

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC126784.4 - phase: 0 /pseudo
(525 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
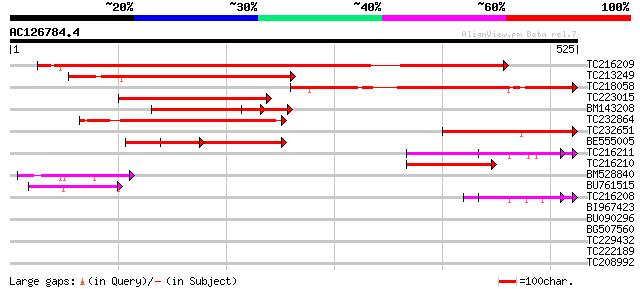
Score E
Sequences producing significant alignments: (bits) Value
TC216209 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partia... 373 e-103
TC213249 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partia... 306 2e-83
TC218058 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partia... 245 3e-65
TC223015 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partia... 233 2e-61
BM143208 152 4e-37
TC232864 weakly similar to UP|Q7GAD9 (Q7GAD9) Expressed protein,... 146 3e-35
TC232651 143 2e-34
BE555005 127 9e-30
TC216211 96 4e-20
TC216210 90 2e-18
BM528840 78 1e-14
BU761515 74 2e-13
TC216208 64 1e-10
BI967423 similar to GP|14532460|gb AT5g13810/MAC12_24 {Arabidops... 31 1.5
BU090296 homologue to GP|14532460|gb| AT5g13810/MAC12_24 {Arabid... 31 1.5
BG507560 similar to GP|1838976|emb| vsf-1 {Lycopersicon esculent... 31 1.5
TC229432 30 2.0
TC222189 similar to UP|ITBX_DROME (P11584) Integrin beta-PS prec... 30 2.0
TC208992 weakly similar to UP|O04925 (O04925) 15.5 kDa oleosin, ... 29 4.5
>TC216209 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partial (36%)
Length = 1457
Score = 373 bits (957), Expect = e-103
Identities = 217/459 (47%), Positives = 283/459 (61%), Gaps = 22/459 (4%)
Frame = +1
Query: 26 KFEIEDSIQDQHAPLNKRHK------------------ATNDILNEPSPLGLSLRKSPSL 67
K E++D I H +KR K A+ + L+EPSPLGL LRKSPSL
Sbjct: 139 KLEVQDEIPPLHT--HKRPKLESPPKCGASDESISIPPASYNPLDEPSPLGLRLRKSPSL 312
Query: 68 LDLIQMTLCQ-ENSVNANTANDNLNSKANKNGRASVEKLKASNFPATHLKIGSWEYKSKY 126
LDLIQM L Q E S + + +S A A+ KLKASNFP T LKIG+WEYKS+Y
Sbjct: 313 LDLIQMRLSQQEESKKKDQKASSSSSVAAAAAAAADSKLKASNFPGTILKIGNWEYKSRY 492
Query: 127 EGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPSTLTLMVARQPLF 186
EGDLVAKCYFAK KLVWEVL+G LK+KIEI WSDI +KAN P+DGP TL +++AR+PLF
Sbjct: 493 EGDLVAKCYFAKHKLVWEVLDGCLKNKIEIPWSDIMAIKANYPEDGPGTLEVVLARRPLF 672
Query: 187 FRETNPQPRKHTLWQSTTDFTGGQASIHRRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQQ 246
FRE NPQPRKHTLWQ+T+DFTGGQASI+RRH LQC QGLL KH+EKL+QC+ RL +LSQ
Sbjct: 673 FREINPQPRKHTLWQATSDFTGGQASINRRHFLQCPQGLLGKHFEKLIQCDPRLNYLSQH 852
Query: 247 PEIMVDSPHFDPRSAAIENPHNLKD-CDLHQGNGSAVSCFQNMGSPHSSLSPSFTTEHSD 305
P++++DSP+F+P + ++ + D D S + Q++ S + S S +EH+
Sbjct: 853 PDLVLDSPYFEPGTTSVHDHIESSDGFDRKSEERSGIFGLQDVESGSAVQSSSSKSEHNL 1032
Query: 306 PSAITLDSVPCEAPSS-SSA*IMK-FSSKICQMSII*NL**LCKSVIVVLAC*FHQ*FNM 363
A+ S +PSS + MK F S+ + +
Sbjct: 1033GKAVENVSQEITSPSSVMNIHAMKDFRSRGAE--------------------------TL 1134
Query: 364 LGSRNWDQIKLPGLRPSMSMSDFLGHIEHHISKEMASGDPSFSAERLEYQQMMDGITQHL 423
N DQIKLPGL PSMSM D + HI H IS++M S +P+F+ + + +++ TQ+L
Sbjct: 1135KFLSNLDQIKLPGLHPSMSMDDLVSHIGHCISEQMGSDNPNFAGDNQYSRSILEEFTQYL 1314
Query: 424 LNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDN 462
ND+Q T SDE+ +MS+VNSL CLLQ DP + N
Sbjct: 1315FNDSQHATTSDEQRVMSKVNSLYCLLQKDPSTAEDMTKN 1431
>TC213249 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partial (40%)
Length = 641
Score = 306 bits (783), Expect = 2e-83
Identities = 152/217 (70%), Positives = 178/217 (81%), Gaps = 7/217 (3%)
Frame = +2
Query: 55 SPLGLSLRKSPSLLDLIQMTLCQENSVNANTANDNLNSKANKNGRASV-------EKLKA 107
SPLGL LRKSPSLLDLIQM L Q ++ +++L+S A + RA+ +KLKA
Sbjct: 2 SPLGLRLRKSPSLLDLIQMKLSQGSA----QPSEDLSSGAKRESRAAAVSGAGGADKLKA 169
Query: 108 SNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKAN 167
SNFPA+ L+IGSWEYKS+YEGDLVAKCYFAK KLVWEVLEG LKSKIEIQWSDI LKA+
Sbjct: 170 SNFPASLLRIGSWEYKSRYEGDLVAKCYFAKHKLVWEVLEGGLKSKIEIQWSDIMALKAH 349
Query: 168 CPDDGPSTLTLMVARQPLFFRETNPQPRKHTLWQSTTDFTGGQASIHRRHVLQCEQGLLI 227
CPDDGPSTLT+++ARQPL+FRETNPQPRKHTLWQ+T DFT GQ+S HR H LQC QGLL
Sbjct: 350 CPDDGPSTLTVVLARQPLYFRETNPQPRKHTLWQATADFTDGQSSKHRLHFLQCPQGLLA 529
Query: 228 KHYEKLVQCNDRLKFLSQQPEIMVDSPHFDPRSAAIE 264
KH+EKL+QC+ RL FLSQQPEI++DSPHFD + +A E
Sbjct: 530 KHFEKLIQCDMRLNFLSQQPEIILDSPHFDTQPSAFE 640
>TC218058 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partial (9%)
Length = 1116
Score = 245 bits (626), Expect = 3e-65
Identities = 146/270 (54%), Positives = 177/270 (65%), Gaps = 5/270 (1%)
Frame = +3
Query: 261 AAIENPHNLKDCDLHQ--GNGSAVSCFQNMGSPHSSLSPSFTTEHSDPSAITLDSVPCEA 318
+A E+P N KD DL Q G GS+ SC+Q+ GSP +SLS SF TEH+DP + LDS+P +A
Sbjct: 3 SAFEDPDNPKDHDLLQVSGKGSSTSCYQDSGSPQASLSSSFKTEHNDPPGMMLDSLPRDA 182
Query: 319 PSSSSA*IMKFSSKICQMSII*NL**LCKSVIVVLAC*FHQ*FNMLGSRNWDQIKLPGLR 378
PS SS +M+ +S S + G RN DQIKLPGLR
Sbjct: 183 PSPSS--VMECTSIEGSTS---------------------SETDSKGPRNGDQIKLPGLR 293
Query: 379 PSMSMSDFLGHIEHHISKEMASGDPSFSAERLEYQQMMDGITQHLLNDNQVTTDSDEKSL 438
PSMS+SDF+G IE +S+++ SG+P FS EY+++++ I QHLLNDNQV SDEKSL
Sbjct: 294 PSMSVSDFIGQIELCLSEQITSGNPPFSDGGSEYKEILEDIAQHLLNDNQVAATSDEKSL 473
Query: 439 MSRVNSLRCLLQMDPPAVPNSH---DNTGFIEGPNDAKVNIDIKATEENSRDVYGGNPAP 495
MSRVNSL CLLQ DP V NSH DNT +EGP+D K D+K E S+D GG A
Sbjct: 474 MSRVNSLCCLLQKDPVTVQNSHFTEDNT--VEGPDDGK---DVKPAVEESKDASGGKQAL 638
Query: 496 GMSRKDSFGDLLLSLPRIASLPKFLFDISE 525
GMSRKDSF DLLL LPRIASLPKFLF+ISE
Sbjct: 639 GMSRKDSFSDLLLHLPRIASLPKFLFNISE 728
>TC223015 similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partial (29%)
Length = 428
Score = 233 bits (593), Expect = 2e-61
Identities = 107/142 (75%), Positives = 126/142 (88%)
Frame = +2
Query: 101 SVEKLKASNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSD 160
S EKLKASNFPA+ L+IGSWEYKSK+EGDLVAKCYFAK KLVWEVLEGELK+K+EIQWSD
Sbjct: 2 SDEKLKASNFPASLLRIGSWEYKSKHEGDLVAKCYFAKHKLVWEVLEGELKNKMEIQWSD 181
Query: 161 ISQLKANCPDDGPSTLTLMVARQPLFFRETNPQPRKHTLWQSTTDFTGGQASIHRRHVLQ 220
I LKANCPD GPS+LT+++ARQPLFF+ETNPQPRKHT+WQ+T+DFT G+A HR+H L+
Sbjct: 182 IMALKANCPDTGPSSLTVVLARQPLFFKETNPQPRKHTIWQATSDFTEGEACKHRQHFLE 361
Query: 221 CEQGLLIKHYEKLVQCNDRLKF 242
QGLL KH+EKL+QC+ L F
Sbjct: 362 FPQGLLAKHFEKLIQCDTHLNF 427
>BM143208
Length = 448
Score = 152 bits (384), Expect = 4e-37
Identities = 71/106 (66%), Positives = 85/106 (79%)
Frame = +1
Query: 132 AKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPSTLTLMVARQPLFFRETN 191
AKCYFAK K VWEVLEG LKSKIEIQWSDI LKA+CPD+GPSTLT+++ARQPL+FRETN
Sbjct: 1 AKCYFAKHKKVWEVLEGGLKSKIEIQWSDIMALKAHCPDNGPSTLTVVLARQPLYFRETN 180
Query: 192 PQPRKHTLWQSTTDFTGGQASIHRRHVLQCEQGLLIKHYEKLVQCN 237
PQP KHTLWQ+T DFT GQ+S HR ++ C+ L+ + + CN
Sbjct: 181 PQPIKHTLWQATADFTDGQSSKHRSYLQGCDV*KLVLWFVGCIFCN 318
Score = 74.7 bits (182), Expect = 9e-14
Identities = 33/48 (68%), Positives = 40/48 (82%)
Frame = +3
Query: 215 RRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQQPEIMVDSPHFDPRSAA 262
R H LQC QGLL KH+EKL+QC+ RL FLSQQPEI++DSPHFD + +A
Sbjct: 300 RLHFLQCPQGLLAKHFEKLIQCDMRLNFLSQQPEIILDSPHFDTQPSA 443
>TC232864 weakly similar to UP|Q7GAD9 (Q7GAD9) Expressed protein, partial
(19%)
Length = 714
Score = 146 bits (368), Expect = 3e-35
Identities = 84/192 (43%), Positives = 118/192 (60%)
Frame = +1
Query: 65 PSLLDLIQMTLCQENSVNANTANDNLNSKANKNGRASVEKLKASNFPATHLKIGSWEYKS 124
P LL L ++T+ E T+ N+K VEKLKA FP L+IG ++ ++
Sbjct: 7 PPLLGL-KITMTPEMLQPTKTSQVAANNK--------VEKLKAVQFPMDMLRIGYFKIEA 159
Query: 125 KYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPSTLTLMVARQP 184
KY +LVAKCY+A+QKL+WE+L LK KIEI +IS ++A ++ P L + + + P
Sbjct: 160 KYPYELVAKCYYARQKLIWEILHDGLKFKIEIHCQNISAIRAVMEENSPGILEIELDKVP 339
Query: 185 LFFRETNPQPRKHTLWQSTTDFTGGQASIHRRHVLQCEQGLLIKHYEKLVQCNDRLKFLS 244
FFRE +P+P+KHT W + DFT GQAS +RRH LQ G+L +HY KL+Q ++RL LS
Sbjct: 340 SFFREIDPKPKKHTTWTISHDFTDGQASEYRRHYLQFPHGVLDQHYIKLLQSDNRLLELS 519
Query: 245 QQPEIMVDSPHF 256
+P S HF
Sbjct: 520 LRP---FPSSHF 546
>TC232651
Length = 712
Score = 143 bits (361), Expect = 2e-34
Identities = 78/140 (55%), Positives = 92/140 (65%), Gaps = 15/140 (10%)
Frame = +2
Query: 401 GDPSFSAERLEYQQMMDGITQHLLNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSH 460
G+PSF R E+Q+M++ I QH LNDN+V T SDEKSLM+RVNSL CLLQ DP A+ +SH
Sbjct: 2 GNPSFCGGRPEFQEMLEEIAQHXLNDNKVITTSDEKSLMTRVNSLCCLLQKDPAALQSSH 181
Query: 461 DNTGFIEGPNDA---------------KVNIDIKATEENSRDVYGGNPAPGMSRKDSFGD 505
D EGP D K+ +D+KA+EE RDV G GM RKDS G+
Sbjct: 182 DKESADEGPADGKSIQLSHDLESMQNNKIKMDVKASEEEFRDVSRGKQTLGMPRKDSLGE 361
Query: 506 LLLSLPRIASLPKFLFDISE 525
LLL LPRIASL KFLFDISE
Sbjct: 362 LLLQLPRIASLSKFLFDISE 421
>BE555005
Length = 431
Score = 127 bits (320), Expect = 9e-30
Identities = 66/117 (56%), Positives = 83/117 (70%)
Frame = +2
Query: 140 KLVWEVLEGELKSKIEIQWSDISQLKANCPDDGPSTLTLMVARQPLFFRETNPQPRKHTL 199
K + VL LKS I W +S+L D +AR+PLFFRE NPQPRKHTL
Sbjct: 110 KSLMVVLRIRLKSHGLISW--LSRLTTLMMDQ--------LARRPLFFREINPQPRKHTL 259
Query: 200 WQSTTDFTGGQASIHRRHVLQCEQGLLIKHYEKLVQCNDRLKFLSQQPEIMVDSPHF 256
WQ+T+DFTGGQA I+RRH LQC QGLL KH+EKL+QC+ RL +LSQQ ++++DSP+F
Sbjct: 260 WQATSDFTGGQAGINRRHFLQCPQGLLGKHFEKLIQCDPRLNYLSQQADLVLDSPYF 430
Score = 113 bits (283), Expect = 2e-25
Identities = 52/73 (71%), Positives = 61/73 (83%)
Frame = +1
Query: 108 SNFPATHLKIGSWEYKSKYEGDLVAKCYFAKQKLVWEVLEGELKSKIEIQWSDISQLKAN 167
++FP T LKIG+WEYKS+YEGDLVAKCYFAK KLVWEVL+G LK+KIEI WSDI +KAN
Sbjct: 1 TSFPGTILKIGNWEYKSRYEGDLVAKCYFAKHKLVWEVLDGCLKNKIEIPWSDIMAIKAN 180
Query: 168 CPDDGPSTLTLMV 180
PDDGP+ +V
Sbjct: 181 YPDDGPACKKTLV 219
>TC216211
Length = 1392
Score = 95.9 bits (237), Expect = 4e-20
Identities = 61/155 (39%), Positives = 84/155 (53%), Gaps = 8/155 (5%)
Frame = +2
Query: 368 NWDQIKLPGLRPSMSMSDFLGHIEHHISKEMASGDPSFSAERLEYQQMMDGITQHLLNDN 427
N DQIKLPGL PSMSM D + HI H IS++M S +PS + + + +++ +Q+L ND+
Sbjct: 425 NLDQIKLPGLHPSMSMDDLVSHIGHCISEQMGSDNPSLAGDSQYSRSILEEFSQYLFNDS 604
Query: 428 QVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDNTGFIEGPNDAKVNIDIK-------- 479
Q T SDE+ +MSRVNSL CLLQ DP + + + T DA +
Sbjct: 605 QHATTSDEQRVMSRVNSLYCLLQKDPSSAEDINTMTKNGNSLRDANALSRVNSLYSLLQH 784
Query: 480 ATEENSRDVYGGNPAPGMSRKDSFGDLLLSLPRIA 514
A + +R+ G A MSR +S LL P A
Sbjct: 785 AEDTMTRNGNGLRDADVMSRVNSLYCLLQKDPSTA 889
Score = 65.9 bits (159), Expect = 4e-11
Identities = 45/108 (41%), Positives = 57/108 (52%), Gaps = 17/108 (15%)
Frame = +2
Query: 435 EKSLMSRVNSLRCLLQMDPPAVPNSHD---NTGFIEGPNDAKVNIDIKATEENSR----- 486
+ +MSRVNSL CLLQ DP ++ N ++ KV + T E S
Sbjct: 824 DADVMSRVNSLYCLLQKDPSTAEDTDTMTRNGNGLDADEGGKVGVSNSNTTELSACKIKV 1003
Query: 487 --------DVYGGNPA-PGMSRKDSFGDLLLSLPRIASLPKFLFDISE 525
D G GMSRK+S GDLLL+LPRIASLP+FLF++SE
Sbjct: 1004PDLEVQPDDASGCKQGVSGMSRKESAGDLLLNLPRIASLPQFLFNMSE 1147
>TC216210
Length = 437
Score = 90.1 bits (222), Expect = 2e-18
Identities = 44/83 (53%), Positives = 59/83 (71%)
Frame = +3
Query: 368 NWDQIKLPGLRPSMSMSDFLGHIEHHISKEMASGDPSFSAERLEYQQMMDGITQHLLNDN 427
N DQIKLPGL PSMSM D + HI H IS++M S +PS + + + +++ +Q+L ND+
Sbjct: 189 NLDQIKLPGLHPSMSMDDLVSHIGHCISEQMGSDNPSLAGDSQYSRSILEEFSQYLFNDS 368
Query: 428 QVTTDSDEKSLMSRVNSLRCLLQ 450
Q T SDE+ +MSRVNSL CLLQ
Sbjct: 369 QHATTSDEQRVMSRVNSLYCLLQ 437
>BM528840
Length = 406
Score = 77.8 bits (190), Expect = 1e-14
Identities = 57/138 (41%), Positives = 74/138 (53%), Gaps = 30/138 (21%)
Frame = +2
Query: 8 KNPRSQWESSSATSSISPKFEIEDSIQDQHAPLNKRHK-------------ATND----- 49
KN R +SS I +EDS++++HAPLNKR K A++D
Sbjct: 8 KNSRKPETASSVKLEI-----VEDSLEEEHAPLNKRCKPSASPLPQPQQWNASDDGSVSS 172
Query: 50 -----ILNEPSPLGLSLRKSPSLLDLIQMTLCQ-------ENSVNANTANDNLNSKANKN 97
IL+EPSPLGL LRKSPSLLDLIQM L Q + S A + ++ A +
Sbjct: 173 PSHLNILDEPSPLGLRLRKSPSLLDLIQMKLSQGSAQQKEDLSSEAKRESRCASAAAASS 352
Query: 98 GRASVEKLKASNFPATHL 115
G + +KLKASNFPA+ L
Sbjct: 353 GAGAADKLKASNFPASLL 406
>BU761515
Length = 437
Score = 73.6 bits (179), Expect = 2e-13
Identities = 47/115 (40%), Positives = 66/115 (56%), Gaps = 28/115 (24%)
Frame = +2
Query: 18 SATSSISPKFEIEDSIQDQHAPLNKRHKATN----------------------DILNEPS 55
S + ++ + E+E+ +Q++HAP NKR K+++ +IL+EPS
Sbjct: 92 SVSKTVPLRLEMEEPLQEEHAPPNKRFKSSSASQDEQWNASNGASSSSSPSQYNILDEPS 271
Query: 56 PLGLSLRKSPSLLDLIQMTLCQENSVNANTANDN-LNSKANKNGR-----ASVEK 104
PLGL LRKSPSLLDLIQM L Q N + ANT ++N ++S K R SVEK
Sbjct: 272 PLGLRLRKSPSLLDLIQMKLSQGNVIIANTQDENFISSGLKKESRGAAASGSVEK 436
>TC216208
Length = 770
Score = 64.3 bits (155), Expect = 1e-10
Identities = 42/109 (38%), Positives = 58/109 (52%), Gaps = 18/109 (16%)
Frame = +2
Query: 435 EKSLMSRVNSLRCLLQMDPPAVPNSHD---NTGFIEGPNDAKVNID-------------- 477
+ +MSRVNSL CLLQ DP +++ N ++ KV +
Sbjct: 233 DADVMSRVNSLYCLLQKDPYTAEDTNTMTRNGNGLDADEGGKVGVSNSNITELSQPCKTK 412
Query: 478 IKATEENSRDVYGGNPA-PGMSRKDSFGDLLLSLPRIASLPKFLFDISE 525
+ E + D G GMSRK+S GDLLL+LPR+ASLP+FLF++SE
Sbjct: 413 VSDLEVQADDASGCKQGGTGMSRKESAGDLLLNLPRVASLPQFLFNMSE 559
Score = 48.9 bits (115), Expect = 5e-06
Identities = 35/99 (35%), Positives = 47/99 (47%), Gaps = 5/99 (5%)
Frame = +2
Query: 421 QHLLNDNQVTTDSDEKSLMSRVNSLRCLLQMDPPAVPNSHDNTGFIEGPNDAKVNIDIKA 480
Q+L ND+Q T SDE+ +MS+VNSL CLLQ DP + N + N + +
Sbjct: 2 QYLFNDSQHATTSDEQRVMSKVNSLYCLLQKDPSTAEDMTKNGNSLGDANALSRVNSLYS 181
Query: 481 TEENSRDVYGGN-----PAPGMSRKDSFGDLLLSLPRIA 514
+N+ D N A MSR +S LL P A
Sbjct: 182 LLQNAEDAMARNGNGLRDADVMSRVNSLYCLLQKDPYTA 298
>BI967423 similar to GP|14532460|gb AT5g13810/MAC12_24 {Arabidopsis
thaliana}, partial (45%)
Length = 683
Score = 30.8 bits (68), Expect = 1.5
Identities = 17/39 (43%), Positives = 24/39 (60%)
Frame = -2
Query: 286 QNMGSPHSSLSPSFTTEHSDPSAITLDSVPCEAPSSSSA 324
++ S H L PS + E+SDPS +LDS P ++ SS A
Sbjct: 580 KSSSSEHQKLKPSKSIENSDPSVKSLDS-PAKSIDSSPA 467
>BU090296 homologue to GP|14532460|gb| AT5g13810/MAC12_24 {Arabidopsis
thaliana}, partial (15%)
Length = 395
Score = 30.8 bits (68), Expect = 1.5
Identities = 17/39 (43%), Positives = 24/39 (60%)
Frame = +2
Query: 286 QNMGSPHSSLSPSFTTEHSDPSAITLDSVPCEAPSSSSA 324
++ S H L PS + E+SDPS +LDS P ++ SS A
Sbjct: 179 KSSSSEHQKLKPSKSIENSDPSVKSLDS-PAKSIDSSPA 292
>BG507560 similar to GP|1838976|emb| vsf-1 {Lycopersicon esculentum}, partial
(12%)
Length = 435
Score = 30.8 bits (68), Expect = 1.5
Identities = 26/93 (27%), Positives = 40/93 (42%)
Frame = +3
Query: 12 SQWESSSATSSISPKFEIEDSIQDQHAPLNKRHKATNDILNEPSPLGLSLRKSPSLLDLI 71
S ++ S + SIS E+S+ + HAPL R A + P G S S L +
Sbjct: 111 SPYKEPSFSDSISTDVSAEESLANSHAPLPNRGHALQLGHSLPPRKGHRRSSSDSPLGIA 290
Query: 72 QMTLCQENSVNANTANDNLNSKANKNGRASVEK 104
S ++ T +D N A++ G + EK
Sbjct: 291 DFIQSVPQSGSSKTWSDREN-LASRGGNSGFEK 386
>TC229432
Length = 1215
Score = 30.4 bits (67), Expect = 2.0
Identities = 28/108 (25%), Positives = 47/108 (42%), Gaps = 6/108 (5%)
Frame = +2
Query: 6 GSKNPRSQWESSSATSSISPKFEIEDSIQDQHAPLNKRHKATNDILNEPS-PLGLSL--- 61
G+ N + ++ +SS+ + H + HK TN + +P+ P G+S
Sbjct: 152 GNANWSTPLNNNHGSSSVQKNHSSNSA--GNHPSTSVEHKVTNTVQKQPTPPKGISFINL 325
Query: 62 -RKSPSLLDLIQMTL-CQENSVNANTANDNLNSKANKNGRASVEKLKA 107
+ SPSL L Q T+ +E+ V T D + +N +KL A
Sbjct: 326 AKLSPSLSSLKQSTVTTKESPVQIGTREDQSSFHKTQNKVDIPKKLPA 469
>TC222189 similar to UP|ITBX_DROME (P11584) Integrin beta-PS precursor
(Position-specific antigen, beta chain) (Myospheroid
protein) (Olfactory C protein), partial (3%)
Length = 1477
Score = 30.4 bits (67), Expect = 2.0
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 2/71 (2%)
Frame = +3
Query: 6 GSKNPRSQWESSSATSSISPKFEIEDSIQDQHAPLNKRHKATNDILNEPSPLGLSLR--K 63
G + +W+S S + +D+ LNKR+K NDIL + G S R +
Sbjct: 435 GLMQKKGKWKSVSRAMMEKGFYVSPQQCEDKFNDLNKRYKRVNDILGK----GTSCRVVE 602
Query: 64 SPSLLDLIQMT 74
+ SLLD + ++
Sbjct: 603 NQSLLDSMDLS 635
>TC208992 weakly similar to UP|O04925 (O04925) 15.5 kDa oleosin, partial
(55%)
Length = 937
Score = 29.3 bits (64), Expect = 4.5
Identities = 21/56 (37%), Positives = 28/56 (49%)
Frame = +2
Query: 253 SPHFDPRSAAIENPHNLKDCDLHQGNGSAVSCFQNMGSPHSSLSPSFTTEHSDPSA 308
SP P SA+ +PH+ + SA SC P+SSLSPS + H + SA
Sbjct: 143 SPSPPPSSASPSSPHS--------SSFSAPSCSPR---PYSSLSPSPASSHPESSA 277
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.322 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,637,995
Number of Sequences: 63676
Number of extensions: 361882
Number of successful extensions: 1866
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 1835
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1856
length of query: 525
length of database: 12,639,632
effective HSP length: 102
effective length of query: 423
effective length of database: 6,144,680
effective search space: 2599199640
effective search space used: 2599199640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC126784.4


