIGVGIESHGLYYLK----PNLSWVCSAVTSPK--LLHERLGHPH------LSKLKIM 361
Query: 822 KGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHIDLFGPVNTASLYGSKY 881
+P+++ D C +CQ GK V+SS + + S P ++H D++GP N S +Y
Sbjct: 362 --VPSLEKIKDLFCESCQLGKHVRSSXRHVESRVDS-PFLVIHXDIWGP-NRVSSMSYRY 529
Query: 882 GLVIVDDYSRWTWVKFIKSKDYACEVFSSFCT-QIQSEKELKILKVRSDHGGEFENEPFE 940
+ +D++S+ T V +K + +S + Q K +KIL RSD+ E+ +
Sbjct: 530 FVTFIDEFSQCTRVFLMKERSEILSFLTSVNKIKTQFGKTIKIL--RSDNAKEYFSSVIS 703
Query: 941 LFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKH 987
F GILH+FS P TPQQN + ERKNR L E ART++ N H
Sbjct: 704 PFXSAQGILHQFSCPHTPQQNDIAERKNRHLVETARTLLLHANEPIH 844
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.131 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 62,891,522
Number of Sequences: 63676
Number of extensions: 824262
Number of successful extensions: 3880
Number of sequences better than 10.0: 164
Number of HSP's better than 10.0 without gapping: 3738
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3844
length of query: 1591
length of database: 12,639,632
effective HSP length: 110
effective length of query: 1481
effective length of database: 5,635,272
effective search space: 8345837832
effective search space used: 8345837832
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC139344.2


