

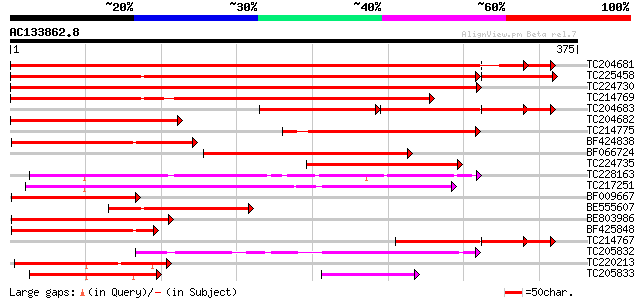
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.8 + phase: 0
(375 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC204681 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein... 599 e-172
TC225458 UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, complete 590 e-169
TC224730 similar to UP|O24074 (O24074) DnaJ-like protein MsJ1, p... 508 e-144
TC214769 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein... 483 e-137
TC204683 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein... 154 3e-71
TC204682 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein... 224 5e-59
TC214775 UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, partia... 223 1e-58
BF424838 197 6e-51
BF066724 195 2e-50
TC224735 similar to UP|O24074 (O24074) DnaJ-like protein MsJ1, p... 168 3e-42
TC228163 similar to UP|Q8LEU4 (Q8LEU4) DnaJ protein-like, partia... 164 8e-41
TC217251 weakly similar to UP|DJB1_HUMAN (P25685) DnaJ homolog s... 163 1e-40
BF009667 similar to GP|5802244|gb|A seed maturation protein PM37... 143 1e-34
BE555607 similar to GP|5802244|gb|A seed maturation protein PM37... 122 3e-28
BE803986 similar to PIR|T43929|T439 DnaJ protein homolog [import... 120 8e-28
BF425848 similar to GP|5802244|gb|A seed maturation protein PM37... 120 1e-27
TC214767 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein... 108 3e-24
TC205832 similar to GB|AAM78044.1|21928031|AY125534 At3g62600/F2... 105 3e-23
TC220213 similar to UP|Q7XYV0 (Q7XYV0) P58IPK, partial (38%) 102 3e-22
TC205833 similar to GB|AAM78044.1|21928031|AY125534 At3g62600/F2... 96 3e-20
>TC204681 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
complete
Length = 1542
Score = 599 bits (1545), Expect = e-172
Identities = 293/343 (85%), Positives = 309/343 (89%)
Frame = +3
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSD+T+YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 111 MFGRAPKKSDSTRYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 290
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YD YGEDALKEGMGGGGGGHDPFDIFSSFFGG F GGSSRGRRQRRGED
Sbjct: 291 LSDPEKREIYDTYGEDALKEGMGGGGGGHDPFDIFSSFFGGSPFGSGGSSRGRRQRRGED 470
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQG+GMK+S+RHLG
Sbjct: 471 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGTGMKVSIRHLG 650
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQHPCNECKGTGETI+D+DRC QCKGEKVVQ+KKVLEV VEKGMQNGQKITFPG
Sbjct: 651 PSMIQQMQHPCNECKGTGETINDRDRCQQCKGEKVVQEKKVLEVVVEKGMQNGQKITFPG 830
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDTVTGDIVFVLQQKEHPKFKRK +DLFVEHTLSLTEALCGFQF L HLD RQLL
Sbjct: 831 EADEAPDTVTGDIVFVLQQKEHPKFKRKADDLFVEHTLSLTEALCGFQFVLAHLDGRQLL 1010
Query: 301 IKSNPGEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
IKSNPGEVVKP + K +EG +RH ++
Sbjct: 1011IKSNPGEVVKP------------DSYKAINDEGMPNYQRHFLK 1103
Score = 60.8 bits (146), Expect = 9e-10
Identities = 30/49 (61%), Positives = 37/49 (75%)
Frame = +1
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNE 361
WMSVRR H MM T +KR G+KLS+RHM RM C+VVL+E + LSSN+
Sbjct: 1222 WMSVRRPHSMM*TWRKR*GGGNKLSRRHMRRMRICMVVLRECSALSSNQ 1368
>TC225458 UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, complete
Length = 1797
Score = 590 bits (1522), Expect = e-169
Identities = 288/312 (92%), Positives = 300/312 (95%), Gaps = 1/312 (0%)
Frame = +2
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNT+YYEILGVSKNAS DDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 116 MFGRAPKKSDNTRYYEILGVSKNASQDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 295
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGG-FPGGGSSRGRRQRRGE 119
LSDPEKRE+YDQYGEDALKEGMGGGGG HDPFDIFSSFFGGG F GGSSRGRRQRRGE
Sbjct: 296 LSDPEKREIYDQYGEDALKEGMGGGGG-HDPFDIFSSFFGGGSPFGSGGSSRGRRQRRGE 472
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVHPLKVSLEDLYLGTSKKLSLSRNV+CSKC+GKGSKSGASM CAGCQG+GMK+S+RHL
Sbjct: 473 DVVHPLKVSLEDLYLGTSKKLSLSRNVICSKCSGKGSKSGASMKCAGCQGTGMKVSIRHL 652
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G +MIQQMQH CNECKGTGETI+D+DRCPQCKGEKVVQ+KKVLEV VEKGMQNGQKITFP
Sbjct: 653 GPSMIQQMQHACNECKGTGETINDRDRCPQCKGEKVVQEKKVLEVIVEKGMQNGQKITFP 832
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
GEADEAPDT+TGDIVFVLQQKEHPKFKRK EDLFVEHTLSLTEALCGFQF LTHLDSRQL
Sbjct: 833 GEADEAPDTITGDIVFVLQQKEHPKFKRKAEDLFVEHTLSLTEALCGFQFVLTHLDSRQL 1012
Query: 300 LIKSNPGEVVKP 311
LIKSNPGEVVKP
Sbjct: 1013LIKSNPGEVVKP 1048
Score = 57.0 bits (136), Expect = 1e-08
Identities = 27/50 (54%), Positives = 35/50 (70%)
Frame = +3
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNEF 362
WM+VR+ H MMST ++R S +RHMMRMM CLVV + Y+ SSN+F
Sbjct: 1227 WMNVRKLHSMMSTWRRRLGGSSNKLRRHMMRMMTCLVVHRGYSAPSSNDF 1376
>TC224730 similar to UP|O24074 (O24074) DnaJ-like protein MsJ1, partial (96%)
Length = 1913
Score = 508 bits (1308), Expect = e-144
Identities = 236/313 (75%), Positives = 276/313 (87%), Gaps = 1/313 (0%)
Frame = +1
Query: 1 MFGRA-PKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYE 59
MFGR P++SDN+KYY+ILG+SKNAS D++KKAY+KAA+KNHPDKGGDPEKFKEL QAYE
Sbjct: 358 MFGRGGPRRSDNSKYYDILGISKNASEDEIKKAYRKAAMKNHPDKGGDPEKFKELGQAYE 537
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGE 119
VLSDPEK+E+YDQYGEDALKEGMGGGG H+PFDIF SFFGG F GGGSSRGRRQ+ GE
Sbjct: 538 VLSDPEKKELYDQYGEDALKEGMGGGGSFHNPFDIFESFFGGASFGGGGSSRGRRQKHGE 717
Query: 120 DVVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHL 179
DVVH LKVSLED+Y GT+KKLSLSRN+LC KC GKGSKSG + C GC+G+GMKI+ R +
Sbjct: 718 DVVHSLKVSLEDVYNGTTKKLSLSRNILCPKCKGKGSKSGTAGRCFGCKGTGMKITRRQI 897
Query: 180 GANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFP 239
G MIQQMQH C +C+G+GE I+++D+CP CKG KV Q+KKVLEVHVEKGMQ GQKI F
Sbjct: 898 GLGMIQQMQHVCPDCRGSGEVINERDKCPLCKGNKVSQEKKVLEVHVEKGMQQGQKIVFE 1077
Query: 240 GEADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
G+ADEAPDT+TGDIVFVLQ K+HPKF+R+ +DL+++H LSLTEALCGFQFA+ HLD RQL
Sbjct: 1078GQADEAPDTITGDIVFVLQVKDHPKFRREQDDLYIDHNLSLTEALCGFQFAVKHLDGRQL 1257
Query: 300 LIKSNPGEVVKPG 312
LIKSNPGEV+KPG
Sbjct: 1258LIKSNPGEVIKPG 1296
Score = 40.8 bits (94), Expect = 0.001
Identities = 29/67 (43%), Positives = 39/67 (57%), Gaps = 5/67 (7%)
Frame = +2
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLS-KRHMMRMMKCLVVLKEYNVLSSNEF----GDILH 367
WM VRR MMSTL++R +E S + RHMM+MM +V E NVL+S+ G I+
Sbjct: 1478 WMIVRRPLCMMSTLRRR*DESSNSNIARHMMKMMMSHLV-NECNVLNSSAIYGSRGSIIS 1654
Query: 368 GCVLALV 374
L+ V
Sbjct: 1655 AWFLSFV 1675
>TC214769 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (64%)
Length = 943
Score = 483 bits (1244), Expect = e-137
Identities = 236/281 (83%), Positives = 253/281 (89%)
Frame = +2
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSDNT+YYEILGVSKNAS DDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 122 MFGRAPKKSDNTRYYEILGVSKNASQDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 301
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGED 120
LSDPEKRE+YDQYGEDALKEGMGGGGG HDPFDIFSSFFGGG SSRGRRQRRGED
Sbjct: 302 LSDPEKREIYDQYGEDALKEGMGGGGG-HDPFDIFSSFFGGG------SSRGRRQRRGED 460
Query: 121 VVHPLKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLG 180
VVHPLKVSLEDLYLGTSKKLSLSRNV+CSKC GKGSKSGASM CAGCQG+GMK+S++HLG
Sbjct: 461 VVHPLKVSLEDLYLGTSKKLSLSRNVICSKCTGKGSKSGASMKCAGCQGTGMKVSIKHLG 640
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
+MIQQMQH CNECKGTGETI+D+DRCPQCKGEKVVQ+KKVLEV V GM NG KITFP
Sbjct: 641 PSMIQQMQHACNECKGTGETINDRDRCPQCKGEKVVQEKKVLEVIVANGMHNGHKITFPD 820
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLT 281
EAD+ P T+TGDIV+VLQQKEH KF R+ +DL +H LT
Sbjct: 821 EADDTPYTITGDIVYVLQQKEHRKFTREADDLCAQHISVLT 943
>TC204683 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (60%)
Length = 1229
Score = 154 bits (389), Expect(2) = 3e-71
Identities = 70/80 (87%), Positives = 77/80 (95%)
Frame = +1
Query: 166 GCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVH 225
GCQG+GMK+S+RHLG +MIQQMQHPCNECKGTGETI+D+DRC QCKGEKVVQ+KKVLEV
Sbjct: 1 GCQGTGMKVSIRHLGPSMIQQMQHPCNECKGTGETINDRDRCQQCKGEKVVQEKKVLEVV 180
Query: 226 VEKGMQNGQKITFPGEADEA 245
VEKGMQNGQKITFPGEADEA
Sbjct: 181 VEKGMQNGQKITFPGEADEA 240
Score = 132 bits (333), Expect(2) = 3e-71
Identities = 68/98 (69%), Positives = 74/98 (75%)
Frame = +3
Query: 246 PDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNP 305
PDTVTGDIVFVLQQKEHPKFKRK +DLFVEHTLSLTEALCGFQF LTHLDSRQLLIKSNP
Sbjct: 315 PDTVTGDIVFVLQQKEHPKFKRKADDLFVEHTLSLTEALCGFQFVLTHLDSRQLLIKSNP 494
Query: 306 GEVVKPGWMSVRRQHFMMSTLKKRREEGSKLSKRHMMR 343
GEVVKP + K +EG +RH ++
Sbjct: 495 GEVVKP------------ESFKAINDEGMPNYQRHFLK 572
Score = 66.2 bits (160), Expect = 2e-11
Identities = 32/49 (65%), Positives = 39/49 (79%)
Frame = +1
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNE 361
WMSVRR H MM T +KR+ G+KLS+RHMMRM C+VVL+E + LSSNE
Sbjct: 691 WMSVRRPHSMM*TWRKRQGGGNKLSRRHMMRMRICMVVLRECSALSSNE 837
>TC204682 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (27%)
Length = 447
Score = 224 bits (571), Expect = 5e-59
Identities = 106/114 (92%), Positives = 109/114 (94%)
Frame = +2
Query: 1 MFGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 60
MFGRAPKKSD+T+YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV
Sbjct: 104 MFGRAPKKSDSTRYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEV 283
Query: 61 LSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRR 114
LSDPEKRE+YD YGEDALKEGMGGGGGGHDPFDIFSSFFGG F GGSSRGRR
Sbjct: 284 LSDPEKREIYDTYGEDALKEGMGGGGGGHDPFDIFSSFFGGSPFGSGGSSRGRR 445
>TC214775 UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37, partial (42%)
Length = 530
Score = 223 bits (568), Expect = 1e-58
Identities = 112/131 (85%), Positives = 117/131 (88%)
Frame = +3
Query: 181 ANMIQQMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPG 240
A +Q MQ P GETI+D+DRCPQCKGEKVVQ+KKVLEV VEKGMQNGQKITFPG
Sbjct: 6 AACLQ*MQSP-------GETINDRDRCPQCKGEKVVQEKKVLEVIVEKGMQNGQKITFPG 164
Query: 241 EADEAPDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLL 300
EADEAPDT+TGDIVFVLQQKEHPKFKRK EDLFVEHTLSLTEALCGFQF LTHLDSRQLL
Sbjct: 165 EADEAPDTITGDIVFVLQQKEHPKFKRKAEDLFVEHTLSLTEALCGFQFVLTHLDSRQLL 344
Query: 301 IKSNPGEVVKP 311
IKSNPGEVVKP
Sbjct: 345 IKSNPGEVVKP 377
Score = 29.3 bits (64), Expect = 3.0
Identities = 12/51 (23%), Positives = 29/51 (56%)
Frame = +1
Query: 186 QMQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKI 236
QMQH CNEC + + + + +++ ++KK L++ ++ + G+++
Sbjct: 1 QMQHACNECNHLEKPSTTEIAAHSAREKRLCRRKKSLKLLWKRECRTGRRL 153
>BF424838
Length = 370
Score = 197 bits (501), Expect = 6e-51
Identities = 97/124 (78%), Positives = 105/124 (84%), Gaps = 1/124 (0%)
Frame = +2
Query: 2 FGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEVL 61
FGRAP KSDNT+YYEILGVS NAS DDLKKAY+ AA+ NHPDKGGDPEKF+ELAQAYE L
Sbjct: 2 FGRAPTKSDNTRYYEILGVSNNASQDDLKKAYQIAALNNHPDKGGDPEKFQELAQAYEAL 181
Query: 62 SDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGG-FPGGGSSRGRRQRRGED 120
SDP KRE+YDQ+G+DAL EGM GGG GHDPFDIFSSFFGGG F GS RG R RGED
Sbjct: 182 SDPVKREIYDQHGDDALTEGM-GGGRGHDPFDIFSSFFGGGSPFGSRGSRRGPRHMRGED 358
Query: 121 VVHP 124
V+HP
Sbjct: 359 VLHP 370
>BF066724
Length = 415
Score = 195 bits (496), Expect = 2e-50
Identities = 94/138 (68%), Positives = 111/138 (80%)
Frame = +2
Query: 129 LEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQ 188
LEDLYLGTS KLSLSRNV+CSKC+GKGSKSGASM CAGCQG+G+K+S+RHLG +MIQQMQ
Sbjct: 2 LEDLYLGTSTKLSLSRNVICSKCSGKGSKSGASMKCAGCQGTGLKVSIRHLGPSMIQQMQ 181
Query: 189 HPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDT 248
CNEC+GTGETI+D DRCPQCKGE VVQ+KKVLEV V+KGM G IT E+DE P+
Sbjct: 182 LACNECEGTGETITD*DRCPQCKGENVVQEKKVLEVIVKKGMSTGATITCACESDETPEH 361
Query: 249 VTGDIVFVLQQKEHPKFK 266
+ + VL + P +K
Sbjct: 362 GLDNCIIVLDIQSFPDYK 415
>TC224735 similar to UP|O24074 (O24074) DnaJ-like protein MsJ1, partial (24%)
Length = 1154
Score = 168 bits (426), Expect = 3e-42
Identities = 76/103 (73%), Positives = 91/103 (87%)
Frame = +1
Query: 197 TGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFV 256
TGE I+++D+CP CKG KV Q+KKVLEVHVEKGMQ GQKI F G+ADEAPDT+TGDIVFV
Sbjct: 844 TGEVINERDKCPLCKGNKVSQEKKVLEVHVEKGMQQGQKIVFEGQADEAPDTITGDIVFV 1023
Query: 257 LQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQL 299
LQ K+HPKF+R+ +DL+++H LSLTEALCGFQFA+ HLD RQL
Sbjct: 1024LQVKDHPKFRREQDDLYIDHNLSLTEALCGFQFAVKHLDGRQL 1152
>TC228163 similar to UP|Q8LEU4 (Q8LEU4) DnaJ protein-like, partial (65%)
Length = 1660
Score = 164 bits (414), Expect = 8e-41
Identities = 109/306 (35%), Positives = 162/306 (52%), Gaps = 7/306 (2%)
Frame = +2
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP----EKFKELAQAYEVLSDPEKREV 69
YY+ILGVSKNAS ++KKAY A K HPD D +KF+E++ AYEVL D E+R+
Sbjct: 401 YYDILGVSKNASSSEIKKAYYGLAKKLHPDTNKDDPQAEKKFQEVSIAYEVLKDEERRQQ 580
Query: 70 YDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSL 129
YDQ G DA G GG F+ F F F S GEDV +++S
Sbjct: 581 YDQLGHDAYVNQQSTGFGGEGGFNPFEQIFRDHDFV---KSFFHENIGGEDVKTFIELSF 751
Query: 130 EDLYLGTSKKLSLSRNVLCSKCNGKGSKSGA-SMTCAGCQGSGMKISMRHLGANMIQQMQ 188
+ G +K L+ +VLC+ C G G G TC C+GS ++ G I +M+
Sbjct: 752 MEAVQGCTKTLTFQTDVLCNACGGSGVPPGTRPETCKPCKGS--RVLFVQAG---IFRME 916
Query: 189 HPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQ--KITFPGEADEAP 246
C CKGTG+ +S D C C+G K+V+ K +++ + G+ + + K+ G AD
Sbjct: 917 STCGTCKGTGKIVS--DYCKSCRGAKIVKGMKSIKLDIMPGIDSNETIKVYRSGGADPDG 1090
Query: 247 DTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPG 306
D GD+ ++ +E P F+R+G D+ V+ LS+T+A+ G + L + +++K PG
Sbjct: 1091DQ-PGDLYVTIKVREDPVFRREGSDIHVDAVLSITQAILGGTIQVPTL-TGDVVLKVRPG 1264
Query: 307 EVVKPG 312
+PG
Sbjct: 1265--TQPG 1276
>TC217251 weakly similar to UP|DJB1_HUMAN (P25685) DnaJ homolog subfamily B
member 1 (Heat shock 40 kDa protein 1) (Heat shock
protein 40) (HSP40) (DnaJ protein homolog 1) (HDJ-1),
partial (21%)
Length = 1669
Score = 163 bits (412), Expect = 1e-40
Identities = 95/290 (32%), Positives = 146/290 (49%), Gaps = 5/290 (1%)
Frame = +2
Query: 11 NTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDP---EKFKELAQAYEVLSDPEKR 67
N YY +LGVS+NAS ++K AY+K A HPD +P +KFKE++ AYEVLSD EKR
Sbjct: 356 NADYYSVLGVSRNASKSEIKSAYRKLARNYHPDVNKEPGAEQKFKEISNAYEVLSDDEKR 535
Query: 68 EVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKV 127
+YD++GE LK G G +PFD+F S F G G GED + L +
Sbjct: 536 SIYDRFGEAGLKGSAMGMGDFSNPFDLFESLFEGMNRGAGSRGSWNGAIDGEDEYYSLVL 715
Query: 128 SLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGASMT-CAGCQGSGMKISMRHLGANMIQQ 186
+ ++ G K++ +SR C CNG G+K G + + C+ C G G +S + QQ
Sbjct: 716 NFKEAVFGIEKEIEISRLESCGTCNGSGAKPGTTPSRCSTCGGQGRVVSSTRTPLGIFQQ 895
Query: 187 MQHPCNECKGTGETISDKDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEA- 245
C+ C GTGE + C C G+ +++ K + + V G+ +G ++ E +
Sbjct: 896 SM-TCSSCNGTGEISTP---CNTCSGDGRLRKSKRISLKVPAGVDSGSRLRVRNEGNAGR 1063
Query: 246 PDTVTGDIVFVLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLD 295
GD+ V++ P KR ++ +S +A+ G + +D
Sbjct: 1064RGGSPGDLFVVIEVIPDPVLKRDDTNILYTCKVSYIDAILGTTIKVPTVD 1213
>BF009667 similar to GP|5802244|gb|A seed maturation protein PM37 {Glycine
max}, partial (20%)
Length = 256
Score = 143 bits (361), Expect = 1e-34
Identities = 66/85 (77%), Positives = 76/85 (88%)
Frame = +2
Query: 2 FGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEVL 61
FGRAPKKSD+T+YYEIL VSKNA+PDDLKKAY AA+ NHPDKGGDP+KF +LAQAY+VL
Sbjct: 2 FGRAPKKSDSTRYYEILWVSKNAAPDDLKKAYMIAAVNNHPDKGGDPDKFTQLAQAYDVL 181
Query: 62 SDPEKREVYDQYGEDALKEGMGGGG 86
S+PEKRE+YD YGE AL EG+G GG
Sbjct: 182SEPEKREIYDTYGEHALTEGLGVGG 256
>BE555607 similar to GP|5802244|gb|A seed maturation protein PM37 {Glycine
max}, partial (23%)
Length = 289
Score = 122 bits (306), Expect = 3e-28
Identities = 64/97 (65%), Positives = 73/97 (74%), Gaps = 1/97 (1%)
Frame = +2
Query: 66 KREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGG-FPGGGSSRGRRQRRGEDVVHP 124
KR++YDQ+GEDAL EGMGGGGG HDPFDIF FFGGG F G SS+ R+QR ED HP
Sbjct: 2 KRDIYDQHGEDALNEGMGGGGG-HDPFDIF*FFFGGGSPFGSGASSQCRKQRLVEDAAHP 178
Query: 125 LKVSLEDLYLGTSKKLSLSRNVLCSKCNGKGSKSGAS 161
L+ +EDLYL TSK L LS V+CSKC+ GS SGAS
Sbjct: 179 LQDFVEDLYLATSKNLYLSIIVVCSKCSVNGSNSGAS 289
>BE803986 similar to PIR|T43929|T439 DnaJ protein homolog [imported] - Salix
gilgiana, partial (15%)
Length = 328
Score = 120 bits (302), Expect = 8e-28
Identities = 63/109 (57%), Positives = 75/109 (68%), Gaps = 2/109 (1%)
Frame = +2
Query: 2 FGRA-PKKSDNTKYYEILGVSKNASPDDLKKA-YKKAAIKNHPDKGGDPEKFKELAQAYE 59
FGR P++SD KYY+ILG+S+NAS D ++ YK + HP KGGDPE KEL QAYE
Sbjct: 2 FGRGGPRRSDIFKYYDILGISRNASEDRNQEGLYKGGY*RTHPYKGGDPE*LKELGQAYE 181
Query: 60 VLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSFFGGGGFPGGG 108
LSDP+K E+YD+YGEDALK+GMGGG P DIF G F GGG
Sbjct: 182 ALSDPDKNELYDRYGEDALKQGMGGGCSLRYPLDIFELLVGRACFGGGG 328
>BF425848 similar to GP|5802244|gb|A seed maturation protein PM37 {Glycine
max}, partial (20%)
Length = 289
Score = 120 bits (301), Expect = 1e-27
Identities = 66/97 (68%), Positives = 69/97 (71%)
Frame = +2
Query: 2 FGRAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPEKFKELAQAYEVL 61
FGRAP K D T YYEIL V NA DDLKKAY KAAI NHPD GGDP KFKELAQAYEVL
Sbjct: 2 FGRAPNKIDYTMYYEILCVFNNAPQDDLKKAYIKAAIYNHPD*GGDPYKFKELAQAYEVL 181
Query: 62 SDPEKREVYDQYGEDALKEGMGGGGGGHDPFDIFSSF 98
DP E+YD +GE AL + M G DP DIFSSF
Sbjct: 182YDPVNLEIYDHFGEFALNDRM-VGRCSLDPRDIFSSF 289
>TC214767 homologue to UP|Q9SWB5 (Q9SWB5) Seed maturation protein PM37,
partial (39%)
Length = 713
Score = 108 bits (271), Expect = 3e-24
Identities = 57/88 (64%), Positives = 62/88 (69%)
Frame = +2
Query: 256 VLQQKEHPKFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKPGWMS 315
VLQQKEHPKFKRK EDLFVEH LSLTEALCGFQF LTHLD RQLLIKSNPGEVVKP
Sbjct: 2 VLQQKEHPKFKRKAEDLFVEHILSLTEALCGFQFVLTHLDGRQLLIKSNPGEVVKP---- 169
Query: 316 VRRQHFMMSTLKKRREEGSKLSKRHMMR 343
+ K +EG + +R M+
Sbjct: 170 --------DSYKAINDEGMPMYQRSFMK 229
Score = 57.8 bits (138), Expect = 8e-09
Identities = 27/49 (55%), Positives = 36/49 (73%)
Frame = +3
Query: 313 WMSVRRQHFMMSTLKKRREEGSKLSKRHMMRMMKCLVVLKEYNVLSSNE 361
WM+VR+QH MMST ++R + +RHMMRMM CLVV + Y+V SSN+
Sbjct: 348 WMNVRKQHSMMSTWRRRLGGSNNKLRRHMMRMMTCLVVHRGYSVPSSND 494
>TC205832 similar to GB|AAM78044.1|21928031|AY125534 At3g62600/F26K9_30
{Arabidopsis thaliana;} , partial (68%)
Length = 1171
Score = 105 bits (262), Expect = 3e-23
Identities = 71/228 (31%), Positives = 110/228 (48%)
Frame = +1
Query: 84 GGGGGHDPFDIFSSFFGGGGFPGGGSSRGRRQRRGEDVVHPLKVSLEDLYLGTSKKLSLS 143
G GGG + DIFS+FFGGG + +G+D+V L +LEDLY+G + K+
Sbjct: 7 GRGGGMNFQDIFSTFFGGGPM-----EEEEKIVKGDDLVVDLDATLEDLYMGGTLKVWRE 171
Query: 144 RNVLCSKCNGKGSKSGASMTCAGCQGSGMKISMRHLGANMIQQMQHPCNECKGTGETISD 203
+NVL K C+ ++ + +G M QQM
Sbjct: 172 KNVL---------KPAPGKRRCNCRN---EVYHKQIGPGMFQQMTEQV------------ 279
Query: 204 KDRCPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHP 263
C QC K V++ + V +EKGMQ+GQ++ F + + D +GD+ F ++ H
Sbjct: 280 ---CEQCPNVKYVREGYFITVDIEKGMQDGQEVLFYEDGEPIIDGESGDLRFRIRTAPHD 450
Query: 264 KFKRKGEDLFVEHTLSLTEALCGFQFALTHLDSRQLLIKSNPGEVVKP 311
F+R+G DL T++L +AL GF+ + HLD L+ + E+ KP
Sbjct: 451 VFRREGNDLHSTVTITLVQALVGFEKTIKHLDEH--LVDISTKEITKP 588
>TC220213 similar to UP|Q7XYV0 (Q7XYV0) P58IPK, partial (38%)
Length = 981
Score = 102 bits (254), Expect = 3e-22
Identities = 58/115 (50%), Positives = 73/115 (63%), Gaps = 11/115 (9%)
Frame = +1
Query: 4 RAPKKSDNTKYYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE-----KFKELAQAY 58
+A K S YY+ILG+SK AS D+K+AYKK A++ HPDK + +F+E+A AY
Sbjct: 190 KALKISKRKDYYKILGISKTASAADIKRAYKKLALQWHPDKNVEKREEAEAQFREIAAAY 369
Query: 59 EVLSDPEKREVYDQYGEDALKEGMGGGGGGHDPFD------IFSSFFGGGGFPGG 107
EVLSD +KR YD+ GED + GMGGGGGG +PF F F GGGF GG
Sbjct: 370 EVLSDEDKRVRYDR-GEDLEEAGMGGGGGGFNPFGGGGYTYTFEGGFPGGGFGGG 531
>TC205833 similar to GB|AAM78044.1|21928031|AY125534 At3g62600/F26K9_30
{Arabidopsis thaliana;} , partial (55%)
Length = 866
Score = 95.9 bits (237), Expect = 3e-20
Identities = 50/93 (53%), Positives = 65/93 (69%), Gaps = 6/93 (6%)
Frame = +1
Query: 14 YYEILGVSKNASPDDLKKAYKKAAIKNHPDKGGDPE----KFKELAQAYEVLSDPEKREV 69
YY+IL +SK AS + +K+AY+K A+K HPDK E KF E++ AYEVLSD EKR +
Sbjct: 283 YYDILQLSKGASDEQIKRAYRKLALKYHPDKNPGNEEANKKFAEISNAYEVLSDSEKRNI 462
Query: 70 YDQYGEDALKE--GMGGGGGGHDPFDIFSSFFG 100
YD+YGE+ LK+ GG GGG + DIFS+F G
Sbjct: 463 YDRYGEEGLKQHAASGGRGGGMNFQDIFSTFGG 561
Score = 51.2 bits (121), Expect = 7e-07
Identities = 23/65 (35%), Positives = 38/65 (58%)
Frame = +3
Query: 207 CPQCKGEKVVQQKKVLEVHVEKGMQNGQKITFPGEADEAPDTVTGDIVFVLQQKEHPKFK 266
C QC K V++ + V +EKGMQ+GQ++ F + + D +GD+ F ++ H F+
Sbjct: 672 CEQCPNVKYVREGYFITVDIEKGMQDGQEVLFYEDGEPIIDGESGDLRFRIRTAPHDVFR 851
Query: 267 RKGED 271
R+G D
Sbjct: 852 REGND 866
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.316 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,160,161
Number of Sequences: 63676
Number of extensions: 229525
Number of successful extensions: 3310
Number of sequences better than 10.0: 258
Number of HSP's better than 10.0 without gapping: 2352
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2835
length of query: 375
length of database: 12,639,632
effective HSP length: 99
effective length of query: 276
effective length of database: 6,335,708
effective search space: 1748655408
effective search space used: 1748655408
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133862.8


