

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC121237.19 + phase: 0 /pseudo
(754 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
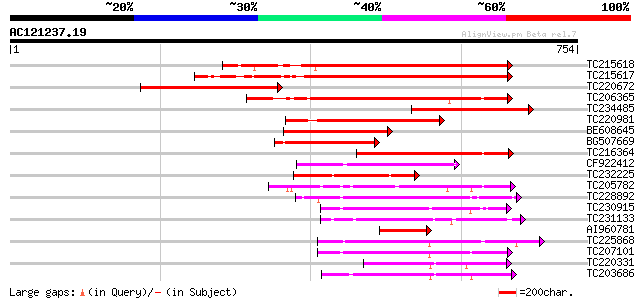
Score E
Sequences producing significant alignments: (bits) Value
TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 428 e-120
TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase ... 425 e-119
TC220672 similar to UP|Q6UY78 (Q6UY78) YDA, partial (15%) 340 1e-93
TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, parti... 320 1e-87
TC234485 similar to UP|Q6UY78 (Q6UY78) YDA, partial (16%) 303 2e-82
TC220981 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, parti... 230 2e-60
BE608645 224 1e-58
BG507669 207 1e-53
TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%) 166 3e-41
CF922412 154 1e-37
TC232225 weakly similar to UP|ST11_YEAST (P23561) Serine/threoni... 135 7e-32
TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, pa... 133 3e-31
TC228892 UP|Q9XF25 (Q9XF25) SNF-1-like serine/threonine protein ... 132 6e-31
TC230915 weakly similar to PIR|T10690|T10690 serine/threonine-sp... 129 6e-30
TC231133 similar to UP|Q7X8K7 (Q7X8K7) CTR1-like kinase kinase k... 128 8e-30
AI960781 127 1e-29
TC225868 UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, complete 127 2e-29
TC207101 similar to UP|O24342 (O24342) Serine/threonine kinase ,... 127 2e-29
TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partia... 125 9e-29
TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete 124 1e-28
>TC215618 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (73%)
Length = 2291
Score = 428 bits (1100), Expect = e-120
Identities = 222/398 (55%), Positives = 276/398 (68%), Gaps = 13/398 (3%)
Frame = +1
Query: 284 HCSSPGSGHNSGHNSMGGDMSGPLFWQPSRGSPEYSPIPSP---------RMTSPGPSSR 334
H GS SG +S+ S F SP +P P+ R SPGP SR
Sbjct: 262 HADPLGSVSVSGGSSVSSSTS---FDDHPISSPNTNPNPNRGHDEVRVNVRSKSPGPGSR 432
Query: 335 IQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSM 394
+ +P+H R +S TG Q H LPLPP + TSPS
Sbjct: 433 GPTSPTSPLHQRLNNLSLDSPTG------SQCHPLPLPP-------------GSPTSPSS 555
Query: 395 PRSPARADSPM----SSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAK 450
S ARA+ + S+ S+W+KGKLLGRGTFGHVY+GFNS++G+MCA+KEV + DD
Sbjct: 556 VLSNARANGHLENATSNVSKWRKGKLLGRGTFGHVYLGFNSENGQMCAIKEVKVVFDDHT 735
Query: 451 SLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGE 510
S E KQL QE++LL++L HPNIVQY+GSE V++ L +YLEYVSGGSIHKLLQEYG F E
Sbjct: 736 SKECLKQLNQEINLLNQLSHPNIVQYHGSELVEESLSVYLEYVSGGSIHKLLQEYGPFKE 915
Query: 511 LAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSF 570
I++YT+QI+SGLAYLH +NT+HRDIKGANILVDPNG +K+ADFGMAKHI LSF
Sbjct: 916 PVIQNYTRQIVSGLAYLHGRNTVHRDIKGANILVDPNGEIKLADFGMAKHINSSASMLSF 1095
Query: 571 KGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELP 630
KGSPYWMAPEV+ N+ SL VDIWSLGCT++EMAT+KPPW+QYEGVAA+FKIGNSK++P
Sbjct: 1096KGSPYWMAPEVVMNTNGYSLPVDIWSLGCTIIEMATSKPPWNQYEGVAAIFKIGNSKDMP 1275
Query: 631 TIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVK 668
IP+HLSN+ K F++ CLQR+P RP+A +LLDHPF++
Sbjct: 1276EIPEHLSNDAKKFIKLCLQRDPLARPTAQKLLDHPFIR 1389
>TC215617 similar to UP|Q75PK5 (Q75PK5) Mitogen-activated kinase kinase
kinase alpha, partial (51%)
Length = 1451
Score = 425 bits (1092), Expect = e-119
Identities = 227/426 (53%), Positives = 290/426 (67%), Gaps = 4/426 (0%)
Frame = +3
Query: 247 LSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGS--GHNSGHNSMGGDMS 304
L PS S ++FG +Q L F +G V SS GS H + H+ +
Sbjct: 255 LPRPSVSSTQSFGIEQGL--VFGSGS-------VSGSSVSSSGSYDDHPTCHSQINA--- 398
Query: 305 GPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQ 364
SRG + R SPGP SR + +P+HP+ +S TGR +
Sbjct: 399 -------SRGQGDTKFYA--RSKSPGPGSRGPTSPTSPLHPKLHVLSLDSPTGRQEG--- 542
Query: 365 QSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSG--SRWKKGKLLGRGTF 422
+ H LPLPP + TSP S S+P + A + ++G S+WKKGKLLGRGTF
Sbjct: 543 ECHPLPLPP--GSPTSP---------SSSLPSTRANGMTEHTTGNLSKWKKGKLLGRGTF 689
Query: 423 GHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETV 482
GHVY+GFNS SG++CA+KEV + DD S E KQL QE+HLLS+L HPNIVQYYGS+
Sbjct: 690 GHVYLGFNSDSGQLCAIKEVRVVCDDQSSKECLKQLNQEIHLLSQLSHPNIVQYYGSDLG 869
Query: 483 DDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANI 542
++ L +YLEYVSGGSIHKLLQEYG F E I++YT+QI+SGL+YLH +NT+HRDIKGANI
Sbjct: 870 EETLSVYLEYVSGGSIHKLLQEYGAFKEPVIQNYTRQIVSGLSYLHGRNTVHRDIKGANI 1049
Query: 543 LVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVL 602
LVDPNG +K+ADFGMAKHI LSFKGSPYWMAPEV+ N+ SL VDIWSLGCT+L
Sbjct: 1050LVDPNGEIKLADFGMAKHINSSSSMLSFKGSPYWMAPEVVMNTNGYSLPVDIWSLGCTIL 1229
Query: 603 EMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELL 662
EMAT+KPPW+QYEG AA+FKIGNS+++P IPDHLS++ K+F++ CLQR+P RP+A +L+
Sbjct: 1230EMATSKPPWNQYEGAAAIFKIGNSRDMPEIPDHLSSDAKNFIQLCLQRDPSARPTAQKLI 1409
Query: 663 DHPFVK 668
+HPF++
Sbjct: 1410EHPFIR 1427
Score = 29.3 bits (64), Expect = 6.8
Identities = 23/90 (25%), Positives = 36/90 (39%)
Frame = +3
Query: 1 MPTWWGKSSSKETKKKAGKESIIDTLHRKFKFPSEGKRSTISGGSRRRSNDTISEKGDRS 60
MPTWWGK SSK ++ ++ H G +++ + + N ++ +S
Sbjct: 6 MPTWWGKKSSKSKDQQPQEDH-----HHPPHGGGGGAATSVLNFNFNKLNKPRPDRPSKS 170
Query: 61 PSESRSPSPSKVARCQSFAERPHAQPLPLP 90
+ SP S A Q LPLP
Sbjct: 171FDDVVRNSPRCSRDFSSSAAAAVDQGLPLP 260
>TC220672 similar to UP|Q6UY78 (Q6UY78) YDA, partial (15%)
Length = 570
Score = 340 bits (872), Expect = 1e-93
Identities = 162/189 (85%), Positives = 170/189 (89%)
Frame = +3
Query: 175 GTRTAAGSPSSLVLKDQSSAVSQPNLREVKKTANILSNHTPSTSPKRKPLRHHVPNLQVP 234
GTRTAAGSPSSL+ KDQSS VSQ N RE KK ANIL NH STSPKR+PL +HV NLQ+P
Sbjct: 3 GTRTAAGSPSSLMQKDQSSTVSQINSREAKKPANILGNHMSSTSPKRRPLSNHVTNLQIP 182
Query: 235 PHGVFYSGPDSSLSSPSRSPLRAFGTDQVLNSAFWAGKPYPEINFVGSGHCSSPGSGHNS 294
PHG F+S PDSS SSPSRSPLRAFGT+QVLNSAFWAGKPYPE+NF GSGHCSSPGSGHNS
Sbjct: 183 PHGAFFSAPDSSRSSPSRSPLRAFGTEQVLNSAFWAGKPYPEVNFGGSGHCSSPGSGHNS 362
Query: 295 GHNSMGGDMSGPLFWQPSRGSPEYSPIPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPTES 354
GHNSMGGDMSG LFWQPSRGSPEYSP+PSPRMTSPGPSSRIQSGAVTPIHPRAGGTP ES
Sbjct: 363 GHNSMGGDMSGQLFWQPSRGSPEYSPVPSPRMTSPGPSSRIQSGAVTPIHPRAGGTPNES 542
Query: 355 QTGRADDGK 363
QTGR DD K
Sbjct: 543 QTGRIDDVK 569
>TC206365 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, partial (37%)
Length = 1866
Score = 320 bits (821), Expect = 1e-87
Identities = 170/363 (46%), Positives = 232/363 (63%), Gaps = 9/363 (2%)
Frame = +1
Query: 315 SPEYSPIPSP--RMTSPGPSSRIQSGAVTPIHPRAGGTPTESQTGRADDGKQQSHRLPLP 372
SP+ SP+ SP + G S V P + P H LPLP
Sbjct: 205 SPDRSPLRSPGSHLNQEGSQLHKFSSRVWPENNHVDANP---------------HPLPLP 339
Query: 373 PLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQ 432
P SP + +S PS+ ++ S +W+KGKL+GRG++G VY N +
Sbjct: 340 P----KASPQTAHSSPQHQPSIVH--LNTENLPSMKGQWQKGKLIGRGSYGSVYHATNLE 501
Query: 433 SGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEY 492
+G CA+KEV LF DD KS + KQL QE+ +L +L HPNIVQYYGSE V D+LYIY+EY
Sbjct: 502 TGASCALKEVDLFPDDPKSADCIKQLEQEIRILRQLHHPNIVQYYGSEIVGDRLYIYMEY 681
Query: 493 VSGGSIHKLLQEY-GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVK 551
V GS+HK + E+ G E +R++T+ ILSGLAYLH T+HRDIKGAN+LVD +G VK
Sbjct: 682 VHPGSLHKFMHEHCGAMTESVVRNFTRHILSGLAYLHGTKTIHRDIKGANLLVDASGSVK 861
Query: 552 VADFGMAKHITGQYCPLSFKGSPYWMAPEVIK------NSKECSLGVDIWSLGCTVLEMA 605
+ADFG++K +T + LS KGSPYWMAPE++K +S + ++ +DIWSLGCT++EM
Sbjct: 862 LADFGVSKILTEKSYELSLKGSPYWMAPELMKAAIKKESSPDIAMAIDIWSLGCTIIEML 1041
Query: 606 TTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHP 665
T KPPWS++EG AMFK+ + P +P+ LS+EG+DF+++C +RNP +RPSA+ LL H
Sbjct: 1042TGKPPWSEFEGPQAMFKVLHKS--PDLPESLSSEGQDFLQQCFRRNPAERPSAAVLLTHA 1215
Query: 666 FVK 668
FV+
Sbjct: 1216FVQ 1224
>TC234485 similar to UP|Q6UY78 (Q6UY78) YDA, partial (16%)
Length = 543
Score = 303 bits (776), Expect = 2e-82
Identities = 144/162 (88%), Positives = 149/162 (91%)
Frame = +1
Query: 535 RDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDI 594
RDIKGANILVDP GRVK+ADFGMAKHITGQ CPLSFKG+PYWMAPEVIKNS C+L VDI
Sbjct: 58 RDIKGANILVDPTGRVKLADFGMAKHITGQSCPLSFKGTPYWMAPEVIKNSNGCNLAVDI 237
Query: 595 WSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRD 654
WSLGCTVLEMATTKPPW QYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNP D
Sbjct: 238 WSLGCTVLEMATTKPPWFQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPHD 417
Query: 655 RPSASELLDHPFVKGAAPLERPIMVPEASDPITGITHGTKAL 696
RPSASELLDHPFVK AAPLERPI PEA DP++GIT G KAL
Sbjct: 418 RPSASELLDHPFVKNAAPLERPIPAPEALDPVSGITQGAKAL 543
>TC220981 similar to UP|Q93ZH4 (Q93ZH4) AT5g66850/MUD21_11, partial (27%)
Length = 646
Score = 230 bits (587), Expect = 2e-60
Identities = 121/215 (56%), Positives = 149/215 (69%), Gaps = 3/215 (1%)
Frame = +1
Query: 367 HRLPLPPLTVTNTSPFS--HSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGH 424
H LPLPP + P + S++ A S S P S+WKKGKL+GRGTFG
Sbjct: 34 HPLPLPPGAALTSPPAAATFSHAVAKSESFPMK-----------SQWKKGKLIGRGTFGS 180
Query: 425 VYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDD 484
VY+ N ++G +CAMKEV LF DD KS E KQL QE+ +LS L+H NIVQYYGSE +D
Sbjct: 181 VYVATNRETGALCAMKEVELFPDDPKSAECIKQLEQEIKVLSNLKHSNIVQYYGSEIEED 360
Query: 485 KLYIYLEYVSGGSIHKLLQEY-GQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANIL 543
+LYIYLEY GSI+K ++E+ G E IR++T+ ILSGLAYLH+K T+HRDIKGAN+L
Sbjct: 361 RLYIYLEYDHPGSINKYVREHCGAITESVIRNFTRHILSGLAYLHSKKTIHRDIKGANLL 540
Query: 544 VDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMA 578
VD G VK+ADFGMAKH+T LS +GSPYWMA
Sbjct: 541 VDSAGVVKLADFGMAKHLTAFEANLSLRGSPYWMA 645
>BE608645
Length = 437
Score = 224 bits (570), Expect = 1e-58
Identities = 104/145 (71%), Positives = 127/145 (86%)
Frame = +3
Query: 365 QSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGH 424
++H+L +PP+T T + PFS + SA T+PS PRSP R+++ S GSRWKKG+LLGRGTFGH
Sbjct: 3 KNHQLAIPPITATKSCPFSPTYSALTTPSAPRSPGRSENSSSPGSRWKKGQLLGRGTFGH 182
Query: 425 VYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDD 484
VY+GFN + GEMCAMKEVTLFSDDAKS ESA+QL QE+ +LS+LRHPNIVQYYGSETVDD
Sbjct: 183 VYLGFNRECGEMCAMKEVTLFSDDAKSRESAQQLGQEIAMLSQLRHPNIVQYYGSETVDD 362
Query: 485 KLYIYLEYVSGGSIHKLLQEYGQFG 509
+LY+YLEYVSGGSI+KL++EYGQ G
Sbjct: 363 RLYVYLEYVSGGSIYKLVKEYGQLG 437
>BG507669
Length = 420
Score = 207 bits (527), Expect = 1e-53
Identities = 105/139 (75%), Positives = 118/139 (84%)
Frame = +2
Query: 353 ESQTGRADDGKQQSHRLPLPPLTVTNTSPFSHSNSAATSPSMPRSPARADSPMSSGSRWK 412
ES T R DD KQ +HRLPLPP+T+ N PFS + SA+T+PS PRSP+ A + GSRWK
Sbjct: 5 ESPTRRPDDVKQ-THRLPLPPITIPNHCPFSPTYSASTTPSAPRSPSIA*NLTYPGSRWK 181
Query: 413 KGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPN 472
KG+LLGRGTFGHVY+GFNS+SGEMCAMKEVTLFSDDAKS ESA+QL QE+ LLS LRHPN
Sbjct: 182 KGQLLGRGTFGHVYLGFNSESGEMCAMKEVTLFSDDAKSRESAQQLGQEIALLSHLRHPN 361
Query: 473 IVQYYGSETVDDKLYIYLE 491
IVQYYGSETVDDKLYIYLE
Sbjct: 362 IVQYYGSETVDDKLYIYLE 418
>TC216364 similar to UP|Q9MAI7 (Q9MAI7) F12M16.4, partial (58%)
Length = 2224
Score = 166 bits (421), Expect = 3e-41
Identities = 88/210 (41%), Positives = 128/210 (60%), Gaps = 1/210 (0%)
Frame = +1
Query: 462 VHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQIL 521
+ +LS+ R P I +YYGS KL+I +EY++GGS+ L+Q E++I + +L
Sbjct: 1 ISVLSQCRCPYITEYYGSYLNQTKLWIIMEYMAGGSVADLIQSGPPLDEMSIACILRDLL 180
Query: 522 SGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYC-PLSFKGSPYWMAPE 580
+ YLH++ +HRDIK ANIL+ NG VKVADFG++ +T +F G+P+WMAPE
Sbjct: 181 HAVDYLHSEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPE 360
Query: 581 VIKNSKECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEG 640
VI+N+ + DIWSLG T +EMA +PP + + +F I P + DH S
Sbjct: 361 VIQNTDGYNEKADIWSLGITAIEMAKGEPPLADLHPMRVLFIIPRENP-PQLDDHFSRPL 537
Query: 641 KDFVRKCLQRNPRDRPSASELLDHPFVKGA 670
K+FV CL++ P +RPSA ELL F++ A
Sbjct: 538 KEFVSLCLKKVPAERPSAKELLKDRFIRNA 627
>CF922412
Length = 701
Score = 154 bits (389), Expect = 1e-37
Identities = 86/220 (39%), Positives = 128/220 (58%), Gaps = 3/220 (1%)
Frame = -2
Query: 382 FSHSNSAATSPSMPRSPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKE 441
FS S+ +T S + + + +++ G +G+G +G VY G + ++G+ A+K+
Sbjct: 649 FSFSDRRSTEMSRQTTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQ 470
Query: 442 VTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIHKL 501
V+L + + E +MQE+ LL L H NIV+Y GS L+I LEYV GS+ +
Sbjct: 469 VSL---ENIAQEDLNIIMQEIDLLKNLNHKNIVKYLGSSKTKSHLHIVLEYVENGSLANI 299
Query: 502 LQ--EYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAK 559
++ ++G F E + Y Q+L GL YLH + +HRDIKGANIL G VK+ADFG+A
Sbjct: 298 IKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVAT 119
Query: 560 HIT-GQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLG 598
+T S G+PYWMAPEVI+ + C+ DIWS+G
Sbjct: 118 KLTEADVNTHSVVGTPYWMAPEVIEMAGVCAAS-DIWSVG 2
>TC232225 weakly similar to UP|ST11_YEAST (P23561) Serine/threonine-protein
kinase STE11 , partial (6%)
Length = 802
Score = 135 bits (340), Expect = 7e-32
Identities = 78/169 (46%), Positives = 103/169 (60%), Gaps = 1/169 (0%)
Frame = +3
Query: 378 NTSPFSHSNSAATSPSMPR-SPARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEM 436
+TS S+S+ T P+ SP + + W+KG+ LG G+FG VY G S G
Sbjct: 294 STSNEDDSSSSTTDPTSNNISPQGRIKRIITAESWQKGEFLGGGSFGSVYEGI-SDDGFF 470
Query: 437 CAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGG 496
A+KEV+L + +S QL QE+ LLS+ H NIVQYYG+E KLYI+LE V+ G
Sbjct: 471 FAVKEVSLLDQGTQGKQSVYQLEQEIALLSQFEHENIVQYYGTEMDQSKLYIFLELVTKG 650
Query: 497 SIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVD 545
S+ L Q+Y + + SYT+QIL GL YLH +N +HRDIK ANILVD
Sbjct: 651 SLRSLYQKY-TLRDSQVSSYTRQILHGLKYLHDRNVVHRDIKCANILVD 794
>TC205782 similar to UP|Q93WR8 (Q93WR8) MEK map kinase kinsae, partial (91%)
Length = 1377
Score = 133 bits (334), Expect = 3e-31
Identities = 105/344 (30%), Positives = 176/344 (50%), Gaps = 16/344 (4%)
Frame = +2
Query: 345 PRAGGTPTESQTGRADDGKQQSHR---LPLPP----LTVTNTSPFSHSNSAATSPSMPRS 397
P +G + T + D Q+ + LPLP L V P S + +AA S +
Sbjct: 71 PPSGSGANPAPTNNSKDRPQRRRKDLTLPLPQRDTNLAVPLPLPPSTAPAAAASGGASQQ 250
Query: 398 PARADSPMSSGSRWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQ 457
A+ P S R + +G G+ G VY + SG + A+K ++ +S+ +Q
Sbjct: 251 AAQQVIPFSELERLNR---IGSGSGGTVYKVVHRTSGRVYALK--VIYGHHEESVR--RQ 409
Query: 458 LMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLEYVSGGSIH-KLLQEYGQFGELAIRSY 516
+ +E+ +L + N+V+ + + ++ + LE++ GGS+ K + + Q +L+
Sbjct: 410 IHREIQILRDVDDANVVKCHEMYDQNSEIQVLLEFMDGGSLEGKHITQEQQLADLS---- 577
Query: 517 TQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVKVADFGMAKHITGQYCPL-SFKGSPY 575
+QIL GLAYLH ++ +HRDIK +N+L++ +VK+ADFG+ + + P S G+
Sbjct: 578 -RQILRGLAYLHRRHIVHRDIKPSNLLINSRKQVKIADFGVGRILNQTMDPCNSSVGTIA 754
Query: 576 WMAPE----VIKNSKECSLGVDIWSLGCTVLEMATTKPPWS---QYEGVAAMFKIGNSKE 628
+M+PE I + + + DIWS G ++LE + P++ Q + + M I S+
Sbjct: 755 YMSPERINTDINDGQYDAYAGDIWSFGVSILEFYMGRFPFAVGRQGDWASLMCAICMSQP 934
Query: 629 LPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAAP 672
P P S KDF+ +CLQR+P R SAS LL+HPF+ P
Sbjct: 935 -PEAPPSASPHFKDFILRCLQRDPSRRWSASRLLEHPFIAPPLP 1063
>TC228892 UP|Q9XF25 (Q9XF25) SNF-1-like serine/threonine protein kinase,
complete
Length = 1931
Score = 132 bits (332), Expect = 6e-31
Identities = 91/309 (29%), Positives = 149/309 (47%), Gaps = 9/309 (2%)
Frame = +1
Query: 381 PFSHSNSAATSPSMPRSPARADSPMSSGS---------RWKKGKLLGRGTFGHVYIGFNS 431
PFS S + +P RS + D G +K GK LG G+FG V I +
Sbjct: 22 PFSFSIHHS-NPRTLRSITKMDRSTGRGGGGSVDMFLRNYKLGKTLGIGSFGKVKIAEHV 198
Query: 432 QSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIVQYYGSETVDDKLYIYLE 491
++G A+K L K++E +++ +E+ +L H +I++ Y +Y+ +E
Sbjct: 199 RTGHKVAIK--ILNRHKIKNMEMEEKVRREIKILRLFMHHHIIRLYEVVETPTDIYVVME 372
Query: 492 YVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVK 551
YV G + + E G+ E R + QQI+SG+ Y H +HRD+K N+L+D +K
Sbjct: 373 YVKSGELFDYIVEKGRLQEDEARHFFQQIISGVEYCHRNMVVHRDLKPENLLLDSKFNIK 552
Query: 552 VADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVDIWSLGCTVLEMATTKPPW 611
+ADFG++ + + + GSP + APEVI VD+WS G + + P+
Sbjct: 553 IADFGLSNIMRDGHFLKTSCGSPNYAAPEVISGKLYAGPEVDVWSCGVILYALLCGTLPF 732
Query: 612 SQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQRNPRDRPSASELLDHPFVKGAA 671
E + +FK + T+P HLS +D + + L +P R + E+ HP+ +
Sbjct: 733 DD-ENIPNLFK-KIKGGIYTLPSHLSPGARDLIPRMLVVDPMKRMTIPEIRQHPWFQ--V 900
Query: 672 PLERPIMVP 680
L R + VP
Sbjct: 901 HLPRYLAVP 927
>TC230915 weakly similar to PIR|T10690|T10690 serine/threonine-specific
protein kinase homolog T16I18.40 - Arabidopsis thaliana
{Arabidopsis thaliana;} , partial (90%)
Length = 1174
Score = 129 bits (323), Expect = 6e-30
Identities = 82/257 (31%), Positives = 136/257 (52%), Gaps = 3/257 (1%)
Frame = +3
Query: 414 GKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNI 473
GK LGRG FG VY+ +S + A+K +F + QL +E+ + + LRH NI
Sbjct: 87 GKPLGRGKFGRVYVAREVKSKFVVALK--VIFKEQIDKYRVHHQLRREMEIQTSLRHANI 260
Query: 474 VQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTL 533
++ YG D++++ LEY G ++K L++ G E +Y + LAY H K+ +
Sbjct: 261 LRLYGGFHDADRVFLILEYAHKGELNKELRKKGHLTEKQAATYIVSLTKALAYCHEKHVI 440
Query: 534 HRDIKGANILVDPNGRVKVADFGMAKHITGQYCPLSFKGSPYWMAPEVIKNSKECSLGVD 593
HRDIK N+L D GR+K+ADFG + + + + G+ ++APE+ +N K VD
Sbjct: 441 HRDIKPENLLPDHEGRLKIADFGWS--VQSRSKRHTMCGTLDYLAPEMAEN-KAHDYAVD 611
Query: 594 IWSLGCTVLEMATTKPPW---SQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRKCLQR 650
W+LG E PP+ SQ + + K+ S P+ P +S E K+ + + L +
Sbjct: 612 NWTLGIRCYESLYGAPPFEAESQSDTFKRIMKVDLS--FPSTPS-VSIEAKNLISRLLVK 782
Query: 651 NPRDRPSASELLDHPFV 667
+ R S ++++HP++
Sbjct: 783 DSSRRLSLQKIMEHPWI 833
>TC231133 similar to UP|Q7X8K7 (Q7X8K7) CTR1-like kinase kinase kinase,
partial (29%)
Length = 1061
Score = 128 bits (322), Expect = 8e-30
Identities = 90/281 (32%), Positives = 141/281 (50%), Gaps = 8/281 (2%)
Frame = +1
Query: 414 GKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNI 473
G+ +G G++G VY G G A+K+ + LE K EV ++ RLRHPN+
Sbjct: 34 GERIGLGSYGEVYRG--EWHGTEVAVKKFLYQDISGELLEEFKS---EVQIMKRLRHPNV 198
Query: 474 VQYYGSETVDDKLYIYLEYVSGGSIHKLLQE-YGQFGELAIRSYTQQILSGLAYLH--AK 530
V + G+ T L I E++ GS+++L+ Q E G+ YLH
Sbjct: 199 VLFMGAVTRPPNLSIVSEFLPRGSLYRLIHRPNNQLDERRRLRMALDAARGMNYLHNCTP 378
Query: 531 NTLHRDIKGANILVDPNGRVKVADFGMA--KHITGQYCPLSFKGSPYWMAPEVIKNS--- 585
+HRD+K N+LVD N VKV DFG++ KH T S G+ WMAPEV++N
Sbjct: 379 VIVHRDLKSPNLLVDKNWVVKVCDFGLSRMKHSTF-LSSRSTAGTAEWMAPEVLRNELSD 555
Query: 586 KECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVR 645
++C D++S G + E++T + PW + + +G IPD++ D +R
Sbjct: 556 EKC----DVFSYGVILWELSTLQQPWGGMNPMQVVGAVGFQHRRLDIPDNVDPAIADIIR 723
Query: 646 KCLQRNPRDRPSASELLDHPFVKGAAPLERPIMVPEASDPI 686
+C Q +P+ RP+ +E++ PL++PI V + PI
Sbjct: 724 QCWQTDPKLRPTFAEIM-----AALKPLQKPITVSQVHRPI 831
>AI960781
Length = 212
Score = 127 bits (320), Expect = 1e-29
Identities = 62/70 (88%), Positives = 67/70 (95%)
Frame = +1
Query: 492 YVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHAKNTLHRDIKGANILVDPNGRVK 551
+ +GGSI+KLLQEYGQ GELAIRSYTQQILSGLAYLHAKNT+HRDIKGANILVD NGRVK
Sbjct: 1 FPAGGSIYKLLQEYGQVGELAIRSYTQQILSGLAYLHAKNTVHRDIKGANILVDTNGRVK 180
Query: 552 VADFGMAKHI 561
+ADFGMAKHI
Sbjct: 181 LADFGMAKHI 210
>TC225868 UP|Q8LK24 (Q8LK24) SOS2-like protein kinase, complete
Length = 2261
Score = 127 bits (318), Expect = 2e-29
Identities = 88/317 (27%), Positives = 154/317 (47%), Gaps = 15/317 (4%)
Frame = +2
Query: 410 RWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLR 469
+++ G+LLG GTF VY + ++G+ AMK V + + +Q+ +E+ ++ ++
Sbjct: 485 KYELGRLLGHGTFAKVYHARHLKTGKSVAMKVVG--KEKVVKVGMMEQIKREISAMNMVK 658
Query: 470 HPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHA 529
HPNIVQ + K+YI +E V GG + + G+ E R Y QQ++S + + H+
Sbjct: 659 HPNIVQLHEVMASKSKIYIAMELVRGGELFNKIAR-GRLREEMARLYFQQLISAVDFCHS 835
Query: 530 KNTLHRDIKGANILVDPNGRVKVADFGM---AKHITGQYCPLSFKGSPYWMAPEVIKNSK 586
+ HRD+K N+L+D +G +KV DFG+ ++H+ + G+P ++APEVI
Sbjct: 836 RGVYHRDLKPENLLLDDDGNLKVTDFGLSTFSEHLRHDGLLHTTCGTPAYVAPEVIGKRG 1015
Query: 587 ECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKI--GNSKELPTIPDHLSNEGKDFV 644
DIWS G + + P+ VA KI G+ K P S+E + +
Sbjct: 1016YDGAKADIWSCGVILYVLLAGFLPFQDDNLVALYKKIYRGDFK----CPPWFSSEARRLI 1183
Query: 645 RKCLQRNPRDRPSASELLDHPFVKGAAP----------LERPIMVPEASDPITGITHGTK 694
K L NP R + S+++D + K P L+ + + ++ +
Sbjct: 1184TKLLDPNPNTRITISKIMDSSWFKKPVPKNLMGKKREELDLEEKIKQHEQEVSTTMNAFH 1363
Query: 695 ALGIGQGRNLSALDSDK 711
+ + +G +LS L +K
Sbjct: 1364IISLSEGFDLSPLFEEK 1414
>TC207101 similar to UP|O24342 (O24342) Serine/threonine kinase , partial
(98%)
Length = 1719
Score = 127 bits (318), Expect = 2e-29
Identities = 75/262 (28%), Positives = 137/262 (51%), Gaps = 3/262 (1%)
Frame = +1
Query: 410 RWKKGKLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLR 469
+++ G+ +G GTF V NS++GE A+K L + + A+Q+ +EV + ++
Sbjct: 217 KYEVGRTIGEGTFAKVKFARNSETGEPVALK--ILDKEKVLKHKMAEQIRREVATMKLIK 390
Query: 470 HPNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLHA 529
HPN+V+ Y K+YI LE+V+GG + + +G+ E R Y QQ+++ + Y H+
Sbjct: 391 HPNVVRLYEVMGSKTKIYIVLEFVTGGELFDKIVNHGRMSENEARRYFQQLINAVDYCHS 570
Query: 530 KNTLHRDIKGANILVDPNGRVKVADFG---MAKHITGQYCPLSFKGSPYWMAPEVIKNSK 586
+ HRD+K N+L+D G +KV+DFG +++ + + G+P ++APEV+ +
Sbjct: 571 RGVYHRDLKPENLLLDTYGNLKVSDFGLSALSQQVRDDGLLHTTCGTPNYVAPEVLNDRG 750
Query: 587 ECSLGVDIWSLGCTVLEMATTKPPWSQYEGVAAMFKIGNSKELPTIPDHLSNEGKDFVRK 646
D+WS G + + P+ + ++K ++ E T P LS + + +
Sbjct: 751 YDGATADLWSCGVILFVLVAGYLPFDD-PNLMNLYKKISAAEF-TCPPWLSFTARKLITR 924
Query: 647 CLQRNPRDRPSASELLDHPFVK 668
L +P R + E+LD + K
Sbjct: 925 ILDPDPTTRITIPEILDDEWFK 990
>TC220331 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, partial (62%)
Length = 669
Score = 125 bits (313), Expect = 9e-29
Identities = 77/208 (37%), Positives = 120/208 (57%), Gaps = 11/208 (5%)
Frame = +1
Query: 471 PNIVQYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYLH-A 529
P +V Y S + + I LEY+ GGS+ LL + E + + +Q+L GL YLH A
Sbjct: 7 PYVVVCYNSFYHNGVISIILEYMDGGSLEDLLSKVKTIPESYLSAICKQVLKGLMYLHYA 186
Query: 530 KNTLHRDIKGANILVDPNGRVKVADFGMA---KHITGQYCPLSFKGSPYWMAPE-VIKNS 585
K+ +HRD+K +N+L++ G VK+ DFG++ ++ +GQ +F G+ +M+PE +I N
Sbjct: 187 KHIIHRDLKPSNLLINHRGEVKITDFGVSVIMENTSGQ--ANTFIGTYSYMSPERIIGNQ 360
Query: 586 KECSLGVDIWSLGCTVLEMAT-----TKPPWSQYEGVAAMFKIGNSKELPTIP-DHLSNE 639
+ DIWSLG +L+ AT T P +E + + ++ K P+ P D S E
Sbjct: 361 HGYNYKSDIWSLGLILLKCATGQFPYTPPDREGWENIFQLIEVIVEKPSPSAPSDDFSPE 540
Query: 640 GKDFVRKCLQRNPRDRPSASELLDHPFV 667
F+ CLQ+NP DRPSA +L++HPF+
Sbjct: 541 FCSFISACLQKNPGDRPSARDLINHPFI 624
>TC203686 similar to UP|Q93WR7 (Q93WR7) MAP kinase kinase, complete
Length = 1502
Score = 124 bits (312), Expect = 1e-28
Identities = 89/272 (32%), Positives = 152/272 (55%), Gaps = 12/272 (4%)
Frame = +2
Query: 415 KLLGRGTFGHVYIGFNSQSGEMCAMKEVTLFSDDAKSLESAKQLMQEVHLLSRLRHPNIV 474
K++G+G G V + + + + A+K + + +++ KQ+ QE+ + + + P +V
Sbjct: 308 KVVGKGNGGVVQLVQHKWTSQFFALKVIQMNIEESMR----KQIAQELKINQQAQCPYVV 475
Query: 475 QYYGSETVDDKLYIYLEYVSGGSIHKLLQEYGQFGELAIRSYTQQILSGLAYL-HAKNTL 533
Y S + + I LEY+ GGS+ LL++ E + + +Q+L GL YL H K+ +
Sbjct: 476 VCYQSFYENGVISIILEYMDGGSLADLLKKVKTIPEDYLAAICKQVLKGLVYLHHEKHII 655
Query: 534 HRDIKGANILVDPNGRVKVADFGMA---KHITGQYCPLSFKGSPYWMAPEVIKNSKE-CS 589
HRD+K +N+L++ G VK+ DFG++ + +GQ +F G+ +M+PE I S+ +
Sbjct: 656 HRDLKPSNLLINHIGEVKITDFGVSAIMESTSGQ--ANTFIGTYNYMSPERINGSQRGYN 829
Query: 590 LGVDIWSLGCTVLEMATTKPPWS---QYEGVAAMFKIGNS--KELPTIP--DHLSNEGKD 642
DIWSLG +LE A + P++ Q E ++F++ + + P IP + S E
Sbjct: 830 YKSDIWSLGLILLECALGRFPYAPPDQSETWESIFELIETIVDKPPPIPPSEQFSTEFCS 1009
Query: 643 FVRKCLQRNPRDRPSASELLDHPFVKGAAPLE 674
F+ CLQ++P+DR SA EL+ HPFV LE
Sbjct: 1010FISACLQKDPKDRLSAQELMAHPFVNMYDDLE 1105
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.313 0.131 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,012,508
Number of Sequences: 63676
Number of extensions: 599371
Number of successful extensions: 5767
Number of sequences better than 10.0: 965
Number of HSP's better than 10.0 without gapping: 4961
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5347
length of query: 754
length of database: 12,639,632
effective HSP length: 104
effective length of query: 650
effective length of database: 6,017,328
effective search space: 3911263200
effective search space used: 3911263200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC121237.19


