

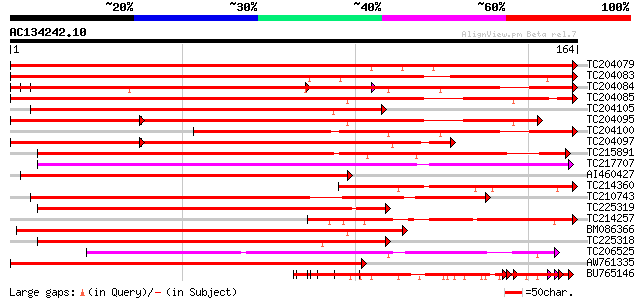
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134242.10 + phase: 0
(164 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding ... 283 2e-77
TC204083 similar to UP|O48567 (O48567) Glycine-rich RNA-binding ... 264 2e-71
TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, co... 257 2e-69
TC204085 similar to UP|Q8VX74 (Q8VX74) Glycine-rich RNA-binding ... 238 1e-63
TC204105 similar to UP|Q8VX74 (Q8VX74) Glycine-rich RNA-binding ... 179 4e-46
TC204095 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding ... 162 5e-41
TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, ... 152 5e-38
TC204097 homologue to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-bindin... 144 2e-35
TC215891 similar to GB|AAM78058.1|21928059|AY125548 AT5g61030/ma... 139 5e-34
TC217707 similar to UP|Q941H8 (Q941H8) RNA-binding protein precu... 134 3e-32
AI460427 similar to GP|5726567|gb|A glycine-rich RNA-binding pro... 130 3e-31
TC214360 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding ... 124 3e-29
TC210743 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1,... 117 2e-27
TC225319 similar to UP|P93486 (P93486) Glycine-rich RNA-binding ... 112 1e-25
TC214257 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 pr... 110 4e-25
BM086366 107 3e-24
TC225318 similar to UP|P93486 (P93486) Glycine-rich RNA-binding ... 104 2e-23
TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (... 102 8e-23
AW761335 similar to GP|5726567|gb|A glycine-rich RNA-binding pro... 101 1e-22
BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvo... 97 3e-21
>TC204079 similar to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
complete
Length = 748
Score = 283 bits (725), Expect = 2e-77
Identities = 137/171 (80%), Positives = 148/171 (86%), Gaps = 7/171 (4%)
Frame = +1
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
MAS+DVEYRCFVGGLAWATD +ALE+AFSQYGEI+++KIINDRETGRSRGFGFVTFA E+
Sbjct: 67 MASSDVEYRCFVGGLAWATDDQALERAFSQYGEIVETKIINDRETGRSRGFGFVTFASEQ 246
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGG-GGGYGGGGR-REGGYN 118
SM+DAIEGMNGQ++DGRNITVNEAQSRG GGGGG GGGGYGGG GGGYGGGGR GGYN
Sbjct: 247 SMKDAIEGMNGQNLDGRNITVNEAQSRGGGGGGGGGGGGYGGGRGGGYGGGGRGGGGGYN 426
Query: 119 RSS-----GGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSWRS 164
RS GGGGGYGGGGGGGYGGGRDRGYG G YSRGG G G WR+
Sbjct: 427 RSGGGGGYGGGGGYGGGGGGGYGGGRDRGYGGGGDRGYSRGGDGGDGGWRN 579
>TC204083 similar to UP|O48567 (O48567) Glycine-rich RNA-binding protein,
partial (95%)
Length = 1019
Score = 264 bits (674), Expect = 2e-71
Identities = 132/177 (74%), Positives = 145/177 (81%), Gaps = 13/177 (7%)
Frame = +2
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
MASADVE+RCFVGGLAW T ++ALEKAFS YG+I++SK+INDRETGRSRGFGFVTFA E+
Sbjct: 38 MASADVEFRCFVGGLAWVTGNDALEKAFSIYGDIVESKVINDRETGRSRGFGFVTFASEQ 217
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQ---SRGSGGGGG--------RGGGGYGGGGGGYGG 109
SM+DAI GMNGQD+DGRNITVNEAQ SRG GGGGG RG GGYGGGGGG G
Sbjct: 218 SMKDAIAGMNGQDLDGRNITVNEAQTRASRGGGGGGGFGSGGGYNRGSGGYGGGGGGGGY 397
Query: 110 GGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRG--GGGDGGSWRS 164
GGRREGGYNR+ GGGGGGGYGGGRDRGYG DGGSRYSRG GGG G+WR+
Sbjct: 398 GGRREGGYNRN-------GGGGGGGYGGGRDRGYGGDGGSRYSRGGEGGGSDGNWRN 547
>TC204084 UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein, complete
Length = 1937
Score = 257 bits (657), Expect = 2e-69
Identities = 127/168 (75%), Positives = 136/168 (80%), Gaps = 4/168 (2%)
Frame = +2
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
MASADVEYRCFVGGLAWATD ALE+AFSQYGEI+++KIINDRETGRSRGFGFVTFA E+
Sbjct: 1220 MASADVEYRCFVGGLAWATDDHALERAFSQYGEIVETKIINDRETGRSRGFGFVTFASEQ 1399
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYG---GGGRREGGY 117
SM+DAI MNGQ++DGRNITVNEAQSRG GGGGG GGGGY GGGGYG GGG GGY
Sbjct: 1400 SMKDAIGAMNGQNLDGRNITVNEAQSRGGGGGGGGGGGGYNRGGGGYGGRSGGGGGGGGY 1579
Query: 118 NRSSGG-GGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSWRS 164
GG GGGYGGGGGGGYGGGRDRG YSRGG G G WR+
Sbjct: 1580 RSRDGGYGGGYGGGGGGGYGGGRDRG--------YSRGGDGGDGGWRN 1699
Score = 81.3 bits (199), Expect = 2e-16
Identities = 39/85 (45%), Positives = 58/85 (67%), Gaps = 1/85 (1%)
Frame = -1
Query: 4 ADVEYRCFVGGLAWATDSEALEKAFSQYGE-IIDSKIINDRETGRSRGFGFVTFADEKSM 62
+D E R VG LAW D ALE F + G+ ++++++I DRE+GRSRGFGFVTF +
Sbjct: 575 SDSENRVHVGNLAWGVDDVALESLFREQGKKVLEARVIYDRESGRSRGFGFVTFGSPDEV 396
Query: 63 RDAIEGMNGQDMDGRNITVNEAQSR 87
+ AI+ ++G D++GR I V+ A S+
Sbjct: 395 KSAIQSLDGVDLNGRAIRVSLADSK 321
Score = 73.9 bits (180), Expect = 3e-14
Identities = 38/102 (37%), Positives = 56/102 (54%), Gaps = 2/102 (1%)
Frame = -1
Query: 7 EYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAI 66
+ + FVG L ++ DS L + F G + ++I D+ TGRSRGFGFVT + + A
Sbjct: 884 DLKLFVGNLPFSVDSARLAELFESAGNVEVVEVIYDKTTGRSRGFGFVTMSSVEEAEAAA 705
Query: 67 EGMNGQDMDGRNITVNEA--QSRGSGGGGGRGGGGYGGGGGG 106
+ NG ++DGR++ VN +R RGG +G GGG
Sbjct: 704 KQFNGYELDGRSLRVNSGPPPARNESAPRFRGGSSFGSRGGG 579
>TC204085 similar to UP|Q8VX74 (Q8VX74) Glycine-rich RNA-binding protein,
partial (87%)
Length = 628
Score = 238 bits (607), Expect = 1e-63
Identities = 124/169 (73%), Positives = 133/169 (78%), Gaps = 5/169 (2%)
Frame = +3
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
MASADVEYRCFVGGLAWATD+ LEKAFSQYG++++SKIINDRETGRSRGFGFVTFA E
Sbjct: 6 MASADVEYRCFVGGLAWATDNYDLEKAFSQYGDVVESKIINDRETGRSRGFGFVTFASED 185
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRG---GGGYGGGGGGYGGGGRREGGY 117
SMRDAIEGMNGQ++DGRNITVNEAQSRGS GGGG G GGGY GG G GGRREG Y
Sbjct: 186 SMRDAIEGMNGQNLDGRNITVNEAQSRGSRGGGGGGYGSGGGYNRSGGAGGYGGRREGAY 365
Query: 118 NRSSGGGGGYGGGGGGGYGGGRDRGYG--DDGGSRYSRGGGGDGGSWRS 164
NR+ GGGYGG RD YG DGGSRYSRGGG GSWR+
Sbjct: 366 NRN-----------GGGYGGDRDHRYGPYGDGGSRYSRGGG--DGSWRN 473
>TC204105 similar to UP|Q8VX74 (Q8VX74) Glycine-rich RNA-binding protein,
partial (61%)
Length = 313
Score = 179 bits (455), Expect = 4e-46
Identities = 87/104 (83%), Positives = 92/104 (87%), Gaps = 1/104 (0%)
Frame = +1
Query: 7 EYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAI 66
EYRCFVGGLAWATD ALEKAFS YG I++SKIINDRETGRSRGFGFVTFA E SM+DAI
Sbjct: 1 EYRCFVGGLAWATDDHALEKAFSHYGNIVESKIINDRETGRSRGFGFVTFASENSMKDAI 180
Query: 67 EGMNGQDMDGRNITVNEAQSRGSGGG-GGRGGGGYGGGGGGYGG 109
EGMNGQ++DGRNITVNEAQSRGS GG GGR GGY GGGGYGG
Sbjct: 181 EGMNGQNLDGRNITVNEAQSRGSRGGYGGRREGGYNRGGGGYGG 312
>TC204095 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding protein,
partial (80%)
Length = 758
Score = 162 bits (411), Expect = 5e-41
Identities = 87/121 (71%), Positives = 90/121 (73%), Gaps = 5/121 (4%)
Frame = +3
Query: 39 IINDRETGRSRGFGFVTFADEKSMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRG-- 96
IINDRETGRSRGFGFVTFA E SMRDAIEGMNGQ++DGRNITVNEAQSRGS GGGG G
Sbjct: 396 IINDRETGRSRGFGFVTFASEDSMRDAIEGMNGQNLDGRNITVNEAQSRGSRGGGGGGYG 575
Query: 97 -GGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYG--DDGGSRYSR 153
GGGY GG G GGRREG YNR+ GGGYGG RD YG DGGSRYSR
Sbjct: 576 SGGGYNRSGGAGGYGGRREGAYNRN-----------GGGYGGDRDHRYGPYGDGGSRYSR 722
Query: 154 G 154
G
Sbjct: 723 G 725
Score = 71.6 bits (174), Expect = 2e-13
Identities = 31/39 (79%), Positives = 37/39 (94%)
Frame = +3
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKI 39
MASADVEYRCFVGGLAWATD+ LEKAFSQYG++++SK+
Sbjct: 9 MASADVEYRCFVGGLAWATDNYDLEKAFSQYGDVVESKV 125
>TC204100 homologue to UP|Q93WA5 (Q93WA5) Ribosomal protein L32, complete
Length = 1145
Score = 152 bits (385), Expect = 5e-38
Identities = 79/115 (68%), Positives = 84/115 (72%), Gaps = 4/115 (3%)
Frame = -3
Query: 54 VTFADEKSMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYG---GG 110
VTFA E+SM+DAI MNGQ++DGRNITVNEAQSRG GGGG GGGGY GGGGYG GG
Sbjct: 492 VTFASEQSMKDAIGAMNGQNLDGRNITVNEAQSRGGGGGG--GGGGYNRGGGGYGGRSGG 319
Query: 111 GRREGGYNRSSGG-GGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSWRS 164
G GGY GG GGGYGGGGGGGYGGGRDRG YSRGG G G WR+
Sbjct: 318 GGGGGGYRSRDGGYGGGYGGGGGGGYGGGRDRG--------YSRGGDGGDGGWRN 178
>TC204097 homologue to UP|Q9SWA8 (Q9SWA8) Glycine-rich RNA-binding protein,
partial (82%)
Length = 764
Score = 144 bits (363), Expect = 2e-35
Identities = 72/93 (77%), Positives = 78/93 (83%), Gaps = 2/93 (2%)
Frame = +1
Query: 39 IINDRETGRSRGFGFVTFADEKSMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGG 98
IINDRETGRSRGFGFVTFA E+SM+DAI MNGQ++DGRNITVNEAQSRG GGGGG GGG
Sbjct: 328 IINDRETGRSRGFGFVTFASEQSMKDAIGAMNGQNLDGRNITVNEAQSRGGGGGGGGGGG 507
Query: 99 GYGGGGGGYGG--GGRREGGYNRSSGGGGGYGG 129
GY GGGGYGG GG R+ GY+R GG GG GG
Sbjct: 508 GYNRGGGGYGGRSGGGRDRGYSR--GGDGGDGG 600
Score = 73.6 bits (179), Expect = 4e-14
Identities = 32/39 (82%), Positives = 37/39 (94%)
Frame = +3
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKI 39
MASADVEYRCFVGGLAWATD ALE+AFSQYGEI+++K+
Sbjct: 60 MASADVEYRCFVGGLAWATDDHALERAFSQYGEIVETKV 176
>TC215891 similar to GB|AAM78058.1|21928059|AY125548 AT5g61030/maf19_30
{Arabidopsis thaliana;} , partial (41%)
Length = 782
Score = 139 bits (351), Expect = 5e-34
Identities = 74/160 (46%), Positives = 97/160 (60%), Gaps = 6/160 (3%)
Frame = +2
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ F+GG++++TD ++L +AFS+YGE++D++II DRETGRSRGFGF+T+ + AI+
Sbjct: 176 KLFIGGVSYSTDEQSLREAFSKYGEVVDARIIMDRETGRSRGFGFITYTSVEEASSAIQA 355
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGG-GGGGYGGGGRREGG-----YNRSSG 122
++GQD+ GR I VN A R G GGG G G YG GGGGY GG GG Y+R+ G
Sbjct: 356 LDGQDLHGRPIRVNYANERPRGYGGG-GFGSYGAVGGGGYEGGSSYRGGYGGDNYSRNDG 532
Query: 123 GGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSW 162
G GYGGG G GY G DGG W
Sbjct: 533 SGYGYGGGNHESSTGFASNGYD---------GSVVDGGCW 625
>TC217707 similar to UP|Q941H8 (Q941H8) RNA-binding protein precursor,
partial (35%)
Length = 1008
Score = 134 bits (336), Expect = 3e-32
Identities = 70/155 (45%), Positives = 92/155 (59%)
Frame = +1
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ FVGG++++TD +L ++ ++YGE+ID K+I DRETGRSRGFGF+TFA + AI+G
Sbjct: 166 KLFVGGISYSTDDMSLRESPARYGEVIDVKVIMDRETGRSRGFGFITFATSEDASYAIQG 345
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYG 128
M+GQD+ GR I VN A R G GG GG GG GY GG GGYN G GY
Sbjct: 346 MDGQDLHGRRIRVNYATERSRPGFGGDGGYRGSGGSDGYNRGGNYGGGYN---SGSDGYN 516
Query: 129 GGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGSWR 163
GG G G DG + S G + G+++
Sbjct: 517 RGGNYGSGNYNVTSSYSDGNAETSYTSGANAGNYQ 621
>AI460427 similar to GP|5726567|gb|A glycine-rich RNA-binding protein
{Glycine max}, partial (60%)
Length = 292
Score = 130 bits (327), Expect = 3e-31
Identities = 68/96 (70%), Positives = 76/96 (78%)
Frame = +2
Query: 4 ADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMR 63
ADVEY FV GLAWATD AL +AFSQYGEI+++KII+ RETGRSRGFGF TFA E M
Sbjct: 5 ADVEYLRFVCGLAWATDHHAL*RAFSQYGEIVETKIISYRETGRSRGFGF*TFASEHPMT 184
Query: 64 DAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGG 99
DAI MNGQ+++ RNITVNEAQSRG GGG G GG
Sbjct: 185 DAI*AMNGQNLN*RNITVNEAQSRGGCSGGGSGLGG 292
>TC214360 similar to UP|Q9MBF3 (Q9MBF3) Glycine-rich RNA-binding protein,
partial (44%)
Length = 630
Score = 124 bits (310), Expect = 3e-29
Identities = 63/77 (81%), Positives = 65/77 (83%), Gaps = 8/77 (10%)
Frame = +2
Query: 96 GGGGYGGGGGGYGGGG-RREGGYNRSSGGGGGYGGGGGG-----GYGGG-RDRGYGDDGG 148
GGGGY GGGGYGGGG RREGGYNR+ GGGGGYGGGGGG GYGGG RDRGYG DGG
Sbjct: 2 GGGGYSRGGGGYGGGGGRREGGYNRN-GGGGGYGGGGGGYGGGGGYGGGGRDRGYGGDGG 178
Query: 149 SRYSRGGGG-DGGSWRS 164
SRYSRGGGG DGGSWR+
Sbjct: 179 SRYSRGGGGSDGGSWRN 229
>TC210743 similar to UP|Q42412 (Q42412) RNA-binding protein RZ-1, partial
(53%)
Length = 452
Score = 117 bits (294), Expect = 2e-27
Identities = 60/133 (45%), Positives = 83/133 (62%)
Frame = +3
Query: 7 EYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAI 66
EYRCF+GGLAW+T L+ F ++G++I++K++ D+ +GRSRGFGFVTF D+K+M +AI
Sbjct: 78 EYRCFIGGLAWSTSDRKLKDTFEKFGKLIEAKVVVDKFSGRSRGFGFVTFDDKKAMDEAI 257
Query: 67 EGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGG 126
+ MNG D+DGR ITV+ AQ + G G Y GR +R GGGGG
Sbjct: 258 DAMNGMDLDGRTITVDRAQPQ---------QGSTRDDGDRYRERGRDR---DRDHGGGGG 401
Query: 127 YGGGGGGGYGGGR 139
G GG + G+
Sbjct: 402 RGSNGGECFKCGK 440
>TC225319 similar to UP|P93486 (P93486) Glycine-rich RNA-binding protein
PsGRBP, partial (88%)
Length = 1167
Score = 112 bits (279), Expect = 1e-25
Identities = 50/102 (49%), Positives = 76/102 (74%)
Frame = +3
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ F+GGL++ D ++L+ AFS +G+++D+K+I DR++GRSRGFGFV F++++S A+
Sbjct: 369 KLFIGGLSYGVDDQSLKDAFSGFGDVVDAKVITDRDSGRSRGFGFVNFSNDESASSALSA 548
Query: 69 MNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGGGGGYGGG 110
M+G+D+DGR+I V+ A R SG G GGGG G GG+GGG
Sbjct: 549 MDGKDLDGRSIRVSYANDRPSGPQSGGGGGG-GYRSGGFGGG 671
>TC214257 weakly similar to UP|Q8L685 (Q8L685) Pherophorin-dz1 protein
precursor, partial (7%)
Length = 607
Score = 110 bits (274), Expect = 4e-25
Identities = 62/86 (72%), Positives = 64/86 (74%), Gaps = 8/86 (9%)
Frame = +2
Query: 87 RGSGG--GGGR---GGGGYG--GGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGR 139
RG GG GGGR GGGGY GGGGGYGGGG GY GGGGG+GGGGG G GGGR
Sbjct: 5 RGGGGYGGGGRREGGGGGYNRNGGGGGYGGGG----GY----GGGGGHGGGGGYG-GGGR 157
Query: 140 DRGYGDDGGSRYSRGGG-GDGGSWRS 164
DRGYG DGGSRYSRGGG DGGSWR+
Sbjct: 158 DRGYGGDGGSRYSRGGGASDGGSWRN 235
>BM086366
Length = 427
Score = 107 bits (267), Expect = 3e-24
Identities = 54/114 (47%), Positives = 76/114 (66%), Gaps = 1/114 (0%)
Frame = +2
Query: 3 SADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSM 62
+A E R FVGGL+W LE AF++YG+I++ +I+ +R+TGR RGFGF+TFAD + M
Sbjct: 74 AAKEENRIFVGGLSWEVTERQLEHAFARYGKILECQIMMERDTGRPRGFGFITFADRRGM 253
Query: 63 RDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRG-GGGYGGGGGGYGGGGRREG 115
DAI+ M+G+++ R I+VN+AQ + G +G GGY GG G G G R G
Sbjct: 254 EDAIKEMHGREIGDRIISVNKAQPKMGGDDVDQGYRGGYSSGGRGSYGAGDRVG 415
>TC225318 similar to UP|P93486 (P93486) Glycine-rich RNA-binding protein
PsGRBP, partial (86%)
Length = 749
Score = 104 bits (259), Expect = 2e-23
Identities = 47/106 (44%), Positives = 74/106 (69%), Gaps = 4/106 (3%)
Frame = +2
Query: 9 RCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEG 68
+ F+GGL++ D ++L+ AFS +G+++D+K+I DR++GRSRGFGFV F++++S A+
Sbjct: 173 KLFIGGLSYGVDDQSLKDAFSGFGDVVDAKVITDRDSGRSRGFGFVNFSNDESASSALSA 352
Query: 69 MNGQDMDGRNITVNEAQSRGS----GGGGGRGGGGYGGGGGGYGGG 110
M+G+D++GR+I V+ A + S GGGGG GG Y G GG
Sbjct: 353 MDGKDLNGRSIRVSYANDKPSAPRPGGGGGYRGGDYDGDFASRSGG 490
>TC206525 weakly similar to UP|Q41042 (Q41042) Pisum sativum L. (clone
na-481-5), partial (20%)
Length = 752
Score = 102 bits (254), Expect = 8e-23
Identities = 61/140 (43%), Positives = 81/140 (57%), Gaps = 3/140 (2%)
Frame = +2
Query: 23 ALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEKSMRDAIEGMNGQDMDGRNITVN 82
+L++ F G+I I D E+G +GF +V F D SM A+E ++ ++ G +TV+
Sbjct: 74 SLQEHFGSCGDITRVSIPKDYESGAVKGFAYVDFGDADSMGKALE-LHETELGGYTLTVD 250
Query: 83 EAQSRGSGGGGGRGGGGYGGGGGGYG--GGGRREGGYNRSSGGGGGYGGGGGGGYGGGRD 140
EA+ R + G GGRG GG GGGG +G GGG GG + GGGG +GG GGGG GG
Sbjct: 251 EAKPRDNQGSGGRGAGGRSGGGGRFGARGGG---GGRFGARGGGGRFGGSGGGGRFGG-- 415
Query: 141 RGYGDDGGSRY-SRGGGGDG 159
GG R+ RGGGG G
Sbjct: 416 -----SGGGRFGGRGGGGRG 460
>AW761335 similar to GP|5726567|gb|A glycine-rich RNA-binding protein
{Glycine max}, partial (38%)
Length = 313
Score = 101 bits (252), Expect = 1e-22
Identities = 56/103 (54%), Positives = 68/103 (65%)
Frame = -1
Query: 1 MASADVEYRCFVGGLAWATDSEALEKAFSQYGEIIDSKIINDRETGRSRGFGFVTFADEK 60
M S+DVEYR V GL + +A+++AFSQYG+I +SKI RS+GFGFVTFA E+
Sbjct: 313 MPSSDVEYRRMVDGLGRVIEHQAVQRAFSQYGDICESKIHQQS*DCRSKGFGFVTFASEQ 134
Query: 61 SMRDAIEGMNGQDMDGRNITVNEAQSRGSGGGGGRGGGGYGGG 103
SM+ AI MNGQ + NITV AQSR G G G G YGGG
Sbjct: 133 SMKAAIAVMNGQTLSRCNITVTTAQSRSEGVGVGVVGVVYGGG 5
>BU765146 similar to GP|21322711|e pherophorin-dz1 protein {Volvox carteri f.
nagariensis}, partial (25%)
Length = 407
Score = 97.1 bits (240), Expect = 3e-21
Identities = 48/80 (60%), Positives = 50/80 (62%), Gaps = 3/80 (3%)
Frame = +3
Query: 87 RGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGY---GGGRDRGY 143
RG GGGGG GGGG GGGGGG+ GG GG GGGGG GGGGGGG GGGR G
Sbjct: 156 RGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGGGGGGGGGXGGGGGGRGEGG 335
Query: 144 GDDGGSRYSRGGGGDGGSWR 163
G GG GGGG+GG R
Sbjct: 336 GGGGGGGGGGGGGGEGGGRR 395
Score = 95.9 bits (237), Expect = 8e-21
Identities = 49/74 (66%), Positives = 49/74 (66%)
Frame = +3
Query: 87 RGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDD 146
RG GGGG GGGG GGGGGG GGGG EGG GGGGG GGGGGGG GGG RG G
Sbjct: 9 RGLPGGGGGGGGGGGGGGGGGGGGGGGEGG-----GGGGGGGGGGGGGGGGGGGRGGGGG 173
Query: 147 GGSRYSRGGGGDGG 160
GG GGGG GG
Sbjct: 174 GGGGGGGGGGGGGG 215
Score = 95.1 bits (235), Expect = 1e-20
Identities = 50/86 (58%), Positives = 51/86 (59%), Gaps = 9/86 (10%)
Frame = +2
Query: 84 AQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGY 143
A+ G GGGGG GGGG GGGGGG GGGGR GG GGGGG GG GGGG GGG G
Sbjct: 17 ARGGGGGGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGG 196
Query: 144 GDDGG---------SRYSRGGGGDGG 160
G GG R RGGGG GG
Sbjct: 197 GGGGGGFFRGERXXGRGGRGGGGGGG 274
Score = 94.0 bits (232), Expect = 3e-20
Identities = 48/85 (56%), Positives = 49/85 (57%), Gaps = 12/85 (14%)
Frame = +3
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGY------------GGGGGGGY 135
G GGGGG GGGG GGGGGG GGGG GG GGGGG+ GGGGGGG
Sbjct: 99 GGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGREXXGGGGEGGGGGGGG 278
Query: 136 GGGRDRGYGDDGGSRYSRGGGGDGG 160
GGG G G GG R GGGG GG
Sbjct: 279 GGGGGGGXGGGGGGRGEGGGGGGGG 353
Score = 93.6 bits (231), Expect = 4e-20
Identities = 48/76 (63%), Positives = 50/76 (65%), Gaps = 3/76 (3%)
Frame = +3
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYG--GGRD-RGYG 144
G GGGGG GGGG GGGGGG GGGG GG GGGGG GGGGGGG G GGR+ G G
Sbjct: 69 GGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGREXXGGG 248
Query: 145 DDGGSRYSRGGGGDGG 160
+GG GGGG GG
Sbjct: 249 GEGGGGGGGGGGGGGG 296
Score = 92.8 bits (229), Expect = 7e-20
Identities = 46/73 (63%), Positives = 48/73 (65%)
Frame = +2
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDG 147
G GGGGGRGGGG GGGGGG GGGG +GG GGGGG GGGGGG + G R G G G
Sbjct: 80 GGGGGGGRGGGG-GGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRG 256
Query: 148 GSRYSRGGGGDGG 160
G GGGG GG
Sbjct: 257 GGGGGGGGGGGGG 295
Score = 92.4 bits (228), Expect = 8e-20
Identities = 47/83 (56%), Positives = 49/83 (58%), Gaps = 10/83 (12%)
Frame = +2
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGG----------RREGGYNRSSGGGGGYGGGGGGGYGG 137
G GGGGG+GGGG GGGGGG GGGG GG GGGGG GGGGGGG GG
Sbjct: 134 GGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGG 313
Query: 138 GRDRGYGDDGGSRYSRGGGGDGG 160
G + G G GG GGGG GG
Sbjct: 314 GGEGGGGGGGGGGGGGGGGGRGG 382
Score = 92.4 bits (228), Expect = 8e-20
Identities = 46/73 (63%), Positives = 46/73 (63%)
Frame = +3
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDG 147
G GGGGG G GG GGGGGG GGGG GG R GGGGG GGGGGGG GGG G G
Sbjct: 63 GGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGGGFLGGREXXG 242
Query: 148 GSRYSRGGGGDGG 160
G GGGG GG
Sbjct: 243 GGGEGGGGGGGGG 281
Score = 92.0 bits (227), Expect = 1e-19
Identities = 51/86 (59%), Positives = 51/86 (59%), Gaps = 10/86 (11%)
Frame = +1
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGG----------RREGGYNRSSGGGGGYGGGGGGGYGG 137
G GGGGG GGGG GGGGGG GGGG REGG R GGGGG GGGGGGG GG
Sbjct: 133 GGGGGGGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGG-ERGGGGGGGGGGGGGGGXGG 309
Query: 138 GRDRGYGDDGGSRYSRGGGGDGGSWR 163
G G G GG GGGG GG R
Sbjct: 310 G--GGGGGRGGGGGGGGGGGGGGEGR 381
Score = 91.3 bits (225), Expect = 2e-19
Identities = 48/76 (63%), Positives = 50/76 (65%), Gaps = 3/76 (3%)
Frame = +1
Query: 88 GSGGGGGRGG--GGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGY-GGGRDRGYG 144
G GGGGGRGG GG GGGGGG GGGG EGG GGGGG GGGGGG + GG R R G
Sbjct: 70 GGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGG 249
Query: 145 DDGGSRYSRGGGGDGG 160
+ GG GGGG GG
Sbjct: 250 ERGGGGGGGGGGGGGG 297
Score = 90.9 bits (224), Expect = 2e-19
Identities = 47/83 (56%), Positives = 49/83 (58%), Gaps = 7/83 (8%)
Frame = +2
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGY-------NRSSGGGGGYGGGGGGGYGGGRD 140
G GGGGG G GG GGGGGG GGGG GG+ R GGGG GGGGGGG GGG
Sbjct: 128 GGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGG 307
Query: 141 RGYGDDGGSRYSRGGGGDGGSWR 163
G G+ GG GGGG GG R
Sbjct: 308 GGGGEGGGGGGGGGGGGGGGGGR 376
Score = 88.6 bits (218), Expect = 1e-18
Identities = 47/84 (55%), Positives = 47/84 (55%), Gaps = 10/84 (11%)
Frame = +2
Query: 87 RGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGG----------GGYGGGGGGGYG 136
RG GGGGG GGGG GGGG G GGGG GG GGG GG GGGGGGG G
Sbjct: 101 RGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGG 280
Query: 137 GGRDRGYGDDGGSRYSRGGGGDGG 160
GG G G GG GGGG GG
Sbjct: 281 GGGGGGGGGGGGGEGGGGGGGGGG 352
Score = 87.8 bits (216), Expect = 2e-18
Identities = 48/85 (56%), Positives = 49/85 (57%), Gaps = 12/85 (14%)
Frame = +1
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGG--GGGGYGGGRDRGYGD 145
G GGGGG GGGG GGGGGG GGG EGG GGGGG GGG GGGG GGG G G
Sbjct: 22 GGGGGGGGGGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGG 201
Query: 146 DGGSRY----------SRGGGGDGG 160
GG + RGGGG GG
Sbjct: 202 GGGGVF*GGERXREGGERGGGGGGG 276
Score = 86.7 bits (213), Expect = 5e-18
Identities = 45/73 (61%), Positives = 45/73 (61%)
Frame = +3
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDG 147
G G G GGGG GGGGGG GGGG GG GGGGG GGGGGGG GGGR G G G
Sbjct: 3 GGRGLPGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGRGGGGGGGG 182
Query: 148 GSRYSRGGGGDGG 160
G GGGG GG
Sbjct: 183 GG----GGGGGGG 209
Score = 86.3 bits (212), Expect = 6e-18
Identities = 47/93 (50%), Positives = 48/93 (51%), Gaps = 16/93 (17%)
Frame = +1
Query: 83 EAQSRGSGGGGGRGGGGYGGGGGGYGGGG----------------RREGGYNRSSGGGGG 126
E G GGGGG GGGG GGGGG GGGG REGG GGGGG
Sbjct: 100 EGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGGERGGGGGGGG 279
Query: 127 YGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDG 159
GGGGGG GGG G G GG GGGG+G
Sbjct: 280 GGGGGGGXGGGGGGGGRGGGGGGGGGGGGGGEG 378
Score = 85.1 bits (209), Expect = 1e-17
Identities = 47/90 (52%), Positives = 47/90 (52%), Gaps = 16/90 (17%)
Frame = +1
Query: 87 RGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGG----------------GYGGG 130
RG GGGG GGGG GGGGGG GGG GG GGGG G GGG
Sbjct: 91 RGGEGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGVF*GGERXREGGERGGGGG 270
Query: 131 GGGGYGGGRDRGYGDDGGSRYSRGGGGDGG 160
GGGG GGG G G GG R GGGG GG
Sbjct: 271 GGGGGGGGGGXGGGGGGGGRGGGGGGGGGG 360
Score = 83.2 bits (204), Expect = 5e-17
Identities = 42/58 (72%), Positives = 43/58 (73%)
Frame = +3
Query: 88 GSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGD 145
G GGGGG GGGG GGGG G GGGGR EGG GGGGG GGGGGGG GGGR RG G+
Sbjct: 249 GEGGGGGGGGGGGGGGGXGGGGGGRGEGG----GGGGGGGGGGGGGGEGGGR-RGGGE 407
Score = 82.0 bits (201), Expect = 1e-16
Identities = 42/69 (60%), Positives = 44/69 (62%)
Frame = +2
Query: 95 RGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRG 154
RGGGG GGGGGG GGGG GG G GGG GGGGGGG GGG +G G GG G
Sbjct: 20 RGGGG-GGGGGGGGGGGGGGGGGGGGGGRGGGGGGGGGGGGGGGGGKGGGGGGGGGGGGG 196
Query: 155 GGGDGGSWR 163
GGG GG +R
Sbjct: 197 GGGGGGFFR 223
Score = 81.6 bits (200), Expect = 1e-16
Identities = 42/71 (59%), Positives = 42/71 (59%)
Frame = +1
Query: 90 GGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGS 149
GGG GGG GGGGGG GGGG GG G GGG GGGGGGG GGG G G GG
Sbjct: 1 GGGEVCPGGGGGGGGGGGGGGGGGGGGGGGRGGEGGGGGGGGGGGGGGGGGEGGGGGGGG 180
Query: 150 RYSRGGGGDGG 160
GGGG GG
Sbjct: 181 GGGGGGGGGGG 213
Score = 80.5 bits (197), Expect = 3e-16
Identities = 44/84 (52%), Positives = 45/84 (53%), Gaps = 14/84 (16%)
Frame = +2
Query: 88 GSGGGGGRGGGGY--------------GGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGG 133
G GGGGG GGGG+ GGGGGG GGGG GG GGGGG GGGGGG
Sbjct: 179 GGGGGGGGGGGGFFRGERXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGG 358
Query: 134 GYGGGRDRGYGDDGGSRYSRGGGG 157
G GGGR G R GGGG
Sbjct: 359 GGGGGR--------GGRREEGGGG 406
Score = 77.4 bits (189), Expect = 3e-15
Identities = 37/58 (63%), Positives = 38/58 (64%)
Frame = +2
Query: 87 RGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYG 144
R G GG GGGG GGGGGG GGGG GG GGGGG GGGGGGG GG R+ G G
Sbjct: 230 RXXGRGGRGGGGGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGRGGRREEGGG 403
Score = 76.6 bits (187), Expect = 5e-15
Identities = 39/65 (60%), Positives = 40/65 (61%)
Frame = +1
Query: 83 EAQSRGSGGGGGRGGGGYGGGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRG 142
E RG GGGGG GGGG GG GGG GGGGR GGGG GGGGGGG G GR+ G
Sbjct: 241 EGGERGGGGGGGGGGGGGGGXGGGGGGGGR----------GGGGGGGGGGGGGGEGREEG 390
Query: 143 YGDDG 147
G G
Sbjct: 391 GGGGG 405
Score = 75.5 bits (184), Expect = 1e-14
Identities = 42/78 (53%), Positives = 43/78 (54%), Gaps = 5/78 (6%)
Frame = +1
Query: 88 GSGGGGGRGGGGYGGG-----GGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRG 142
G GGGGG GGG + GG GG GGGG GG GGGG GGGGGGG GGG G
Sbjct: 181 GGGGGGGGGGGVF*GGERXREGGERGGGG---GGGGGGGGGGGXGGGGGGGGRGGGGGGG 351
Query: 143 YGDDGGSRYSRGGGGDGG 160
G GG GGG GG
Sbjct: 352 GGGGGGGEGREEGGGGGG 405
Score = 59.7 bits (143), Expect = 6e-10
Identities = 33/62 (53%), Positives = 33/62 (53%)
Frame = +3
Query: 102 GGGGGYGGGGRREGGYNRSSGGGGGYGGGGGGGYGGGRDRGYGDDGGSRYSRGGGGDGGS 161
GG G GGGG GGGGG GGGGGGG GGG G G GG GGGG GG
Sbjct: 3 GGRGLPGGGG----------GGGGGGGGGGGGGGGGGGGEGGGGGGGGGGGGGGGGGGGG 152
Query: 162 WR 163
R
Sbjct: 153 GR 158
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.312 0.144 0.444
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,174,901
Number of Sequences: 63676
Number of extensions: 149655
Number of successful extensions: 30761
Number of sequences better than 10.0: 1858
Number of HSP's better than 10.0 without gapping: 4115
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12888
length of query: 164
length of database: 12,639,632
effective HSP length: 90
effective length of query: 74
effective length of database: 6,908,792
effective search space: 511250608
effective search space used: 511250608
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 55 (25.8 bits)
Medicago: description of AC134242.10


