

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135161.1 - phase: 0
(820 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
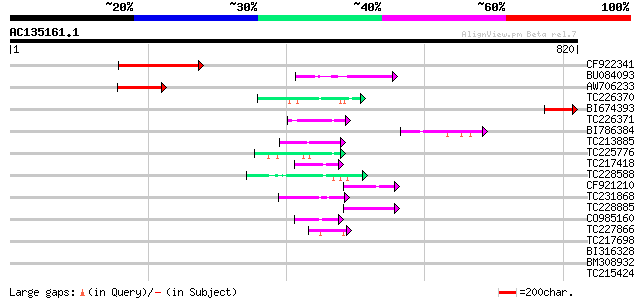
Score E
Sequences producing significant alignments: (bits) Value
CF922341 125 1e-28
BU084093 72 8e-13
AW706233 62 8e-10
TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 55 1e-07
BI674393 53 5e-07
TC226371 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pr... 51 2e-06
BI786384 49 9e-06
TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partia... 47 3e-05
TC225776 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, par... 46 8e-05
TC217418 weakly similar to GB|AAA29531.1|160178|PFACSAZ circumsp... 45 2e-04
TC228588 similar to UP|Q6NLV4 (Q6NLV4) At5g13480, partial (29%) 44 2e-04
CF921210 43 5e-04
TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein... 43 5e-04
TC228885 43 6e-04
CO985160 43 6e-04
TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine prot... 42 8e-04
TC217698 similar to UP|Q89GY6 (Q89GY6) Blr6209 protein, partial ... 42 0.001
BI316328 42 0.001
BM308932 42 0.001
TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%) 41 0.002
>CF922341
Length = 675
Score = 125 bits (313), Expect = 1e-28
Identities = 60/124 (48%), Positives = 84/124 (67%), Gaps = 1/124 (0%)
Frame = +1
Query: 158 KYDELRRDMKALREKGKFG-KTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMYV 216
K D L ++A+ + +L LVP++ P KFK+ DF+KYKG++CP+ HLKMY
Sbjct: 304 KLDHLEEGLRAIEGGEDYAFANLEELFLVPNIITPPKFKVLDFDKYKGTTCPKNHLKMYC 483
Query: 217 RRMPAYAQDDQILIYYFQESLTGPASKWYTNLDKTRVQTFRDLCEAFVEQYSYNVDMTPD 276
++M AYA+D+++LI+ FQESLTG A WYTNL+ +RV +++DL AFV QY YN DM
Sbjct: 484 QKMGAYAKDEELLIHSFQESLTGVAVTWYTNLEPSRVHSWKDLMVAFVRQYQYNFDMVLS 663
Query: 277 RSDL 280
R L
Sbjct: 664 RMQL 675
>BU084093
Length = 421
Score = 72.4 bits (176), Expect = 8e-13
Identities = 43/148 (29%), Positives = 64/148 (43%)
Frame = -1
Query: 414 NNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPY 473
N P P+ P + PQ P+P +P V N +NF +P+
Sbjct: 364 NASSSTPVQPKAPAQREAPQVPTPNTTRP--VGNSN--------------TTRNFPPRPF 233
Query: 474 QQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHE 533
Q+ P+P+TY +LLP L+ +L + P WY + TC +H
Sbjct: 232 QE-----------FTPLPMTYEDLLPSLIANHLAVVTPGRVLQPPFPKWYDPNATCKYHG 86
Query: 534 GGRGHNIETCYAFKSTVQRLINDGKITF 561
G GH++E C A K VQ L++ G +TF
Sbjct: 85 GVPGHSVEKCLALKYKVQHLMDAGWLTF 2
>AW706233
Length = 376
Score = 62.4 bits (150), Expect = 8e-10
Identities = 30/72 (41%), Positives = 47/72 (64%), Gaps = 1/72 (1%)
Frame = -3
Query: 157 EKYDELRRDMKALREKGKFG-KTAYDLCLVPSVQVPHKFKIPDFEKYKGSSCPEEHLKMY 215
EK D L+ KA+ + +L LV ++ P KFK+ +F+KYKG++CP+ HLKMY
Sbjct: 347 EKLDHLKERFKAIEGGQDYAFANLEELFLVXNIISPPKFKVLNFDKYKGTTCPKNHLKMY 168
Query: 216 VRRMPAYAQDDQ 227
++M AYA+D++
Sbjct: 167 CQKMGAYAKDEK 132
>TC226370 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (43%)
Length = 1810
Score = 55.1 bits (131), Expect = 1e-07
Identities = 56/194 (28%), Positives = 73/194 (36%), Gaps = 38/194 (19%)
Frame = +2
Query: 359 KNTTPASGTKKTGNHFPRKKEQEVGMVTHG-GPQQTYPAYQHIAAI---------TPTSH 408
+ ++P + KKT N +Q + H PQ PA Q A P
Sbjct: 530 ERSSPTTDPKKTDNASDANNQQLALALPHQIAPQPQPPAPQVQAQAQAPAPNVTQVPQQP 709
Query: 409 PFQQTNN---HPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYP-YQQYP 464
P+ +P +PQ+PQ P Q +PQ+V PQ Q P QQ+ YQQ
Sbjct: 710 PYYMPPTPLPNPAVPQHPQNQYLPSDQQYRTPQHVAPQPTPSQ-VTPSPVQQFSHYQQQQ 886
Query: 465 QQNFQQQPYQQR-------PQQPRP-----------------PRMPINPIPVTYAELLPG 500
QQ QQQP QQ+ P QP P P NP P AE LP
Sbjct: 887 QQPQQQQPQQQQQWSQQVLPSQPPPMQSQVRPPSPNVYPPYQPNQATNPSP---AETLPN 1057
Query: 501 LLKKNLVQTRTAPP 514
+ + + PP
Sbjct: 1058SMAMQMPYSGVPPP 1099
>BI674393
Length = 152
Score = 53.1 bits (126), Expect = 5e-07
Identities = 22/47 (46%), Positives = 39/47 (82%)
Frame = +1
Query: 774 EFDEILKLIKKSEYKVVDQLMQTPSKISIMSLLLNSEAHKDALMKVL 820
E E L++I++SE+KV++QL +TP+++S++ LL++SE H+ L+KVL
Sbjct: 7 EASEFLRIIQQSEFKVIEQLNKTPARVSLLELLMSSEPHRALLVKVL 147
>TC226371 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (10%)
Length = 1099
Score = 50.8 bits (120), Expect = 2e-06
Identities = 34/90 (37%), Positives = 42/90 (45%)
Frame = +1
Query: 403 ITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQ 462
I PT P + +PQ+PQ P Q +PQ V PQ Q P QQ+ + Q
Sbjct: 19 IPPTPMP------NSALPQHPQNQYLPSDQQYRTPQLVAPQPTPSQVTPSPPVQQFSHYQ 180
Query: 463 YPQQNFQQQPYQQRPQQPRPPRMPINPIPV 492
PQQ QQQP QQ+ QQ P P P+
Sbjct: 181 QPQQ--QQQPPQQQQQQWSQQVQPSQPPPM 264
>BI786384
Length = 421
Score = 48.9 bits (115), Expect = 9e-06
Identities = 38/143 (26%), Positives = 69/143 (47%), Gaps = 18/143 (12%)
Frame = +2
Query: 566 PNVQTNPLPNHGAATVNMIEDCQ--KTRPILNVQHIRTPLVPLHAKLCKVDLFEHDHDLC 623
PNV+TNPL NHG VN +E + +++P+ + + TP + L K + H
Sbjct: 2 PNVRTNPLANHGGGAVNAVESDRPHRSKPL---RDVATPRRFIFEALQKGGVIPHSGCKE 172
Query: 624 EICLMNSG------GCQKVRNDIQGLLDRGELVV---ERKCDDVCVITPEG-------PL 667
+ CL++SG C +V +Q ++D+G L V ++ +C+ + EG PL
Sbjct: 173 DSCLLHSGEMHDMETCLEVEELLQRMIDQGRLEVGIEGKEEQHICMQSTEGSGVAKPKPL 352
Query: 668 EVFYDSRKSTITPLVICLPGPLP 690
+++ ++ P + P+P
Sbjct: 353 VIYFTKSAASQKPGHPLMAKPVP 421
>TC213885 weakly similar to UP|Q9VUB7 (Q9VUB7) CG32133-PA, partial (4%)
Length = 540
Score = 47.4 bits (111), Expect = 3e-05
Identities = 37/98 (37%), Positives = 45/98 (45%), Gaps = 3/98 (3%)
Frame = +2
Query: 391 QQTYPAYQHIAAITPTSH-PFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQN 449
QQ QH+ + P Q PQ+ Q Q+PQ Q Q PQ Q Q +QQQ
Sbjct: 26 QQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQ--QQQLPQLQQQQQLQQQQ 199
Query: 450 FQQ-QPYQQYP-YQQYPQQNFQQQPYQQRPQQPRPPRM 485
Q Q QQ P QQ PQ QQQ Q + QP+ +M
Sbjct: 200 LPQLQQQQQLPQLQQLPQLQQQQQLPQLQQLQPQQQQM 313
Score = 40.8 bits (94), Expect = 0.002
Identities = 30/77 (38%), Positives = 35/77 (44%), Gaps = 2/77 (2%)
Frame = +2
Query: 411 QQTNNHPQIPQYPQ-IPQYPQFPQNPSPQNVQ-PQNVQQQNFQQQPYQQYPYQQYPQQNF 468
QQ H Q+ Q +PQ Q Q P Q Q PQ QQQ Q QQ Q QQ
Sbjct: 8 QQQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQLPQLQQQQQLPQLQQQQQL 187
Query: 469 QQQPYQQRPQQPRPPRM 485
QQQ Q QQ + P++
Sbjct: 188 QQQQLPQLQQQQQLPQL 238
>TC225776 similar to UP|Q93YH8 (Q93YH8) HMG I/Y like protein, partial (26%)
Length = 1663
Score = 45.8 bits (107), Expect = 8e-05
Identities = 44/152 (28%), Positives = 62/152 (39%), Gaps = 21/152 (13%)
Frame = +1
Query: 355 GRLVKNTTPASGTKKTGN--------HFPRKKEQEVGMV----THGGPQQTYPAYQHIAA 402
GR K +T AS T+ N HF K ++ + ++ H P A Q +
Sbjct: 835 GRPKKGSTSASTTQNAANEDLRRKLEHFQSKVKESLAVLKPHFNHESPVTAIAAIQELEV 1014
Query: 403 I--TPTSHPFQQTNNHPQIPQYP----QIPQYPQFP---QNPSPQNVQPQNVQQQNFQQQ 453
+ + P + PQ Q P Q P QFP Q P Q++ PQ + Q
Sbjct: 1015LGTMDLNAPLRDETLPPQEEQPPLQQQQQPPQQQFPPPQQQPPQQHLAPQQLPPQPQAPI 1194
Query: 454 PYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRM 485
Q YP PQ + QP Q QQP+PP++
Sbjct: 1195FQQTYPPFHLPQFH-HHQPTLQFQQQPQPPQL 1287
Score = 31.2 bits (69), Expect = 1.9
Identities = 21/67 (31%), Positives = 26/67 (38%), Gaps = 4/67 (5%)
Frame = +1
Query: 390 PQQTYPAYQHIAAITPTSHP----FQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNV 445
P Q P QH+A P FQQT +PQ+ QF Q P P +
Sbjct: 1123 PPQQQPPQQHLAPQQLPPQPQAPIFQQTYPPFHLPQFHHHQPTLQFQQQPQPPQLFQHQA 1302
Query: 446 QQQNFQQ 452
Q + QQ
Sbjct: 1303 QPPSHQQ 1323
>TC217418 weakly similar to GB|AAA29531.1|160178|PFACSAZ circumsporozoite
protein {Plasmodium berghei;} , partial (12%)
Length = 773
Score = 44.7 bits (104), Expect = 2e-04
Identities = 27/73 (36%), Positives = 35/73 (46%), Gaps = 1/73 (1%)
Frame = +3
Query: 412 QTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQ-YPQQNFQQ 470
Q++N PQ QY P Q P P ++ PQ Q +Q QP P QQ PQ FQ
Sbjct: 21 QSHNEPQPLQYQPQPHPHLLSQPPPPPHLHPQPQPQPQYQPQP---GPIQQPQPQSQFQN 191
Query: 471 QPYQQRPQQPRPP 483
Q ++ + P PP
Sbjct: 192 QHFEYPARYPPPP 230
Score = 29.6 bits (65), Expect = 5.6
Identities = 24/79 (30%), Positives = 32/79 (40%), Gaps = 1/79 (1%)
Frame = +3
Query: 405 PTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSP-QNVQPQNVQQQNFQQQPYQQYPYQQY 463
P HP + P +PQ PQ+ P P Q QPQ+ FQ Q + +YP +Y
Sbjct: 57 PQPHPHLLSQPPPPPHLHPQPQPQPQYQPQPGPIQQPQPQS----QFQNQHF-EYP-ARY 218
Query: 464 PQQNFQQQPYQQRPQQPRP 482
P + P Q P
Sbjct: 219 PPPPWAATPGYANYQNNSP 275
>TC228588 similar to UP|Q6NLV4 (Q6NLV4) At5g13480, partial (29%)
Length = 1178
Score = 44.3 bits (103), Expect = 2e-04
Identities = 53/198 (26%), Positives = 71/198 (35%), Gaps = 23/198 (11%)
Frame = +1
Query: 343 GMGVQLEEGVREGRLVKNTTPASGTKKTGNHFPRKKEQEVGMVTHGGPQQTYPAYQHIAA 402
GM E+ GR N T A G G P G+ + G T P +
Sbjct: 400 GMQGYAEQSPVAGRTGGNFTIAEGPTTPGPFAP-------GLTRNEG---TIPG---VGV 540
Query: 403 ITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQ--NVQPQNVQQQNFQQQPYQQYPY 460
P S P Q +P P P P P N Q QQN QQ P Q+ +
Sbjct: 541 AMPLSIPSLDMPQGEQKQPHPVSMGAPPLPPGPHPSLINANQQQPYQQNPQQMP--QHQH 714
Query: 461 QQYPQQ--------NFQQQPYQQR---PQQPR-PPRMP---------INPIPVTYAELLP 499
Q PQQ N QQ + P PR PP+MP I+ +P ++ +P
Sbjct: 715 QALPQQMGSLPMPPNMQQLQHPSHSPFPNMPRPPPQMPIGMPGSIPGISSVPTSHPMPMP 894
Query: 500 GLLKKNLVQTRTAPPIPE 517
G + + P+P+
Sbjct: 895 GPMGMQGAMNQMGTPMPQ 948
>CF921210
Length = 790
Score = 43.1 bits (100), Expect = 5e-04
Identities = 31/83 (37%), Positives = 42/83 (50%), Gaps = 2/83 (2%)
Frame = +2
Query: 484 RMPIN--PIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHNIE 541
R P+ PIPV+YA+LL LL ++V A L Y + TC G H+IE
Sbjct: 326 RKPVEFTPIPVSYADLLSYLLDNSMVAITLAKVHQPPLF*GYDSNATCG---GALRHSIE 496
Query: 542 TCYAFKSTVQRLINDGKITFTDS 564
C A K VQ LI+ G + F ++
Sbjct: 497 HCRALKRKVQGLIDAGWLKFEEN 565
>TC231868 similar to UP|Q93424 (Q93424) C. elegans GRL-23 protein
(Corresponding sequence E02A10.2), partial (23%)
Length = 790
Score = 43.1 bits (100), Expect = 5e-04
Identities = 35/103 (33%), Positives = 43/103 (40%), Gaps = 1/103 (0%)
Frame = -3
Query: 390 PQQTYPAYQHIAAITPTSHPFQQTNNHPQ-IPQYPQIPQYPQFPQNPSPQNVQPQNVQQQ 448
PQ P ++ P P PQ PQ P + ++PQ P P PQ QP + +
Sbjct: 446 PQPPQPLLSNLFPHPPPQPPQPPPQPPPQPAPQPPLLIKFPQPPPQPPPQPPQPPPLIK- 270
Query: 449 NFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRMPINPIP 491
F Q P Q P Q F Q P Q PQ P+PP P P
Sbjct: 269 -FPQPPPQPEP-QPPLLIIFPQPPPQPAPQPPQPPLSIAFPQP 147
>TC228885
Length = 901
Score = 42.7 bits (99), Expect = 6e-04
Identities = 29/83 (34%), Positives = 40/83 (47%), Gaps = 2/83 (2%)
Frame = -3
Query: 484 RMPIN--PIPVTYAELLPGLLKKNLVQTRTAPPIPEKLPSWYRLDQTCDFHEGGRGHNIE 541
R P+ PIPV+YA+LLP LL ++V A Y + C H G +IE
Sbjct: 632 RKPVEFTPIPVSYADLLPYLLDNSMVAITLAKVHQPPFLREYDSNAMCACHGEAPGRSIE 453
Query: 542 TCYAFKSTVQRLINDGKITFTDS 564
A K VQ LI+ G + F ++
Sbjct: 452 HYRALKRKVQGLIDAGWLKFEEN 384
>CO985160
Length = 777
Score = 42.7 bits (99), Expect = 6e-04
Identities = 32/75 (42%), Positives = 36/75 (47%), Gaps = 4/75 (5%)
Frame = -2
Query: 412 QTNNHPQIPQYPQIP--QYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQ 469
Q + PQ+ Q Q+P Q Q PQ Q PQ QQQ QQ P Q QQ PQ Q
Sbjct: 716 QQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQ--QQLPQLQQQQQQLPQLQQQ 543
Query: 470 QQ--PYQQRPQQPRP 482
QQ P Q+ QQ P
Sbjct: 542 QQQLPQLQQQQQQLP 498
Score = 41.2 bits (95), Expect = 0.002
Identities = 36/111 (32%), Positives = 47/111 (41%), Gaps = 4/111 (3%)
Frame = -2
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQN---VQQQNFQQQPYQQYPYQQYPQ-Q 466
Q N +PQ Q Q P Q PQ Q Q QQ QQ P Q QQ PQ Q
Sbjct: 728 QMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQQQLPQLQQQQQQLPQLQ 549
Query: 467 NFQQQPYQQRPQQPRPPRMPINPIPVTYAELLPGLLKKNLVQTRTAPPIPE 517
QQQ Q + QQ + P++ V +L L ++ L Q + +P+
Sbjct: 548 QQQQQLPQLQQQQQQLPQLQQQQQQVQQQQLAQ-LQQQQLPQIQQQQQLPQ 399
Score = 41.2 bits (95), Expect = 0.002
Identities = 36/112 (32%), Positives = 46/112 (40%), Gaps = 1/112 (0%)
Frame = -2
Query: 375 PRKKEQEVGMVTHGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQ-IPQYPQFPQ 433
P ++Q++ + QQ P Q P QQ PQ+ Q Q +PQ Q Q
Sbjct: 674 PHMQQQQLPQLQQ--QQQQLPQLQQQQQQLPQLQ--QQQQQLPQLQQQQQQLPQLQQQQQ 507
Query: 434 NPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRPPRM 485
Q Q VQQQ Q QQ P Q QQ Q Q Q QQ + P++
Sbjct: 506 QLPQLQQQQQQVQQQQLAQLQQQQLPQIQQQQQLPQLQQLPQLQQQQQLPQL 351
Score = 33.5 bits (75), Expect = 0.39
Identities = 25/73 (34%), Positives = 30/73 (40%), Gaps = 3/73 (4%)
Frame = -2
Query: 417 PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQ--QQPYQ 474
PQ+ Q Q Q + Q PQ QQQ QQ P Q QQ QQ Q
Sbjct: 776 PQLSSQQQQQMQQQHQQMQNQQQHLPQLQQQQQLPHMQQQQLPQLQQQQQQLPQLQQQQQ 597
Query: 475 QRPQ-QPRPPRMP 486
Q PQ Q + ++P
Sbjct: 596 QLPQLQQQQQQLP 558
Score = 31.6 bits (70), Expect = 1.5
Identities = 21/57 (36%), Positives = 26/57 (44%), Gaps = 1/57 (1%)
Frame = -2
Query: 391 QQTYPAYQHIAAITPTSHP-FQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQ 446
QQ Q +A + P QQ PQ+ Q PQ+ Q Q PQ Q +QPQ Q
Sbjct: 485 QQQQVQQQQLAQLQQQQLPQIQQQQQLPQLQQLPQLQQQQQLPQ---LQQLQPQQQQ 324
>TC227866 similar to UP|P2B1_DROME (P48456) Serine/threonine protein
phosphatase 2B catalytic subunit 1
(Calmodulin-dependent calcineurin A1 subunit) , partial
(3%)
Length = 1013
Score = 42.4 bits (98), Expect = 8e-04
Identities = 27/75 (36%), Positives = 36/75 (48%), Gaps = 13/75 (17%)
Frame = +3
Query: 433 QNPSPQNVQPQNVQQQ------NFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPR----- 481
Q P+P N+Q Q QQQ N QQQ QQ YP +P+ + Q P
Sbjct: 516 QQPNPYNMQQQQQQQQLMAMMMNQQQQQQQQANMNMYPPHMMYGRPHPHQHQHPSMNYFP 695
Query: 482 PPRMPINPI--PVTY 494
PP MP +P+ P+T+
Sbjct: 696 PPPMPSHPMADPITH 740
>TC217698 similar to UP|Q89GY6 (Q89GY6) Blr6209 protein, partial (5%)
Length = 865
Score = 42.0 bits (97), Expect = 0.001
Identities = 36/104 (34%), Positives = 41/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 387 HGGPQQTYPAYQHIAAITPTSHPFQQTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQ 446
+G Q YP Q+ + P Q N P YP +Y Q PQN P
Sbjct: 53 YGQASQNYPPQQNYGMTSQNYTP--QQNYGQASPNYPPQQKYGQAPQNYPP--------- 199
Query: 447 QQNFQQQPYQQYPYQQ----YPQQNFQQQPYQQRPQQPRPPRMP 486
QQNF Q P Q YP QQ Q QQQ + Q R MP
Sbjct: 200 QQNFDQAP-QNYPQQQRYGPSSPQYAQQQSFGPPGQGERRNYMP 328
>BI316328
Length = 407
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/64 (42%), Positives = 33/64 (51%)
Frame = +2
Query: 423 PQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRP 482
PQ+ Q Q Q Q QPQ +QQQ QQQP Q Q + Q +QQ+ QQ +
Sbjct: 248 PQLLQQQQQQQ----QQPQPQQLQQQLIQQQPQQ--------QSHLQVSVHQQQQQQQQS 391
Query: 483 PRMP 486
PRMP
Sbjct: 392 PRMP 403
>BM308932
Length = 429
Score = 42.0 bits (97), Expect = 0.001
Identities = 28/60 (46%), Positives = 31/60 (51%)
Frame = +1
Query: 423 PQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQPYQQRPQQPRP 482
PQ+ Q Q Q Q QPQ +QQQ QQQP QQ Q Q QQQ QQ P+ P P
Sbjct: 244 PQLLQQQQQQQ----QQPQPQQLQQQLIQQQPQQQSHLQVSVHQ--QQQQQQQSPRMPGP 405
Score = 40.0 bits (92), Expect = 0.004
Identities = 32/85 (37%), Positives = 39/85 (45%), Gaps = 9/85 (10%)
Frame = +1
Query: 411 QQTNNHPQIPQYPQIPQYPQFPQNPSPQNV--QPQNVQQQNFQQ-----QPYQQYPYQQY 463
QQ + Q+ Q Q Q Q QPQ +QQQ QQ Q QQ QQ
Sbjct: 145 QQLGSSTQLQQNSMTLNQQQLSQLMQQQKSMGQPQLLQQQQQQQQQPQPQQLQQQLIQQQ 324
Query: 464 PQQ--NFQQQPYQQRPQQPRPPRMP 486
PQQ + Q +QQ+ QQ + PRMP
Sbjct: 325 PQQQSHLQVSVHQQQQQQQQSPRMP 399
>TC215424 similar to UP|Q9MA04 (Q9MA04) F20B17.16, partial (26%)
Length = 1030
Score = 40.8 bits (94), Expect = 0.002
Identities = 29/100 (29%), Positives = 37/100 (37%), Gaps = 3/100 (3%)
Frame = +3
Query: 395 PAYQHIAAITPTSHPFQQTNNH-PQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQ 453
P+ I P S+ Q +H P P P P YP P P P + ++Q
Sbjct: 27 PSTSSFPQIPPNSNYHQNQQHHVPYAPPNPHHPHYPYPPPPPPPP-------PEASYQPP 185
Query: 454 PYQQYPYQQYPQQN--FQQQPYQQRPQQPRPPRMPINPIP 491
P YP N Q P P P PP P++P P
Sbjct: 186 PPPPPAPMYYPSNNQYSNQPPPPPPPLSPPPPPPPVSPPP 305
Score = 30.4 bits (67), Expect = 3.3
Identities = 16/69 (23%), Positives = 30/69 (43%)
Frame = +3
Query: 412 QTNNHPQIPQYPQIPQYPQFPQNPSPQNVQPQNVQQQNFQQQPYQQYPYQQYPQQNFQQQ 471
Q +N P P P P P P +P P QN ++++F++ ++ +
Sbjct: 228 QYSNQPPPPPPPLSPPPPPPPVSPPPPPPVTQNNEERHFKEPSTSGRREYEHSNHGIAHK 407
Query: 472 PYQQRPQQP 480
++Q+P P
Sbjct: 408 QHKQQPPVP 434
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.133 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,709,026
Number of Sequences: 63676
Number of extensions: 586900
Number of successful extensions: 9077
Number of sequences better than 10.0: 296
Number of HSP's better than 10.0 without gapping: 6077
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7885
length of query: 820
length of database: 12,639,632
effective HSP length: 105
effective length of query: 715
effective length of database: 5,953,652
effective search space: 4256861180
effective search space used: 4256861180
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC135161.1


