

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139344.1 + phase: 0
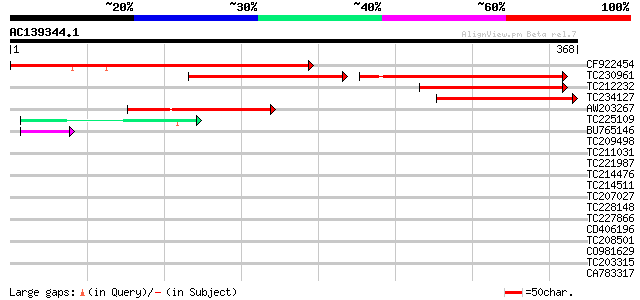
(368 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CF922454 310 7e-85
TC230961 weakly similar to UP|Q9FG54 (Q9FG54) Root cap protein 2... 131 7e-63
TC212232 similar to UP|Q9FG54 (Q9FG54) Root cap protein 2-like p... 105 3e-23
TC234127 similar to UP|Q9FG54 (Q9FG54) Root cap protein 2-like p... 100 8e-22
AW203267 96 3e-20
TC225109 weakly similar to UP|Q9V5V8 (Q9V5V8) CG13214-PA, partia... 44 2e-04
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 42 3e-04
TC209498 similar to UP|Q8L8Z3 (Q8L8Z3) Structural protein-like p... 40 0.002
TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 39 0.005
TC221987 nodulin 37 0.011
TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 37 0.018
TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protei... 37 0.018
TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%) 36 0.024
TC228148 similar to UP|Q7YXC8 (Q7YXC8) C. elegans GRD-1 protein ... 36 0.024
TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine prot... 36 0.024
CD406196 36 0.031
TC208501 similar to GB|AAN31116.1|23506209|AY149962 At3g06130/F2... 35 0.041
CO981629 35 0.041
TC203315 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, complete 35 0.054
CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycopro... 35 0.054
>CF922454
Length = 785
Score = 310 bits (794), Expect = 7e-85
Identities = 147/208 (70%), Positives = 167/208 (79%), Gaps = 11/208 (5%)
Frame = +2
Query: 1 MEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDK--------EEGKEKGKEKEKA 52
MEA IAQGQGNGNGN NGNG GNGNG GN+ K +K E+ ++K KEKEK
Sbjct: 161 MEAVIAQGQGNGNGNNGNGNGNGNGNGKGNDKEEKEREKREKEKEKREKKEKKEKEKEKK 340
Query: 53 PKKKKAPKE--ESSNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDK 110
K+K+A K+ E++NY +L LP+GQER FC+ N C F T+VCP+EC RKPKKNKK K
Sbjct: 341 EKEKQAKKQQGEATNYDKLSPLPTGQERGFCRKNTACEFKTIVCPSECAYRKPKKNKKQK 520
Query: 111 GCFIDCSSK-CEATCKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEF 169
CFIDCSS CEATCK R+ANCDGYG+LCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEF
Sbjct: 521 ACFIDCSSSTCEATCKVRKANCDGYGALCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEF 700
Query: 170 QINAHFIGTRPQGRTRDYTWVQALSLMF 197
QINAHFIGTRP+G+TRDYTWVQALS+MF
Sbjct: 701 QINAHFIGTRPKGKTRDYTWVQALSVMF 784
>TC230961 weakly similar to UP|Q9FG54 (Q9FG54) Root cap protein 2-like
protein, partial (67%)
Length = 962
Score = 131 bits (330), Expect(2) = 7e-63
Identities = 57/103 (55%), Positives = 70/103 (67%)
Frame = +2
Query: 117 SSKCEATCKFRRANCDGYGSLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQINAHFI 176
S CE CK R+ NC+G GS C DPRFVG DG++FYFHG + NFA+VSD QINA FI
Sbjct: 5 SPTCETKCKTRKPNCNGRGSACLDPRFVGADGIVFYFHGRRNENFALVSDANLQINARFI 184
Query: 177 GTRPQGRTRDYTWVQALSLMFDTHTLVIAANRVTQWNDNVDSL 219
G RP RTRDYTW+QAL +++ +H I A W+D +D L
Sbjct: 185 GLRPATRTRDYTWIQALGILYGSHQFTIEATPSPTWDDEIDHL 313
Score = 127 bits (319), Expect(2) = 7e-63
Identities = 58/135 (42%), Positives = 90/135 (65%)
Frame = +3
Query: 228 VIIPTDDDAEWKTNGDEREVVVERTDDANSVRVTVSGLLEMDIRVRPIGEKENKAHNYQL 287
++IP + W+ E ++ +ERT NSV +T+ + ++ I V P+ +++++ HNYQ+
Sbjct: 339 LVIPDSFLSTWQC--PENKLRIERTSSKNSVSITLQEVADISINVVPVTKEDSRIHNYQI 512
Query: 288 PSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYVSPVKRGVAMPMMGGEDKYQTPSLFS 347
P DD FAHLE QFKF ++ +EGVLG+TY+P + +P K GVAMP++GGED+Y+T SL S
Sbjct: 513 PDDDCFAHLEVQFKFHGLSNKVEGVLGKTYQPDFQNPAKLGVAMPVVGGEDRYRTTSLVS 692
Query: 348 TTCKLCRFQRPSTSQ 362
C +C F S+
Sbjct: 693 ADCGVCLFDAREGSE 737
>TC212232 similar to UP|Q9FG54 (Q9FG54) Root cap protein 2-like protein,
partial (30%)
Length = 620
Score = 105 bits (262), Expect = 3e-23
Identities = 48/96 (50%), Positives = 70/96 (72%)
Frame = +3
Query: 267 EMDIRVRPIGEKENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYVSPVK 326
E+ + V PI +++++ HNYQ+PSDD FAHLE QF+F + ++GVLG+TYR + +P K
Sbjct: 3 EILVNVVPITKEDDRIHNYQVPSDDCFAHLEVQFRFFALSQKVDGVLGRTYRLDFENPAK 182
Query: 327 RGVAMPMMGGEDKYQTPSLFSTTCKLCRFQRPSTSQ 362
GVAMP++GGEDKY+T SL S C C F + S+++
Sbjct: 183 PGVAMPVVGGEDKYRTNSLLSPHCGSCVFSQGSSTE 290
>TC234127 similar to UP|Q9FG54 (Q9FG54) Root cap protein 2-like protein,
partial (23%)
Length = 573
Score = 100 bits (250), Expect = 8e-22
Identities = 45/91 (49%), Positives = 65/91 (70%)
Frame = +1
Query: 278 KENKAHNYQLPSDDAFAHLETQFKFKNPTDSIEGVLGQTYRPSYVSPVKRGVAMPMMGGE 337
++++ HNYQ+PSDD FAHLE QF+F + ++GVLG+TYR + +P K GVAMP++GGE
Sbjct: 1 EDDRIHNYQVPSDDCFAHLEVQFRFFALSQKVDGVLGRTYRLDFENPAKPGVAMPVVGGE 180
Query: 338 DKYQTPSLFSTTCKLCRFQRPSTSQGLIAQY 368
DKY+T SL S C C F + S+++ +Y
Sbjct: 181 DKYRTNSLLSPDCVSCVFSQESSAEKETMEY 273
>AW203267
Length = 392
Score = 95.9 bits (237), Expect = 3e-20
Identities = 43/97 (44%), Positives = 59/97 (60%), Gaps = 1/97 (1%)
Frame = +2
Query: 77 ERAFCKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSK-CEATCKFRRANCDGYG 135
+ +C C+ + CPAEC + N K K C I+C+ C+A C+ R+ NC+ G
Sbjct: 104 QNLYCGRGTRCYGKYITCPAECPNSETN-NPKTKVCQIECNKPTCKAVCRSRKPNCNAPG 280
Query: 136 SLCYDPRFVGGDGVMFYFHGAKGGNFAIVSDDEFQIN 172
S CYDPRF+GGDG +FYFHG +F++VSD QIN
Sbjct: 281 SGCYDPRFIGGDGRVFYFHGKTNEHFSLVSDSNLQIN 391
>TC225109 weakly similar to UP|Q9V5V8 (Q9V5V8) CG13214-PA, partial (8%)
Length = 952
Score = 43.5 bits (101), Expect = 2e-04
Identities = 33/123 (26%), Positives = 40/123 (31%), Gaps = 6/123 (4%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQ 67
G GNG GNG G G G+G G N G+ KG
Sbjct: 353 GFGNGFGNGIIGGGYGSGYGGPNGGSSKGG------------------------------ 442
Query: 68 QLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNK------KDKGCFIDCSSKCE 121
+ CK C + CPA+C + + K GC IDC KC
Sbjct: 443 ------IIRPTVVCKDKGPCFQKKVTCPAKCFSSFSRSGKGYGGGGGGGGCTIDCKKKCI 604
Query: 122 ATC 124
A C
Sbjct: 605 AYC 613
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/35 (48%), Positives = 19/35 (53%)
Frame = +1
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
G G G G G G G+G G G G G G G +EEG
Sbjct: 286 GGGGGXGGGGGGGGRGGGGGGGGGGGGGGEGREEG 390
Score = 42.4 bits (98), Expect = 3e-04
Identities = 17/35 (48%), Positives = 19/35 (53%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
G G G G G G G G G G G G G+G +EEG
Sbjct: 293 GGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREEG 397
Score = 36.2 bits (82), Expect = 0.024
Identities = 15/37 (40%), Positives = 17/37 (45%)
Frame = +3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKE 44
G G G G G G G G G G G G+G + G E
Sbjct: 297 GXGGGGGGRGEGGGGGGGGGGGGGGGGEGGGRRGGGE 407
Score = 33.9 bits (76), Expect = 0.12
Identities = 13/29 (44%), Positives = 15/29 (50%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G+G G G G G G G G G G G+G
Sbjct: 239 GRGGRGGGGGGGGGGGGGGGGGGGGGGEG 325
>TC209498 similar to UP|Q8L8Z3 (Q8L8Z3) Structural protein-like protein,
partial (51%)
Length = 795
Score = 39.7 bits (91), Expect = 0.002
Identities = 19/53 (35%), Positives = 28/53 (51%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPK 60
G G G+G G + G G+G+G+GN+G G +DK G G + + P K
Sbjct: 416 GYGYGSGGGAHAGGYGSGSGSGNSGGGGYDDKNGG---GDDNNRLPSSSSRGK 565
Score = 31.6 bits (70), Expect = 0.59
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 1/41 (2%)
Frame = +2
Query: 4 AIAQGQGNGNGNGNN-GNGKGNGNGNGNNGNGKGNDKEEGK 43
A A G G+G+G+GN+ G G + NG G++ N + GK
Sbjct: 443 AHAGGYGSGSGSGNSGGGGYDDKNGGGDDNNRLPSSSSRGK 565
>TC211031 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (11%)
Length = 678
Score = 38.5 bits (88), Expect = 0.005
Identities = 15/36 (41%), Positives = 20/36 (54%)
Frame = -3
Query: 1 MEAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
+++ I G+G G GN G G G G G+ GNG G
Sbjct: 307 VDSGIGGGKGGGGDGGNGGGGDGGNGGGGDGGNGGG 200
Score = 33.1 bits (74), Expect = 0.20
Identities = 16/37 (43%), Positives = 17/37 (45%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKE 44
G G GNG +G G G GNG G G D G E
Sbjct: 466 GPFGGIGNGGSGGGGGASGGNGGGGASGGGDGRFGGE 356
Score = 31.6 bits (70), Expect = 0.59
Identities = 16/44 (36%), Positives = 19/44 (42%), Gaps = 9/44 (20%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNG---------NGNGNNGNGKGNDKEEG 42
G+G+G G GKG G G G+ GNG G D G
Sbjct: 361 GEGDGGKGGGGDGGKGGGVDSGIGGGKGGGGDGGNGGGGDGGNG 230
Score = 31.6 bits (70), Expect = 0.59
Identities = 14/41 (34%), Positives = 18/41 (43%)
Frame = -3
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
A G G G G+G+ G G+G G G K G + G
Sbjct: 418 ASGGNGGGGASGGGDGRFGGEGDGGKGGGGDGGKGGGVDSG 296
Score = 31.6 bits (70), Expect = 0.59
Identities = 12/25 (48%), Positives = 13/25 (52%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNG 32
G G G G+ GNG G GNG G
Sbjct: 271 GDGGNGGGGDGGNGGGGDGGNGGGG 197
Score = 31.6 bits (70), Expect = 0.59
Identities = 14/37 (37%), Positives = 16/37 (42%)
Frame = -3
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G G G+G G G+G NG G G G
Sbjct: 298 GIGGGKGGGGDGGNGGGGDGGNGGGGDGGNGGGGASG 188
Score = 31.6 bits (70), Expect = 0.59
Identities = 16/41 (39%), Positives = 20/41 (48%), Gaps = 4/41 (9%)
Frame = -3
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGN----GKGNDKEEGKEKG 46
G G +G NG G +G G+G G GKG + GK G
Sbjct: 430 GGGGASGGNGGGGASGGGDGRFGGEGDGGKGGGGDGGKGGG 308
Score = 29.6 bits (65), Expect = 2.3
Identities = 17/46 (36%), Positives = 22/46 (46%), Gaps = 4/46 (8%)
Frame = -3
Query: 10 GNGNGNGNNGNGKGNGNGNGN---NGNGK-GNDKEEGKEKGKEKEK 51
G GNG G G GNG G G+G+ G + + GK G + K
Sbjct: 454 GIGNGGSGGGGGASGGNGGGGASGGGDGRFGGEGDGGKGGGGDGGK 317
Score = 29.3 bits (64), Expect = 2.9
Identities = 14/29 (48%), Positives = 15/29 (51%), Gaps = 2/29 (6%)
Frame = -3
Query: 10 GNGNGN--GNNGNGKGNGNGNGNNGNGKG 36
GNG G GN G G G G G +G G G
Sbjct: 262 GNGGGGDGGNGGGGDGGNGGGGASGIGGG 176
Score = 29.3 bits (64), Expect = 2.9
Identities = 15/43 (34%), Positives = 19/43 (43%), Gaps = 5/43 (11%)
Frame = -3
Query: 10 GNGNGNGNNGN-----GKGNGNGNGNNGNGKGNDKEEGKEKGK 47
GNG G + G G+G+G G GKG + G GK
Sbjct: 409 GNGGGGASGGGDGRFGGEGDGGKGGGGDGGKGGGVDSGIGGGK 281
Score = 28.5 bits (62), Expect = 5.0
Identities = 16/41 (39%), Positives = 19/41 (46%)
Frame = -3
Query: 6 AQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
A G G+G G GKG G G+G G G + GK G
Sbjct: 391 ASGGGDGRFGGEGDGGKGGG-GDGGKGGGVDSGIGGGKGGG 272
Score = 27.7 bits (60), Expect = 8.6
Identities = 13/30 (43%), Positives = 16/30 (53%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
G G GNG G+G GNG G + G G+
Sbjct: 256 GGGGDGGNGGGGDG-GNGGGGASGIGGGGH 170
>TC221987 nodulin
Length = 844
Score = 37.4 bits (85), Expect = 0.011
Identities = 15/31 (48%), Positives = 23/31 (73%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGND 38
G G G+G+ N G G+G G G+G +G+G+G+D
Sbjct: 536 GSGEGDGDDNEGEGEGEGEGDG-DGDGEGDD 447
Score = 32.3 bits (72), Expect = 0.35
Identities = 14/33 (42%), Positives = 21/33 (63%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKE 40
G+G+G+ N G G+G G+G+G +G G D E
Sbjct: 530 GEGDGDDNEGEGEGEGEGDGDG-DGEGDDGDGE 435
Score = 28.1 bits (61), Expect = 6.6
Identities = 11/29 (37%), Positives = 20/29 (68%)
Frame = -3
Query: 7 QGQGNGNGNGNNGNGKGNGNGNGNNGNGK 35
+G+G G G G+ G+G+G G++G+G+
Sbjct: 506 EGEGEGEGEGD-----GDGDGEGDDGDGE 435
>TC214476 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (14%)
Length = 642
Score = 36.6 bits (83), Expect = 0.018
Identities = 15/39 (38%), Positives = 21/39 (53%)
Frame = -3
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G+G+G+ G +G G G G+G+ G G G G E G
Sbjct: 502 GEGSGSQGGGSGGGGSQGGGSGSGGGGYGGGGSGGSEGG 386
Score = 33.1 bits (74), Expect = 0.20
Identities = 18/45 (40%), Positives = 24/45 (53%)
Frame = -3
Query: 2 EAAIAQGQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
E + +QG G+G G G+ G G G+G G G G G G + G G
Sbjct: 499 EGSGSQGGGSG-GGGSQGGGSGSG-GGGYGGGGSGGSEGGGSSGG 371
Score = 31.6 bits (70), Expect = 0.59
Identities = 17/49 (34%), Positives = 22/49 (44%), Gaps = 8/49 (16%)
Frame = -3
Query: 6 AQGQGNGNGNGNNGNGKGNGN--------GNGNNGNGKGNDKEEGKEKG 46
+QG G+G+G G G G G+ G G G G G G +KG
Sbjct: 457 SQGGGSGSGGGGYGGGGSGGSEGGGSSGGGGGGGGGGGGGGYGGGGDKG 311
>TC214511 weakly similar to PIR|C85356|C85356 glycine-rich protein [imported]
- Arabidopsis thaliana {Arabidopsis thaliana;} , partial
(88%)
Length = 1085
Score = 36.6 bits (83), Expect = 0.018
Identities = 15/39 (38%), Positives = 20/39 (50%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G+G G G G G G G G G+G G+ + G +G
Sbjct: 686 GSGSGRSQGRGGGGGGGGGGGGGGGSGYGHGEGYGHGEG 802
Score = 27.7 bits (60), Expect = 8.6
Identities = 13/30 (43%), Positives = 17/30 (56%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
G G G G G+G+G G+G G G G G+
Sbjct: 737 GGGGGGGGSGYGHGEGYGHGEG-YGQGGGD 823
Score = 27.7 bits (60), Expect = 8.6
Identities = 12/40 (30%), Positives = 21/40 (52%), Gaps = 1/40 (2%)
Frame = +2
Query: 8 GQGNGNGNGNN-GNGKGNGNGNGNNGNGKGNDKEEGKEKG 46
G G G+G+G G G G+G+G+G++ + + G
Sbjct: 512 GIGGGSGSGAGAGAGSGSGSGSGSSSSSASSSASSSSSSG 631
>TC207027 weakly similar to UP|Q9LW95 (Q9LW95) KED, partial (17%)
Length = 1005
Score = 36.2 bits (82), Expect = 0.024
Identities = 29/98 (29%), Positives = 44/98 (44%), Gaps = 3/98 (3%)
Frame = +3
Query: 16 GNNGNGKGNGNGNGNN---GNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQLQTL 72
G +GK GN + G+G G +K+E K+K KEK+ K+KK EE+ ++
Sbjct: 285 GKEDDGKDEGNKEKKDKEKGDGDGEEKKEKKDKEKEKK---KEKKDKDEETDTLKEKGKN 455
Query: 73 PSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKKDK 110
G++ K + K + KK KKDK
Sbjct: 456 DEGEDDEGNKKKKK--------DKKEKEKDHKKEKKDK 545
Score = 27.7 bits (60), Expect = 8.6
Identities = 22/75 (29%), Positives = 35/75 (46%)
Frame = +3
Query: 36 GNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQLQTLPSGQERAFCKANNTCHFATLVCP 95
G+D EE KEK K+++K KK+ K + ++ ++++ + E
Sbjct: 3 GDDGEEKKEKDKKEKK--KKENKDKGDKTDVEKVKGKENDGE-----------------D 125
Query: 96 AECKTRKPKKNKKDK 110
E K K KK KKDK
Sbjct: 126 DEEKKEKKKKEKKDK 170
>TC228148 similar to UP|Q7YXC8 (Q7YXC8) C. elegans GRD-1 protein
(Corresponding sequence R08B4.1b), partial (3%)
Length = 1180
Score = 36.2 bits (82), Expect = 0.024
Identities = 25/109 (22%), Positives = 42/109 (37%), Gaps = 8/109 (7%)
Frame = +3
Query: 8 GQGNGNGNGNNGNGKGNGNGNG--------NNGNGKGNDKEEGKEKGKEKEKAPKKKKAP 59
G G+G G G++ + KG G GNG NNG G + + K+
Sbjct: 510 GGGSGGGGGSSSSSKGGGGGNGVKEQKKMVNNGKGDKGKERASRHNNVGGGSGSGDKRNS 689
Query: 60 KEESSNYQQLQTLPSGQERAFCKANNTCHFATLVCPAECKTRKPKKNKK 108
K +N Q +++ ++A + + K ++PKK K
Sbjct: 690 KNVENNSQSKRSVGRPPKKAAETNAGSAKRGRESSASAGKDKRPKKRSK 836
Score = 35.4 bits (80), Expect = 0.041
Identities = 17/49 (34%), Positives = 26/49 (52%), Gaps = 8/49 (16%)
Frame = +3
Query: 9 QGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGK--------EKGKEK 49
+G G+G G+G G G+ + + G G GN +E K +KGKE+
Sbjct: 486 KGGSGGSGGGGSGGGGGSSSSSKGGGGGNGVKEQKKMVNNGKGDKGKER 632
>TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine protein
phosphatase 2B catalytic subunit 1
(Calmodulin-dependent calcineurin A1 subunit) , partial
(3%)
Length = 1013
Score = 36.2 bits (82), Expect = 0.024
Identities = 18/56 (32%), Positives = 26/56 (46%)
Frame = +3
Query: 10 GNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSN 65
G G NG N N KG+G+ NGN G+ G + G ++ P + P + N
Sbjct: 264 GKGGNNGKNNNDKGHGSNNGNVGHHMG----PMGQMGPMVQRGPMNQMGPMDHMRN 419
>CD406196
Length = 529
Score = 35.8 bits (81), Expect = 0.031
Identities = 15/48 (31%), Positives = 24/48 (49%)
Frame = +1
Query: 81 CKANNTCHFATLVCPAECKTRKPKKNKKDKGCFIDCSSKCEATCKFRR 128
C+ + ++ TL C E K + PK+NK CF++ + C F R
Sbjct: 67 CRNQSV*NYQTLECFVELKYKDPKRNKTRNLCFVNSNLSCAKNSSFNR 210
>TC208501 similar to GB|AAN31116.1|23506209|AY149962 At3g06130/F28L1_7
{Arabidopsis thaliana;} , partial (26%)
Length = 1081
Score = 35.4 bits (80), Expect = 0.041
Identities = 23/72 (31%), Positives = 28/72 (37%), Gaps = 12/72 (16%)
Frame = +3
Query: 1 MEAAIAQGQGNGNGNGNNGNGKGNGN-----------GNGNNG-NGKGNDKEEGKEKGKE 48
M ++ Q GNG GNNG G G+ G GN G NG G KE G +
Sbjct: 297 MSNSMMNAQKGGNGGGNNGKKGGGGSGGPVPVQVHNMGGGNEGKNGNGGKKEGGGGGNNQ 476
Query: 49 KEKAPKKKKAPK 60
+ K K
Sbjct: 477 TQGGGNKNNGGK 512
Score = 35.0 bits (79), Expect = 0.054
Identities = 18/37 (48%), Positives = 18/37 (48%), Gaps = 4/37 (10%)
Frame = +3
Query: 11 NGNGNGNNGNGKGN----GNGNGNNGNGKGNDKEEGK 43
NGNG G G GN G GN NNG G EGK
Sbjct: 429 NGNGGKKEGGGGGNNQTQGGGNKNNGGKNGGGVPEGK 539
Score = 34.3 bits (77), Expect = 0.092
Identities = 15/29 (51%), Positives = 15/29 (51%)
Frame = +3
Query: 9 QGNGNGNGNNGNGKGNGNGNGNNGNGKGN 37
Q G GN NNG G G G NGN K N
Sbjct: 474 QTQGGGNKNNGGKNGGGVPEGKNGNNKNN 560
Score = 33.5 bits (75), Expect = 0.16
Identities = 15/34 (44%), Positives = 16/34 (46%)
Frame = +3
Query: 9 QGNGNGNGNNGNGKGNGNGNGNNGNGKGNDKEEG 42
+G N NNG G G N NGN G N EG
Sbjct: 531 EGKNGNNKNNGGGGGIPNNNGNGGKKGNNGMAEG 632
>CO981629
Length = 844
Score = 35.4 bits (80), Expect = 0.041
Identities = 24/72 (33%), Positives = 32/72 (44%), Gaps = 10/72 (13%)
Frame = -3
Query: 6 AQGQGNGNGN------GNNGNGKGNGNGN--GNNGNGKGNDKEEGKEKGKE--KEKAPKK 55
A+ NGN N G +GNGK N N N G N GND + K + EK
Sbjct: 479 AKSNANGNNNSSCKSDGASGNGKSNYNRNSDGPKSNAIGNDNQNSGAKSSDNGNEKINSN 300
Query: 56 KKAPKEESSNYQ 67
+ + +S+N Q
Sbjct: 299 RSSDGAKSNNIQ 264
>TC203315 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, complete
Length = 778
Score = 35.0 bits (79), Expect = 0.054
Identities = 18/55 (32%), Positives = 30/55 (53%), Gaps = 3/55 (5%)
Frame = +2
Query: 18 NGNGKGN---GNGNGNNGNGKGNDKEEGKEKGKEKEKAPKKKKAPKEESSNYQQL 69
NG+GKG G + G G G ++ EG E+ E + ++++A +EE + Q L
Sbjct: 86 NGSGKGREEAGGEEASGGEGTGGEEAEGGEEDLEGRREREEEEAHEEERGDLQDL 250
>CA783317 similar to GP|6523547|emb| hydroxyproline-rich glycoprotein
DZ-HRGP {Volvox carteri f. nagariensis}, partial (23%)
Length = 159
Score = 35.0 bits (79), Expect = 0.054
Identities = 13/29 (44%), Positives = 15/29 (50%)
Frame = +2
Query: 8 GQGNGNGNGNNGNGKGNGNGNGNNGNGKG 36
G+G G G G G G G G G G G+G
Sbjct: 68 GEGGGGGGGGGGGGXXXGEGGGGGGGGRG 154
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.314 0.133 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,321,819
Number of Sequences: 63676
Number of extensions: 269861
Number of successful extensions: 2901
Number of sequences better than 10.0: 215
Number of HSP's better than 10.0 without gapping: 1954
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2567
length of query: 368
length of database: 12,639,632
effective HSP length: 99
effective length of query: 269
effective length of database: 6,335,708
effective search space: 1704305452
effective search space used: 1704305452
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139344.1


