

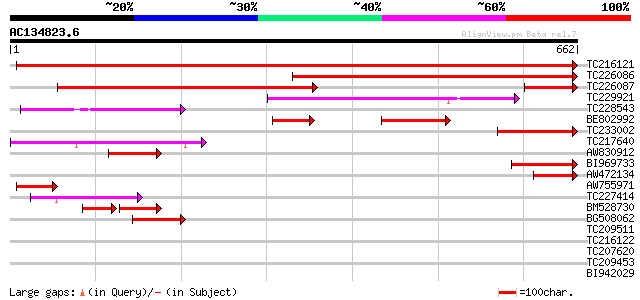
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134823.6 + phase: 0
(662 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC216121 similar to UP|Q9ZPI5 (Q9ZPI5) MFP2 (Fatty acid multifun... 777 0.0
TC226086 similar to UP|Q9ZPI6 (Q9ZPI6) AIM1 protein, partial (51%) 559 e-159
TC226087 similar to UP|Q9ZPI6 (Q9ZPI6) AIM1 protein, partial (55%) 518 e-147
TC229921 similar to GB|BAB02158.1|9294256|AP000413 3-hydroxybuty... 144 2e-34
TC228543 similar to UP|Q6NL24 (Q6NL24) At4g16210, partial (97%) 110 2e-24
BE802992 similar to GP|4337027|gb| MFP2 {Arabidopsis thaliana}, ... 104 1e-22
TC233002 similar to UP|Q9ZPI5 (Q9ZPI5) MFP2 (Fatty acid multifun... 101 9e-22
TC217640 similar to UP|Q9FHR8 (Q9FHR8) Enoyl CoA hydratase-like ... 91 2e-18
AW830912 weakly similar to PIR|T08956|T089 AIM1 protein - Arabid... 88 1e-17
BI969733 weakly similar to GP|17105223|gb| peroxisomal multifunc... 85 9e-17
AW472134 weakly similar to GP|17105223|gb| peroxisomal multifunc... 85 1e-16
AW755971 weakly similar to GP|17065148|gb| fatty acid multifunct... 64 3e-10
TC227414 similar to UP|Q9LKJ1 (Q9LKJ1) CoA-thioester hydrolase C... 54 3e-07
BM528730 52 8e-07
BG508062 52 1e-06
TC209511 41 0.001
TC216122 similar to UP|MFPA_BRANA (O49809) Glyoxysomal fatty aci... 32 0.69
TC207620 similar to UP|Q8LK74 (Q8LK74) Cyclin D3.1 protein, part... 31 1.5
TC209453 similar to UP|O81335 (O81335) Geranylgeranyl hydrogenas... 31 2.0
BI942029 30 2.6
>TC216121 similar to UP|Q9ZPI5 (Q9ZPI5) MFP2 (Fatty acid multifunctional
protein) (AtMFP2), partial (97%)
Length = 2449
Score = 777 bits (2007), Expect = 0.0
Identities = 384/656 (58%), Positives = 499/656 (75%), Gaps = 2/656 (0%)
Frame = +3
Query: 9 EVGNDGVAVITMCNPPVNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDIS 68
EVG DGVAVIT+ NPPVN+L+ ++R LK F++A +R+DVKAIV+TG G+FSGGFDIS
Sbjct: 132 EVGPDGVAVITIVNPPVNSLSFDVLRSLKESFDQAIQRDDVKAIVVTGAKGKFSGGFDIS 311
Query: 69 VMQKVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPK 128
+ + + +SVE++ ++IE ++KP VAA++GLALGGGLE+AM C+AR++ P
Sbjct: 312 AFGGIQEAKERPKPGWISVEIITDTIEAARKPSVAAIDGLALGGGLEVAMACNARLSTPT 491
Query: 129 AQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAE 188
AQLGLPEL LGIIPGFGGTQRLPRLVG +K +EM+L SKP+ +E LGL+D +VSP +
Sbjct: 492 AQLGLPELQLGIIPGFGGTQRLPRLVGLTKGLEMILASKPVKGKEAFSLGLVDGLVSPND 671
Query: 189 LLKLSRQWALEIAEQRRPWIRSLHITDKLG--SDAREVLRTARQHVKKTAPHLPQQQACI 246
L+ +RQWAL++ RRPWI SL+ T+KL +ARE+L+ AR +K AP+L CI
Sbjct: 672 LVNTARQWALDMLGHRRPWIASLYKTEKLEPLGEAREILKFARAQARKRAPNLQHPLVCI 851
Query: 247 DVIEHGILHGGYSGVLREAEVFKKLVLSETAKGLIHVFFAQRTISKIPGVTDIGLKPRNV 306
DVIE GI+ G +G+ +EAE F+ LV S+T K L+HVFFAQR SK+PGVTD GL PR V
Sbjct: 852 DVIEAGIVAGPRAGLWKEAEAFEGLVRSDTCKSLVHVFFAQRGTSKVPGVTDCGLAPRQV 1031
Query: 307 RKAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANVRGLVTRKKLTQQK 366
+K A+IGGGLMGSGIATALIL N VILKEVN ++L GI I+AN++ V + KLT++
Sbjct: 1032KKVAIIGGGLMGSGIATALILSNYPVILKEVNEKFLDAGINRIKANLQSRVKKGKLTKEN 1211
Query: 367 AEGALSLLKGVLDYAEFKDVDMVIEAVIEKVSLKQDIFSDLEKICPPHCILASNTSTIDL 426
E +SLLKG LDY F+DVDMVIEAVIE +SLKQ IFSDLEK CPPHCILASNTSTIDL
Sbjct: 1212FEKTISLLKGSLDYESFRDVDMVIEAVIENISLKQQIFSDLEKYCPPHCILASNTSTIDL 1391
Query: 427 NIIGEKISSQDRVIGAHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKSPVVVGN 486
N+IGEK +QDR++GAHFFSPAH+MPLLEIVRT +TSAQVI+D++ + K IKK+PVVVGN
Sbjct: 1392NLIGEKTKTQDRIVGAHFFSPAHVMPLLEIVRTKQTSAQVIVDVLDISKKIKKTPVVVGN 1571
Query: 487 CTGFAVNRTFFPYAQGAHFLANLGVDVFRIDRLISNFGLPMGPFQLQDLSGYGVAVAVGK 546
CTGFAVNR FFPY Q L G DV++IDR+I+ FG+PMGPF+L DL G+GVA+A G
Sbjct: 1572CTGFAVNRMFFPYTQAGLLLVERGADVYQIDRVITKFGMPMGPFRLMDLVGFGVAIATGM 1751
Query: 547 EFAGSFAGRTFPTPLLDLLIKSGRNGKNNGKGYYIYEKGSKPKPDPSVLPIVEESRRLSN 606
+F +F RT+ + L+ LL + R G+ KG+Y+Y K PDP + +E++R +S
Sbjct: 1752QFIQNFPERTYKSMLISLLQEDKRAGETTHKGFYLYNDKRKASPDPELKNYIEKARSISG 1931
Query: 607 IMPNGKPISITDQEIVEMILFPVVNEACRVLEEGIVIRASDLDIASVLGMSFPSYR 662
+ + K + +++I+EMI FPVVNEACRVL+EGI ++A+DLDI++++GM FP YR
Sbjct: 1932VSVDPKLAKLQEKDIIEMIFFPVVNEACRVLDEGIAVKAADLDISAIMGMGFPPYR 2099
>TC226086 similar to UP|Q9ZPI6 (Q9ZPI6) AIM1 protein, partial (51%)
Length = 1490
Score = 559 bits (1440), Expect = e-159
Identities = 274/332 (82%), Positives = 306/332 (91%)
Frame = +3
Query: 331 RVILKEVNSEYLQKGIKTIEANVRGLVTRKKLTQQKAEGALSLLKGVLDYAEFKDVDMVI 390
+VILKE+N E+L KGIKTIEANV GLV R KLT+QKA+ ALSLL+GVLDY+EFKDVD+VI
Sbjct: 3 QVILKEINPEFLLKGIKTIEANVNGLVRRGKLTKQKADAALSLLRGVLDYSEFKDVDLVI 182
Query: 391 EAVIEKVSLKQDIFSDLEKICPPHCILASNTSTIDLNIIGEKISSQDRVIGAHFFSPAHI 450
EAVIE +SLKQ IFSDLEKICPPHCILASNTSTIDL+++G+ SSQ+R+ GAHFFSPAHI
Sbjct: 183 EAVIENISLKQTIFSDLEKICPPHCILASNTSTIDLDVVGKTTSSQNRIAGAHFFSPAHI 362
Query: 451 MPLLEIVRTNKTSAQVILDLVTVGKIIKKSPVVVGNCTGFAVNRTFFPYAQGAHFLANLG 510
MPLLEI+RT+KTSAQVILDL+TVGKIIKK+PVVVGNCTGFAVN+TFFPY+QGAH L NLG
Sbjct: 363 MPLLEIIRTDKTSAQVILDLITVGKIIKKTPVVVGNCTGFAVNKTFFPYSQGAHLLVNLG 542
Query: 511 VDVFRIDRLISNFGLPMGPFQLQDLSGYGVAVAVGKEFAGSFAGRTFPTPLLDLLIKSGR 570
VDVFRID LI NFG P+GPFQLQDL+GYGVAVA K FA +F+ R F +PLLDLLIKSGR
Sbjct: 543 VDVFRIDTLIRNFGFPIGPFQLQDLAGYGVAVATSKVFADAFSDRIFKSPLLDLLIKSGR 722
Query: 571 NGKNNGKGYYIYEKGSKPKPDPSVLPIVEESRRLSNIMPNGKPISITDQEIVEMILFPVV 630
NGKNNGKGYYIYEK KPKPDPS+LPI+EESRRL NIMPNGKPIS+TDQEIVEMILFPVV
Sbjct: 723 NGKNNGKGYYIYEKAGKPKPDPSILPIIEESRRLCNIMPNGKPISVTDQEIVEMILFPVV 902
Query: 631 NEACRVLEEGIVIRASDLDIASVLGMSFPSYR 662
NEACRVLE+G+VIRASDLDIASVLGMSFP+YR
Sbjct: 903 NEACRVLEDGVVIRASDLDIASVLGMSFPNYR 998
>TC226087 similar to UP|Q9ZPI6 (Q9ZPI6) AIM1 protein, partial (55%)
Length = 1223
Score = 518 bits (1335), Expect = e-147
Identities = 264/306 (86%), Positives = 288/306 (93%), Gaps = 2/306 (0%)
Frame = +3
Query: 56 GKGGRFSGGFDISVMQKVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLE 115
GKGGRFSGGFDISVMQKVH+TGD + +PDVSVELVVNSIEDSKKPVVAAV GLALGGGLE
Sbjct: 6 GKGGRFSGGFDISVMQKVHRTGDSSHLPDVSVELVVNSIEDSKKPVVAAVAGLALGGGLE 185
Query: 116 LAMGCHARVAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQ 175
LAMGCHARVAAP+AQLGLPELTLGIIPGFGGTQRLPRL+G SKAVEMMLTSKPIT+EEG+
Sbjct: 186 LAMGCHARVAAPRAQLGLPELTLGIIPGFGGTQRLPRLIGLSKAVEMMLTSKPITSEEGR 365
Query: 176 KLGLIDAIVSPAELLKLSRQWALEIAEQRRPWIRSLHITDKLG--SDAREVLRTARQHVK 233
KLGLIDAIVS ELLK SR WALEI E+R+PWIRSLH TDK+G S+AR VL+TARQ VK
Sbjct: 366 KLGLIDAIVSSEELLKASRLWALEIGERRKPWIRSLHRTDKIGSLSEARAVLKTARQQVK 545
Query: 234 KTAPHLPQQQACIDVIEHGILHGGYSGVLREAEVFKKLVLSETAKGLIHVFFAQRTISKI 293
KTAPHLPQQQAC+DVIEHGI+HGGYSGVL+EAEVFK+LV+S+T+KGLI+VFFAQR ISK+
Sbjct: 546 KTAPHLPQQQACVDVIEHGIVHGGYSGVLKEAEVFKQLVISDTSKGLINVFFAQRAISKV 725
Query: 294 PGVTDIGLKPRNVRKAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANV 353
PGVTDIGLKPRNV+KAAVIGGGLMGSGIATALILGNIRVILKE+NSE+L KGIKTIEANV
Sbjct: 726 PGVTDIGLKPRNVKKAAVIGGGLMGSGIATALILGNIRVILKEINSEFLLKGIKTIEANV 905
Query: 354 RGLVTR 359
GLV R
Sbjct: 906 NGLVRR 923
Score = 116 bits (290), Expect = 4e-26
Identities = 57/61 (93%), Positives = 60/61 (97%)
Frame = +3
Query: 602 RRLSNIMPNGKPISITDQEIVEMILFPVVNEACRVLEEGIVIRASDLDIASVLGMSFPSY 661
RRL NIMPNGKPISITDQEIVEMILFPVVNEACRVLE+G+VIRASDLDIASVLGMSFP+Y
Sbjct: 918 RRLCNIMPNGKPISITDQEIVEMILFPVVNEACRVLEDGVVIRASDLDIASVLGMSFPNY 1097
Query: 662 R 662
R
Sbjct: 1098R 1100
>TC229921 similar to GB|BAB02158.1|9294256|AP000413 3-hydroxybutyryl-CoA
dehydrogenase-like protein {Arabidopsis thaliana;} ,
partial (96%)
Length = 1199
Score = 144 bits (362), Expect = 2e-34
Identities = 93/299 (31%), Positives = 150/299 (50%), Gaps = 5/299 (1%)
Frame = +2
Query: 302 KPRNVRKAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANVRGLVTRKK 361
K V++ V+G G MGSGIA + I V L +++ + L K +I +++ V++
Sbjct: 137 KMLEVKRIGVVGSGQMGSGIAQVAAMHGIDVWLHDLDPQALSKASSSISSSINRFVSKNH 316
Query: 362 LTQQKAEGALSLLKGVLDYAEFKDVDMVIEAVIEKVSLKQDIFSDLEKICPPHCILASNT 421
++Q AL L+ D + + D++IEA++E +K+ +F+ L++I ILASNT
Sbjct: 317 ISQVAGADALKRLRLTTDLEDLQLADLIIEAIVESEDVKKSLFAQLDRIAKSSAILASNT 496
Query: 422 STIDLNIIGEKISSQDRVIGAHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKSP 481
S+I + + S +VIG HF +P +M L+EIVR TS + + + + + K
Sbjct: 497 SSISVTRLATSTSRPRQVIGMHFMNPPPVMKLIEIVRGADTSEETFVATKALSERLGKVV 676
Query: 482 VVVGNCTGFAVNRTFFPYAQGAHFLANLGV----DVFRIDRLISNFGLPMGPFQLQDLSG 537
+ + +GF VNR P A F GV D+ +L +N PMGP +L D G
Sbjct: 677 ITSHDYSGFIVNRILMPMINEAFFALYTGVATKEDIDTGMKLGTNH--PMGPLELADFIG 850
Query: 538 YGVAVAVGKEF-AGSFAGRTFPTPLLDLLIKSGRNGKNNGKGYYIYEKGSKPKPDPSVL 595
V +++ + AG + P PLL + +GR G+ G G Y Y K + S L
Sbjct: 851 LDVCLSIMRVLHAGLGDNKYAPCPLLVQYVDAGRLGRKRGIGVYEYSKDPRSTKSSSRL 1027
>TC228543 similar to UP|Q6NL24 (Q6NL24) At4g16210, partial (97%)
Length = 1060
Score = 110 bits (275), Expect = 2e-24
Identities = 68/194 (35%), Positives = 111/194 (57%), Gaps = 1/194 (0%)
Frame = +3
Query: 13 DGVAVITMCNP-PVNALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGRFSGGFDISVMQ 71
+GVA++T+ P +N+L P++ L F+ R V+ I+LTG G F G D++ +
Sbjct: 132 NGVALVTINRPGSLNSLTRPMMVDLAQAFKRLDRDELVRVIILTGSGRSFCSGVDLTAAE 311
Query: 72 KVHQTGDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARVAAPKAQL 131
V + GD V D + VV +E +KP++ A+ G A+ G E+A+ C VAA ++
Sbjct: 312 DVFK-GD---VKDPESDPVVQ-MELCRKPIIGAIRGFAVTAGFEIALACDILVAAKGSKF 476
Query: 132 GLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAELLK 191
GI P +G +Q+L R++G +KA E+ L++ P+TAE +KLG ++ +V ELLK
Sbjct: 477 MDTHARFGIFPSWGLSQKLSRVIGANKAREVSLSATPLTAEVAEKLGFVNHVVEEGELLK 656
Query: 192 LSRQWALEIAEQRR 205
SR+ A I + +
Sbjct: 657 KSREIADAIVKNNQ 698
>BE802992 similar to GP|4337027|gb| MFP2 {Arabidopsis thaliana}, partial
(16%)
Length = 416
Score = 104 bits (260), Expect = 1e-22
Identities = 51/80 (63%), Positives = 60/80 (74%)
Frame = +1
Query: 435 SQDRVIGAHFFSPAHIMPLLEIVRTNKTSAQVILDLVTVGKIIKKSPVVVGNCTGFAVNR 494
SQ R++GAHFFSPAH+M LLEIVRT +TS VI+D++ V K IKK+PVVVGNC FAVNR
Sbjct: 175 SQGRIVGAHFFSPAHVMLLLEIVRTKQTSP*VIVDVLDVSKKIKKTPVVVGNCARFAVNR 354
Query: 495 TFFPYAQGAHFLANLGVDVF 514
FFPY Q L G DV+
Sbjct: 355 MFFPYTQAGLLLVEHGADVY 414
Score = 59.3 bits (142), Expect = 5e-09
Identities = 30/49 (61%), Positives = 36/49 (73%)
Frame = +3
Query: 308 KAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQKGIKTIEANVRGL 356
K A++GGGLMGSGIAT LILG+ VILKEVN ++L GI I+ R L
Sbjct: 3 KVAILGGGLMGSGIATTLILGSYPVILKEVNEKFLDAGINRIKGENRFL 149
>TC233002 similar to UP|Q9ZPI5 (Q9ZPI5) MFP2 (Fatty acid multifunctional
protein) (AtMFP2), partial (21%)
Length = 611
Score = 101 bits (252), Expect = 9e-22
Identities = 43/93 (46%), Positives = 66/93 (70%)
Frame = +1
Query: 570 RNGKNNGKGYYIYEKGSKPKPDPSVLPIVEESRRLSNIMPNGKPISITDQEIVEMILFPV 629
R G+ KG+Y+Y+ KP PDP + +E++R + + + K + + +++I+EM FPV
Sbjct: 13 RAGETTRKGFYLYDDKCKPSPDPELKNYIEKARSFTGVSVDPKLVKLQEKDIIEMTFFPV 192
Query: 630 VNEACRVLEEGIVIRASDLDIASVLGMSFPSYR 662
VNEACRVL+EGI ++ SDLDI++V+GM FP YR
Sbjct: 193 VNEACRVLDEGIAVKGSDLDISAVMGMGFPPYR 291
>TC217640 similar to UP|Q9FHR8 (Q9FHR8) Enoyl CoA hydratase-like protein,
partial (83%)
Length = 1203
Score = 90.5 bits (223), Expect = 2e-18
Identities = 65/244 (26%), Positives = 117/244 (47%), Gaps = 16/244 (6%)
Frame = +1
Query: 2 ASVKVDFEVGNDGVAVITMCNPPV-NALAIPIIRGLKNKFEEAARRNDVKAIVLTGKGGR 60
+S+++ E N GV + + P NAL+ +VK IVL+G G
Sbjct: 19 SSLEIVEESPNSGVFFLILNRPSRRNALSREFFSEFPKALHALDHNPEVKVIVLSGAGDH 198
Query: 61 FSGGFDISVMQKVHQT------GDITLVPDVSVELVVNSIEDSKKPVVAAVEGLALGGGL 114
F G D+S++ + G+ ++++ V ++E +KPV+A+V G +GGG+
Sbjct: 199 FCSGIDLSLLGSTAASSGSSGSGETLRREIMAMQDAVTALERCRKPVIASVHGACIGGGI 378
Query: 115 ELAMGCHARVAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEG 174
++ C R+ + +A + E+ L + G QRLP +VG A+E+ LT + + +E
Sbjct: 379 DIVTACDIRMCSEEAFFSVKEVDLALAADLGTLQRLPLIVGFGNAMELALTGRTFSGKEA 558
Query: 175 QKLGLIDAI-VSPAELLKLSRQWALEIAEQ--------RRPWIRSLHITDKLGSDAREVL 225
++LGL+ + +S +L + R A IA + + ++S +T G D L
Sbjct: 559 KELGLVSRVFLSKHDLHQAVRDVAQAIATKSPLAVVGTKTVLLKSRDLTVDQGLDYVATL 738
Query: 226 RTAR 229
+AR
Sbjct: 739 NSAR 750
>AW830912 weakly similar to PIR|T08956|T089 AIM1 protein - Arabidopsis
thaliana, partial (8%)
Length = 187
Score = 87.8 bits (216), Expect = 1e-17
Identities = 46/62 (74%), Positives = 49/62 (78%)
Frame = +2
Query: 116 LAMGCHARVAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQ 175
LAMGC ARV A K QLGLPELTLGII G GTQ L RL+ S+A EMMLTSKPIT+EEG
Sbjct: 2 LAMGCRARVEAQKTQLGLPELTLGIIRGL*GTQSLTRLIELSEAAEMMLTSKPITSEEGT 181
Query: 176 KL 177
KL
Sbjct: 182 KL 187
>BI969733 weakly similar to GP|17105223|gb| peroxisomal multifunctional
protein {Oryza sativa}, partial (12%)
Length = 533
Score = 85.1 bits (209), Expect = 9e-17
Identities = 40/76 (52%), Positives = 52/76 (67%)
Frame = -3
Query: 587 KPKPDPSVLPIVEESRRLSNIMPNGKPISITDQEIVEMILFPVVNEACRVLEEGIVIRAS 646
KPKP PS+LPI+ E + N PNGKP + Q++ EM FPVVNEA VL++G+VI AS
Sbjct: 531 KPKPGPSILPILGEFKXXCNFRPNGKPFFVAGQKVXEMXXFPVVNEASXVLKDGVVIGAS 352
Query: 647 DLDIASVLGMSFPSYR 662
L +A V G FP++R
Sbjct: 351 GLALAFVFGXXFPNFR 304
>AW472134 weakly similar to GP|17105223|gb| peroxisomal multifunctional
protein {Oryza sativa}, partial (11%)
Length = 319
Score = 84.7 bits (208), Expect = 1e-16
Identities = 42/51 (82%), Positives = 47/51 (91%)
Frame = +2
Query: 612 KPISITDQEIVEMILFPVVNEACRVLEEGIVIRASDLDIASVLGMSFPSYR 662
KPIS+TDQEI EM+LFPV NEACRVLE+G+ IRASDLDIASVLGMS P+YR
Sbjct: 2 KPISMTDQEIGEMMLFPVGNEACRVLEDGVGIRASDLDIASVLGMSCPNYR 154
>AW755971 weakly similar to GP|17065148|gb| fatty acid multifunctional
protein (AtMFP2) {Arabidopsis thaliana}, partial (14%)
Length = 433
Score = 63.5 bits (153), Expect = 3e-10
Identities = 28/48 (58%), Positives = 39/48 (80%)
Frame = +1
Query: 9 EVGNDGVAVITMCNPPVNALAIPIIRGLKNKFEEAARRNDVKAIVLTG 56
EVG DGVA+IT+ NPPVN+L+ ++R K F++A +R+DVKAIV+TG
Sbjct: 289 EVGPDGVAIITIVNPPVNSLSYDVLRSFKENFDQALQRDDVKAIVVTG 432
>TC227414 similar to UP|Q9LKJ1 (Q9LKJ1) CoA-thioester hydrolase CHY1
(3-hydroxyisobutyryl-coenzyme A hydrolase), partial
(81%)
Length = 1538
Score = 53.5 bits (127), Expect = 3e-07
Identities = 37/138 (26%), Positives = 64/138 (45%), Gaps = 7/138 (5%)
Frame = +3
Query: 25 VNALAIPIIRGLKNKFEEAARRNDVKAIV-----LTGKGGRFSGGFDISVMQKVHQTGDI 79
+NAL+ ++ L F E + +D+K +V + G G F G D++ + + GD
Sbjct: 117 LNALSFYMVSRLLEIFSEDEKDSDIKLVVVKSSVIKGNGRAFCAGGDVAAVARDGSKGDW 296
Query: 80 TLVPDV-SVELVVNSIEDS-KKPVVAAVEGLALGGGLELAMGCHARVAAPKAQLGLPELT 137
+ E +N + + KP V+ + G+ +GGG +++ RV +PE
Sbjct: 297 RFGANFFQSEFKLNYLMATYSKPQVSILNGIVMGGGAGVSVHGRFRVVTENTVFAMPETA 476
Query: 138 LGIIPGFGGTQRLPRLVG 155
LG+ P G + L RL G
Sbjct: 477 LGLFPDIGSSYFLSRLPG 530
>BM528730
Length = 424
Score = 52.0 bits (123), Expect = 8e-07
Identities = 26/49 (53%), Positives = 33/49 (67%)
Frame = +2
Query: 129 AQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKL 177
A +GLPE L IIPG GGTQRLPRLVG + A +++ T + I +E L
Sbjct: 278 ALMGLPETGLAIIPGAGGTQRLPRLVGKAIAKDIIFTGRKIDGKEALSL 424
Score = 42.7 bits (99), Expect = 5e-04
Identities = 17/39 (43%), Positives = 27/39 (68%)
Frame = +1
Query: 86 SVELVVNSIEDSKKPVVAAVEGLALGGGLELAMGCHARV 124
S+ + +ED + P +A +EG+ALGGGLE+A+ C R+
Sbjct: 28 SLRSTFSFLEDVRVPTIAVIEGVALGGGLEMALACDIRI 144
>BG508062
Length = 429
Score = 51.6 bits (122), Expect = 1e-06
Identities = 24/62 (38%), Positives = 44/62 (70%)
Frame = +1
Query: 144 FGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAIVSPAELLKLSRQWALEIAEQ 203
+G +Q+L +++G +KA E+ L++ P+TAE ++LGL++ ++ AELLK SR+ A I +
Sbjct: 7 WGLSQKLSQIIGANKAREVSLSATPLTAEVAERLGLVNHVIEEAELLKKSREIADAIVKN 186
Query: 204 RR 205
+
Sbjct: 187 NQ 192
>TC209511
Length = 652
Score = 41.2 bits (95), Expect = 0.001
Identities = 28/88 (31%), Positives = 41/88 (45%)
Frame = +3
Query: 124 VAAPKAQLGLPELTLGIIPGFGGTQRLPRLVGTSKAVEMMLTSKPITAEEGQKLGLIDAI 183
+AA A G +G G+ + RLVG KA EM ++ A E +K+GL++ +
Sbjct: 9 IAADNAIFGQTGPKVGSFDAGYGSSIMSRLVGPKKAREMWFLTRFYDAVEAEKMGLVNTV 188
Query: 184 VSPAELLKLSRQWALEIAEQRRPWIRSL 211
V L K + +W EI IR L
Sbjct: 189 VPLENLEKETIKWCREILRNSPTAIRVL 272
>TC216122 similar to UP|MFPA_BRANA (O49809) Glyoxysomal fatty acid
beta-oxidation multifunctional protein MFP-a [Includes:
Enoyl-CoA hydratase ; 3-2-trans-enoyl-CoA isomerase ;
3-hydroxybutyryl-CoA epimerase ; 3-hydroxyacyl-CoA
dehydrogenase , partial (10%)
Length = 599
Score = 32.3 bits (72), Expect = 0.69
Identities = 11/20 (55%), Positives = 18/20 (90%)
Frame = +1
Query: 643 IRASDLDIASVLGMSFPSYR 662
++A+DLDI++++GM FP YR
Sbjct: 4 VKAADLDISAIMGMGFPPYR 63
>TC207620 similar to UP|Q8LK74 (Q8LK74) Cyclin D3.1 protein, partial (51%)
Length = 1405
Score = 31.2 bits (69), Expect = 1.5
Identities = 19/57 (33%), Positives = 30/57 (52%)
Frame = -3
Query: 164 LTSKPITAEEGQKLGLIDAIVSPAELLKLSRQWALEIAEQRRPWIRSLHITDKLGSD 220
+T KP ++EG+K L A PAE++ S+ I ++ R W +H+ K G D
Sbjct: 719 ITHKPRISKEGEKNALRLAEELPAEMILRSKPSCEVIKKRERGW--GVHLPSKSGKD 555
>TC209453 similar to UP|O81335 (O81335) Geranylgeranyl hydrogenase , partial
(36%)
Length = 745
Score = 30.8 bits (68), Expect = 2.0
Identities = 17/37 (45%), Positives = 20/37 (53%)
Frame = +1
Query: 308 KAAVIGGGLMGSGIATALILGNIRVILKEVNSEYLQK 344
+AAVIGGG GS A AL G + L E N + K
Sbjct: 214 RAAVIGGGPAGSSAAEALASGGVETFLFERNPPSVAK 324
>BI942029
Length = 424
Score = 30.4 bits (67), Expect = 2.6
Identities = 15/30 (50%), Positives = 18/30 (60%)
Frame = +1
Query: 100 PVVAAVEGLALGGGLELAMGCHARVAAPKA 129
PV+A V G A+GGG L M C +AA A
Sbjct: 328 PVIAMVAGYAVGGGHVLHMVCDLTIAADNA 417
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.139 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,699,065
Number of Sequences: 63676
Number of extensions: 297649
Number of successful extensions: 1373
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 1360
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1370
length of query: 662
length of database: 12,639,632
effective HSP length: 103
effective length of query: 559
effective length of database: 6,081,004
effective search space: 3399281236
effective search space used: 3399281236
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC134823.6


