

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126785.11 + phase: 0
(538 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
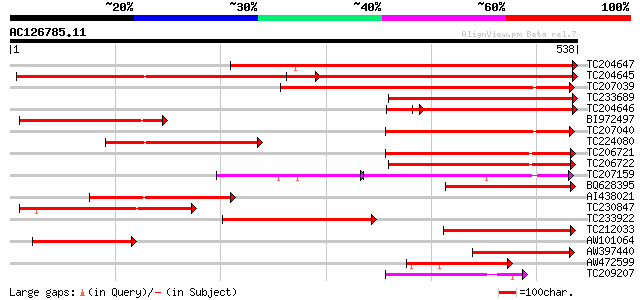
Score E
Sequences producing significant alignments: (bits) Value
TC204647 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (58%) 486 e-137
TC204645 similar to UP|Q9FM07 (Q9FM07) Permease 1, complete 460 e-130
TC207039 366 e-101
TC233689 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (34%) 331 4e-91
TC204646 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (34%) 254 9e-68
BI972497 238 7e-63
TC207040 237 1e-62
TC224080 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (28%) 222 3e-58
TC206721 200 2e-51
TC206722 195 4e-50
TC207159 similar to GB|AAR18374.1|38350523|AY444865 nucleobase-a... 122 9e-47
BQ628395 similar to GP|15983805|gb At1g49960/F2J10_14 {Arabidops... 167 1e-41
AI438021 weakly similar to GP|15983805|gb At1g49960/F2J10_14 {Ar... 157 9e-39
TC230847 153 2e-37
TC233922 weakly similar to UP|O97576 (O97576) Yolk sac permease-... 147 1e-35
TC212033 weakly similar to UP|Q9XIJ8 (Q9XIJ8) T10O24.16, partial... 135 6e-32
AW101064 121 7e-28
AW397440 similar to GP|10177467|dbj permease 1 {Arabidopsis thal... 120 2e-27
AW472599 similar to GP|10177467|dbj permease 1 {Arabidopsis thal... 117 1e-26
TC209207 weakly similar to UP|Q6SZ87 (Q6SZ87) Nucleobase-ascorba... 81 1e-15
>TC204647 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (58%)
Length = 1404
Score = 486 bits (1250), Expect = e-137
Identities = 239/332 (71%), Positives = 275/332 (81%), Gaps = 3/332 (0%)
Frame = +1
Query: 210 FLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAI-ILTGCGAYKNAEHETQDTCRTDRAG 268
F+SQF+PH++ G+H+F RFAV+F++ IVW+YA +LT GAY+ + + +
Sbjct: 1 FISQFVPHVLHAGKHVFDRFAVLFTIAIVWLYAXYLLTVGGAYQPCCNLKTQSTLSHLID 180
Query: 269 L--IHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASATPVPP 326
L I A WI P PF+WGAP+FDAGEAFAMM ASFVA +ES+G FIAV R+ASATP+PP
Sbjct: 181 LV*IESAPWIRVPYPFQWGAPSFDAGEAFAMMMASFVALVESSGAFIAVYRYASATPLPP 360
Query: 327 SVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSIL 386
S+LSRGIGWQGVGILLSG+FGT NGSSVS+ENAGLLALTRVGSRRVVQISAGFMIFFSIL
Sbjct: 361 SILSRGIGWQGVGILLSGLFGTVNGSSVSVENAGLLALTRVGSRRVVQISAGFMIFFSIL 540
Query: 387 GKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQ 446
GKFGAVFASIP PI+AALYCL F+ VG+ GLSFLQFCNLNSFRTKFI+GFSIF+G SVPQ
Sbjct: 541 GKFGAVFASIPPPIIAALYCLFFAYVGAGGLSFLQFCNLNSFRTKFILGFSIFVGLSVPQ 720
Query: 447 YFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGM 506
YF EYTAI YGPVHT ARWFND+INVPF S FVAG++A F D TL K + RKDRG
Sbjct: 721 YFNEYTAINGYGPVHTGARWFNDIINVPFQSKPFVAGVVAYFLDNTLFKREAAIRKDRGK 900
Query: 507 HWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
HWWD++ SFK DTRSEEFYSLPFNLNK+FPSV
Sbjct: 901 HWWDKYKSFKGDTRSEEFYSLPFNLNKYFPSV 996
>TC204645 similar to UP|Q9FM07 (Q9FM07) Permease 1, complete
Length = 1930
Score = 460 bits (1183), Expect = e-130
Identities = 222/276 (80%), Positives = 243/276 (87%)
Frame = +3
Query: 263 RTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASAT 322
RTDRAGLI A WI P PF+WGAPTFDAGEAFAMM ASFVA +ES+G FIAV R+ASAT
Sbjct: 825 RTDRAGLIESAPWIRVPYPFQWGAPTFDAGEAFAMMMASFVALVESSGAFIAVYRYASAT 1004
Query: 323 PVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIF 382
P+PPS+LSRGIGWQGVGILLSG+FGTGNGSSVS+ENAGLLALTRVGSRRVVQI+AGFMIF
Sbjct: 1005 PLPPSILSRGIGWQGVGILLSGLFGTGNGSSVSVENAGLLALTRVGSRRVVQIAAGFMIF 1184
Query: 383 FSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGF 442
FSILGKFGAVFASIP PIVAALYCL F+ VG+ GLSFLQFCNLNSFRT F++G+SIF+G
Sbjct: 1185 FSILGKFGAVFASIPPPIVAALYCLFFAYVGAGGLSFLQFCNLNSFRTIFVLGYSIFIGL 1364
Query: 443 SVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRK 502
SV QYF EYTAI YGPVHT ARWFND+INVPF S AFVAG +A F D TLHK + RK
Sbjct: 1365 SVSQYFNEYTAINGYGPVHTKARWFNDIINVPFQSKAFVAGCVAYFLDNTLHKKEAAIRK 1544
Query: 503 DRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
DRG HWWD++ SFKTDTRSEEFYSLPFNLNK+FPSV
Sbjct: 1545 DRGKHWWDKYRSFKTDTRSEEFYSLPFNLNKYFPSV 1652
Score = 394 bits (1011), Expect = e-110
Identities = 195/289 (67%), Positives = 232/289 (79%)
Frame = +2
Query: 7 GGGATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTAL 66
GGG P PK DE QPHP KDQLPNVSYCITSPPPWPEAI+LGFQHYLVMLGTTVLIPTAL
Sbjct: 59 GGGGAPAPKIDEPQPHPPKDQLPNVSYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTAL 238
Query: 67 VSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYD 126
V QMGGGNEEKA +IQ LFVAGINTL+QTLFGTRLPAVIGGS+T+V TTISIIL+ R+
Sbjct: 239 VPQMGGGNEEKAKVIQTLLFVAGINTLLQTLFGTRLPAVIGGSYTYVATTISIILSGRFS 418
Query: 127 DDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGL 186
D+ P EKFKRIMR TQGALIVAS+LQI++GFSGLW +V RF+SPLSAVPLV+L GFGL
Sbjct: 419 DE-PDPIEKFKRIMRATQGALIVASTLQIVLGFSGLWRNVARFLSPLSAVPLVSLVGFGL 595
Query: 187 YELGFPMLAKCIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILT 246
YELGFP +AKC+EIGLPE+++LVF+SQF+PH++ G+H+F RFAV+F++ IVW+YA +LT
Sbjct: 596 YELGFPGVAKCVEIGLPELILLVFISQFVPHVLHAGKHVFDRFAVLFTIAIVWLYAYLLT 775
Query: 247 GCGAYKNAEHETQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAF 295
GAY +A +TQ T H +SW + P + + G +
Sbjct: 776 VGGAYNHAAPKTQSTWS-------HRSSWFNRICPMDTSSISISMGSTY 901
>TC207039
Length = 1026
Score = 366 bits (939), Expect = e-101
Identities = 177/279 (63%), Positives = 212/279 (75%)
Frame = +3
Query: 258 TQDTCRTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVAR 317
TQ +CRTDRA LI A WI P P WGAPTFDAG AF MMAA V+ +ESTG + A +R
Sbjct: 6 TQHSCRTDRANLISSAPWIKIPYPLEWGAPTFDAGHAFGMMAAVLVSLVESTGAYKAASR 185
Query: 318 FASATPVPPSVLSRGIGWQGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISA 377
ASATP P VLSRGIGWQG+GILL+G+FGT GS+VS+EN GLL R+GSRRV+Q+SA
Sbjct: 186 LASATPPPAHVLSRGIGWQGIGILLNGLFGTLTGSTVSVENVGLLGSNRIGSRRVIQVSA 365
Query: 378 GFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFS 437
GFMIFFS+LGKFGA+FASIP P+ AA+YC+LF V S GLSFLQF N+NS R FI G S
Sbjct: 366 GFMIFFSMLGKFGALFASIPFPMFAAVYCVLFGIVASVGLSFLQFTNMNSMRNLFICGVS 545
Query: 438 IFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSD 497
+F+G S+P+YF+EYT +GP HTNA WFND +N F S VA I+A+F D TL D
Sbjct: 546 LFLGLSIPEYFREYTIRAFHGPAHTNAGWFNDFLNTIFFSSPTVALIVAVFLDNTLDYKD 725
Query: 498 NQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
+ KDRGM WW +F +FK D+R+EEFY+LPFNLN+FFP
Sbjct: 726 --SAKDRGMPWWAKFRTFKGDSRNEEFYTLPFNLNRFFP 836
>TC233689 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (34%)
Length = 708
Score = 331 bits (849), Expect = 4e-91
Identities = 161/179 (89%), Positives = 165/179 (91%)
Frame = +3
Query: 360 GLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSF 419
GLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIP PIVAALYCL F+ VGSAGLSF
Sbjct: 6 GLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPAPIVAALYCLFFAYVGSAGLSF 185
Query: 420 LQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGA 479
LQFCNLNSFRTKFI+GFSIFMGFS+PQYF EYTA K YGPVHT ARWFNDMINVPF S A
Sbjct: 186 LQFCNLNSFRTKFILGFSIFMGFSIPQYFNEYTAFKGYGPVHTRARWFNDMINVPFQSEA 365
Query: 480 FVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
FVAG+LAL DVTL K DNQTRKDRGMHWWDRF SFKTDTRSEEFYSLPFNLNKFFPSV
Sbjct: 366 FVAGMLALLLDVTLRKKDNQTRKDRGMHWWDRFLSFKTDTRSEEFYSLPFNLNKFFPSV 542
>TC204646 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (34%)
Length = 768
Score = 254 bits (648), Expect = 9e-68
Identities = 119/156 (76%), Positives = 130/156 (83%)
Frame = +3
Query: 383 FSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGF 442
F GKFGAVFASIP PIVAALYCL F+ VG+ GLSFLQFCNLNSFRT F++G+SIFMG
Sbjct: 78 FQFWGKFGAVFASIPPPIVAALYCLFFAYVGAGGLSFLQFCNLNSFRTIFVLGYSIFMGL 257
Query: 443 SVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQTRK 502
SV QYF EYTAI YGPVHT ARWFND+INVPF S AFVAG +A F D TLHK + RK
Sbjct: 258 SVSQYFNEYTAINGYGPVHTKARWFNDIINVPFQSKAFVAGCVAYFLDNTLHKKEAAIRK 437
Query: 503 DRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPSV 538
DRG HWWD++ SFKTDTRSEEFYSLPFNLNK+FPSV
Sbjct: 438 DRGKHWWDKYRSFKTDTRSEEFYSLPFNLNKYFPSV 545
Score = 60.5 bits (145), Expect = 2e-09
Identities = 31/36 (86%), Positives = 33/36 (91%)
Frame = +2
Query: 358 NAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVF 393
NAGLLALTRVGSRRVVQISAGFMIFFSILG+ + F
Sbjct: 2 NAGLLALTRVGSRRVVQISAGFMIFFSILGEIWSRF 109
>BI972497
Length = 421
Score = 238 bits (606), Expect = 7e-63
Identities = 116/140 (82%), Positives = 127/140 (89%)
Frame = +3
Query: 10 ATPPPKQDEFQPHPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQ 69
A P PKQDE QPHPVKDQLPNVS+CITSPPPWPEAI+LGFQHYLVMLGTTVLIP++LV Q
Sbjct: 3 AAPQPKQDEHQPHPVKDQLPNVSFCITSPPPWPEAILLGFQHYLVMLGTTVLIPSSLVPQ 182
Query: 70 MGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDI 129
MGGGNEEKA +IQ LFVAGINT QT FGTRLPAVIGGS+TFVPTTISIILA RY D+
Sbjct: 183 MGGGNEEKAKVIQTLLFVAGINTFFQTFFGTRLPAVIGGSYTFVPTTISIILAGRY-SDV 359
Query: 130 MHPREKFKRIMRGTQGALIV 149
++P+EKF+RIMRGTQGALIV
Sbjct: 360 VNPQEKFERIMRGTQGALIV 419
>TC207040
Length = 718
Score = 237 bits (604), Expect = 1e-62
Identities = 115/180 (63%), Positives = 139/180 (76%)
Frame = +3
Query: 357 ENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAG 416
EN GLL TRVGSRRV+QISAGFMIFFS+LGKFGA+FASIP PI AA+YC+LF V S G
Sbjct: 15 ENVGLLGSTRVGSRRVIQISAGFMIFFSMLGKFGALFASIPFPIFAAVYCVLFGLVASVG 194
Query: 417 LSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFS 476
LSFLQF N+NS R FI+G ++F+GFSVP+YF+EYT+ +GP HT A WF+D +N F
Sbjct: 195 LSFLQFTNMNSMRNLFIVGVALFLGFSVPEYFREYTSKALHGPTHTRAGWFDDXLNTIFF 374
Query: 477 SGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
S VA I+A+F D TL D + KDRGM WW RF +F D+R+EEFY+LPFNLN+FFP
Sbjct: 375 SSPTVALIVAVFLDNTLDYKD--SAKDRGMPWWARFRTFNGDSRNEEFYTLPFNLNRFFP 548
>TC224080 similar to UP|Q9FM07 (Q9FM07) Permease 1, partial (28%)
Length = 445
Score = 222 bits (566), Expect = 3e-58
Identities = 104/149 (69%), Positives = 133/149 (88%)
Frame = +1
Query: 92 TLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREKFKRIMRGTQGALIVAS 151
T +QT+FGTRLPAVIGGS+TFVPTTISIILA R+ D+ P EKFKRIMR QGALIVAS
Sbjct: 1 TRLQTMFGTRLPAVIGGSYTFVPTTISIILAGRFSDE-PDPIEKFKRIMRSIQGALIVAS 177
Query: 152 SLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLAKCIEIGLPEIVILVFL 211
+LQI++GFSGLW +V RF+SPLS+VPLV+L GFGLYELGFP +AKC+EIGLP++++LVF+
Sbjct: 178 TLQIVLGFSGLWRNVARFLSPLSSVPLVSLVGFGLYELGFPGVAKCVEIGLPQLILLVFV 357
Query: 212 SQFMPHMMKGGRHIFARFAVIFSVIIVWV 240
SQ++PH++ G+HIF RFAV+F+++IVW+
Sbjct: 358 SQYVPHVLHSGKHIFDRFAVLFTIVIVWI 444
>TC206721
Length = 1013
Score = 200 bits (508), Expect = 2e-51
Identities = 97/181 (53%), Positives = 123/181 (67%)
Frame = +2
Query: 357 ENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAG 416
EN GLL LT +GSRRVVQIS GFMIFFSI GKFGA FASIP+PI AA+YC+LF V + G
Sbjct: 83 ENVGLLGLTHIGSRRVVQISCGFMIFFSIFGKFGAFFASIPLPIFAAIYCVLFGIVAATG 262
Query: 417 LSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFS 476
+SF+QF N NS R +++G ++F+ S+PQYF TA +GPV T WFND++N FS
Sbjct: 263 ISFIQFANTNSIRNIYVLGLTLFLAISIPQYFVMNTAPDGHGPVRTGGGWFNDILNTIFS 442
Query: 477 SGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
S VA I+ D TL QT DRG+ WW F + K D R++EFY LP +N++ P
Sbjct: 443 SAPTVAIIVGTLVDNTL--EGKQTAVDRGLPWWGPFQNRKGDVRNDEFYRLPLRINEYMP 616
Query: 537 S 537
+
Sbjct: 617 T 619
>TC206722
Length = 749
Score = 195 bits (496), Expect = 4e-50
Identities = 94/178 (52%), Positives = 122/178 (67%)
Frame = +2
Query: 360 GLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAGLSF 419
GLL LT +GSRRVVQIS G+MIFFSI GKFGA FASIP+PI AA+YC+LF V + G+SF
Sbjct: 14 GLLGLTHIGSRRVVQISCGYMIFFSIFGKFGAFFASIPLPIFAAIYCVLFGIVAATGISF 193
Query: 420 LQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGA 479
+QF N NS R +++G ++F+ S+PQYF TA +GPV T+ WFND++N FSS
Sbjct: 194 IQFANTNSIRNIYVLGLTLFLAISIPQYFVMNTAPDGHGPVRTDGGWFNDILNTIFSSAP 373
Query: 480 FVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFPS 537
VA I+ D TL QT DRG+ WW F + K D R++EFY LP +N++ P+
Sbjct: 374 TVAIIVGTLIDNTL--EGKQTAVDRGLPWWGPFQNRKGDVRNDEFYRLPLRINEYMPT 541
>TC207159 similar to GB|AAR18374.1|38350523|AY444865 nucleobase-ascorbate
transporter 12 {Arabidopsis thaliana;} , partial (52%)
Length = 1445
Score = 122 bits (305), Expect(2) = 9e-47
Identities = 66/216 (30%), Positives = 114/216 (52%), Gaps = 16/216 (7%)
Frame = +3
Query: 336 QGVGILLSGIFGTGNGSSVSIENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFAS 395
+G+ +L+G++GTG GS+ EN +A+T++GSR+ VQ+ A F+I S++GK G AS
Sbjct: 468 EGLSSVLAGLWGTGTGSTTLTENVHTIAVTKMGSRKAVQLGACFLIVLSLVGKVGGFIAS 647
Query: 396 IPMPIVAALYCLLFSQVGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEY---- 451
IP +VA L C +++ + + GLS L++ S R I+G S+F S+P YF++Y
Sbjct: 648 IPEVMVAGLLCFMWAMLTALGLSNLRYSEAGSSRNIIIVGLSLFFSLSIPAYFQQYGISP 827
Query: 452 ------------TAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQ 499
+ +GP H+ N ++N FS +A ++A D T+ S
Sbjct: 828 NSNLSVPSYFQPYIVTSHGPFHSKYGGLNYVLNTLFSLHMVIAFLVAFILDNTVPGS--- 998
Query: 500 TRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFF 535
+++RG++ W + + + Y LP + + F
Sbjct: 999 -KQERGVYVWSKAEVARREPAVANDYELPLKVGRIF 1103
Score = 84.0 bits (206), Expect(2) = 9e-47
Identities = 45/152 (29%), Positives = 74/152 (48%), Gaps = 11/152 (7%)
Frame = +1
Query: 197 CIEIGLPEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKN--- 253
CIEIG +I++++ S ++ + G IF +AV + I W +A +LT G Y
Sbjct: 10 CIEIGAVKILVVIVFSLYLHKISVLGHRIFLIYAVPLGLAITWAFAFLLTEAGVYNYKGC 189
Query: 254 -----AEHETQDTCRTDRAGLIH---GASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQ 305
A + + C+ + + H ++W P P +WG P F A M S ++
Sbjct: 190 DVNIPASNMVSEHCKKHFSRMRHCRVSSTWFRFPYPLQWGTPVFHWKMAIVMCVVSLISS 369
Query: 306 IESTGGFIAVARFASATPVPPSVLSRGIGWQG 337
++S G + A + ++ P P VLSRGIG +G
Sbjct: 370 VDSVGSYHASSLLVASRPPTPGVLSRGIGLEG 465
>BQ628395 similar to GP|15983805|gb At1g49960/F2J10_14 {Arabidopsis
thaliana}, partial (23%)
Length = 440
Score = 167 bits (423), Expect = 1e-41
Identities = 74/124 (59%), Positives = 95/124 (75%)
Frame = +3
Query: 414 SAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINV 473
SAGL FLQFCNLNS+R+ FI+GFS+FMG SVPQYF EY + +GPVHT FN+++ V
Sbjct: 63 SAGLGFLQFCNLNSYRSMFIVGFSLFMGLSVPQYFNEYVLLSGHGPVHTGTTAFNNIVQV 242
Query: 474 PFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNK 533
FSS A VA I+A F D+T+ + + TR+D G HWW++F +F DTR+E+FYSLP NLN+
Sbjct: 243 IFSSPATVAIIVAYFLDLTMSRGEGSTRRDSGRHWWEKFRTFNQDTRTEDFYSLPLNLNR 422
Query: 534 FFPS 537
FFPS
Sbjct: 423 FFPS 434
>AI438021 weakly similar to GP|15983805|gb At1g49960/F2J10_14 {Arabidopsis
thaliana}, partial (26%)
Length = 416
Score = 157 bits (398), Expect = 9e-39
Identities = 83/139 (59%), Positives = 98/139 (69%)
Frame = +1
Query: 76 EKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYDDDIMHPREK 135
EKA IQ LFVA INTL+QT FGTRLP V+G S+ F+ S+ +SR + P ++
Sbjct: 1 EKAETIQTLLFVAAINTLLQTWFGTRLPVVVGASYAFLIPAFSVAFSSRMSI-FLDPHQR 177
Query: 136 FKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVPLVALTGFGLYELGFPMLA 195
FK+ MR QGALIVAS QIIVGF G W RF+SPLS VPLV LTG GL+ LGFP LA
Sbjct: 178 FKQSMRAIQGALIVASFFQIIVGFFGFWRIFARFLSPLSVVPLVTLTGLGLFVLGFPRLA 357
Query: 196 KCIEIGLPEIVILVFLSQF 214
C+EIGLP +VILV LSQ+
Sbjct: 358 DCVEIGLPALVILVILSQY 414
>TC230847
Length = 627
Score = 153 bits (387), Expect = 2e-37
Identities = 84/171 (49%), Positives = 109/171 (63%), Gaps = 3/171 (1%)
Frame = +3
Query: 10 ATPPPKQDEFQPHPV---KDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTAL 66
A PPP PV +QL + YCI S P WP AI+LGFQHY+VMLGTTVLI T L
Sbjct: 117 AGPPPPNLALSRGPVWNPAEQLSQLHYCIHSNPSWPVAILLGFQHYIVMLGTTVLIATTL 296
Query: 67 VSQMGGGNEEKAMLIQNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISIILASRYD 126
V MGG + +KA +IQ+ LF++G+NTL+QT FG+RLP V+GGSF F+ +SII D
Sbjct: 297 VPAMGGDHGDKARVIQSLLFMSGVNTLLQTWFGSRLPTVMGGSFAFLLPVLSII-NDYTD 473
Query: 127 DDIMHPREKFKRIMRGTQGALIVASSLQIIVGFSGLWCHVVRFISPLSAVP 177
E+F +R QG+LIV+S + I +GFS W ++ R SP+ VP
Sbjct: 474 RTFPSEHERFTYTIRTIQGSLIVSSFVNIFLGFSKTWGNLTRLFSPIIIVP 626
>TC233922 weakly similar to UP|O97576 (O97576) Yolk sac permease-like
molecule 2, partial (8%)
Length = 449
Score = 147 bits (371), Expect = 1e-35
Identities = 67/146 (45%), Positives = 92/146 (62%)
Frame = +1
Query: 203 PEIVILVFLSQFMPHMMKGGRHIFARFAVIFSVIIVWVYAIILTGCGAYKNAEHETQDTC 262
P +++LV Q++ + + RFA++ + ++W +A ILT GAY A+ +TQ +C
Sbjct: 1 PMLILLVITQQYLKSLHHAAHQVLERFALLLCIAVIWAFAAILTVAGAYNTAKPQTQVSC 180
Query: 263 RTDRAGLIHGASWISPPIPFRWGAPTFDAGEAFAMMAASFVAQIESTGGFIAVARFASAT 322
RTDR+ L+ A WI P PF+WG P F A F MM A+ V+ ESTG F A AR + AT
Sbjct: 181 RTDRSYLMSSAPWIKVPYPFQWGTPIFRASHVFGMMGAALVSSAESTGAFFAAARLSGAT 360
Query: 323 PVPPSVLSRGIGWQGVGILLSGIFGT 348
P P VLSR IG QG+G+LL GIFG+
Sbjct: 361 PPPAHVLSRSIGMQGIGMLLEGIFGS 438
>TC212033 weakly similar to UP|Q9XIJ8 (Q9XIJ8) T10O24.16, partial (5%)
Length = 569
Score = 135 bits (339), Expect = 6e-32
Identities = 63/126 (50%), Positives = 83/126 (65%)
Frame = +3
Query: 412 VGSAGLSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMI 471
V S GL F QFC S+R+ FI+ FS+FMG SVPQYF EY + +GPV FN+++
Sbjct: 9 VASTGLGFPQFCXXXSYRSMFIVAFSLFMGXSVPQYFSEYVVLSXHGPVXXGTTAFNNIV 188
Query: 472 NVPFSSGAFVAGILALFFDVTLHKSDNQTRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNL 531
V FSS A VA I+A F +T+ + + T + G WW++F +F DTRSE+FYSLP NL
Sbjct: 189 QVFFSSTAXVAIIVAYFLYLTMSRGEGSTLRVSGRLWWEKFRTFNQDTRSEDFYSLPLNL 368
Query: 532 NKFFPS 537
+FFPS
Sbjct: 369 TRFFPS 386
>AW101064
Length = 417
Score = 121 bits (304), Expect = 7e-28
Identities = 57/99 (57%), Positives = 71/99 (71%)
Frame = +2
Query: 22 HPVKDQLPNVSYCITSPPPWPEAIMLGFQHYLVMLGTTVLIPTALVSQMGGGNEEKAMLI 81
HP DQL + YCI S P W E I LGFQHY++ LGT V+IP+ LV MG +++K ++
Sbjct: 119 HPPMDQLQGLEYCIDSNPSWAETIALGFQHYILALGTAVMIPSFLVPVMGESDDDKVRVV 298
Query: 82 QNHLFVAGINTLIQTLFGTRLPAVIGGSFTFVPTTISII 120
Q LFV GINTL+QTLFGTRLP V+GGS+ F+ ISII
Sbjct: 299 QTLLFVEGINTLLQTLFGTRLPTVVGGSYAFMVXVISII 415
>AW397440 similar to GP|10177467|dbj permease 1 {Arabidopsis thaliana},
partial (15%)
Length = 295
Score = 120 bits (301), Expect = 2e-27
Identities = 53/97 (54%), Positives = 66/97 (67%)
Frame = +2
Query: 440 MGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSSGAFVAGILALFFDVTLHKSDNQ 499
+G S P+Y+ EYTAI YG VHT ARWFND+INVP+ S FVAG+ A T+ K +
Sbjct: 5 VGLSAPKYYNEYTAINGYGAVHTGARWFNDIINVPYESKPFVAGVEAYILHNTVFKREAA 184
Query: 500 TRKDRGMHWWDRFSSFKTDTRSEEFYSLPFNLNKFFP 536
DRG HWWD++ SF DT S+EFY LPFN++K P
Sbjct: 185 LMNDRGKHWWDKYKSFNGDT*SDEFYDLPFNIHKHSP 295
>AW472599 similar to GP|10177467|dbj permease 1 {Arabidopsis thaliana},
partial (13%)
Length = 328
Score = 117 bits (293), Expect = 1e-26
Identities = 60/105 (57%), Positives = 72/105 (68%), Gaps = 4/105 (3%)
Frame = +2
Query: 377 AGF--MIFFSILGKFGAVFASIPMPIVAALYC--LLFSQVGSAGLSFLQFCNLNSFRTKF 432
AGF + F S G FGA F ++ YC L F+ VG+ GLSFLQFCNLNS+RT F
Sbjct: 2 AGFYDVYFSSSRGNFGARFCMPFHTAISCRYCTSLFFAYVGAGGLSFLQFCNLNSYRTIF 181
Query: 433 IIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPFSS 477
++G+SIFMG S QYF EYTA YGPVHT ARWF+D+I+V F S
Sbjct: 182 VLGYSIFMGLSASQYFNEYTAFNGYGPVHTKARWFDDIISVAFRS 316
>TC209207 weakly similar to UP|Q6SZ87 (Q6SZ87) Nucleobase-ascorbate
transporter 11, partial (20%)
Length = 419
Score = 81.3 bits (199), Expect = 1e-15
Identities = 49/138 (35%), Positives = 75/138 (53%), Gaps = 3/138 (2%)
Frame = +3
Query: 357 ENAGLLALTRVGSRRVVQISAGFMIFFSILGKFGAVFASIPMPIVAALYCLLFSQVGSAG 416
EN + +T+V S++VV + A F+I FS +GK GA+ ASIP + A + + + + G
Sbjct: 3 ENTHTIDITKVASQKVVVVGATFVILFSFIGKVGALLASIPQALAAPVLSFM*ALTAALG 182
Query: 417 LSFLQFCNLNSFRTKFIIGFSIFMGFSVPQYFKEYTAIKQYGPVHTNARWFNDMINVPF- 475
LS LQ+ SFR I+G ++F+G S+P YF++Y A + VP+
Sbjct: 183 LSNLQYSKSASFRNITIVGVTLFLGMSIPAYFQQYQA---------ESSLILPRYLVPYA 335
Query: 476 --SSGAFVAGILALFFDV 491
SSG F +GI L F +
Sbjct: 336 ATSSGPFRSGIKQLDFAI 389
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.327 0.142 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,990,583
Number of Sequences: 63676
Number of extensions: 367178
Number of successful extensions: 4940
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 3649
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4758
length of query: 538
length of database: 12,639,632
effective HSP length: 102
effective length of query: 436
effective length of database: 6,144,680
effective search space: 2679080480
effective search space used: 2679080480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126785.11


