

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC119416.7 - phase: 0 /pseudo
(447 letters)
Database: GMGI
63,676 sequences; 37,918,896 total letters
Searching..................................................done
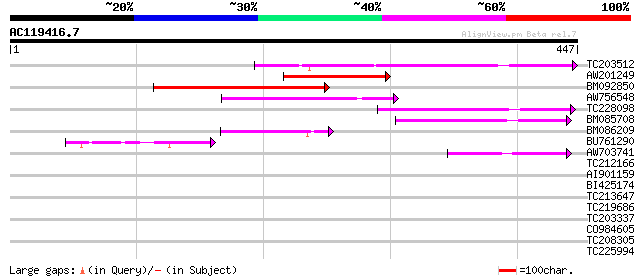
Score E
Sequences producing significant alignments: (bits) Value
TC203512 weakly similar to PIR|C86287|C86287 F9L1.24 protein - A... 114 1e-25
AW201249 108 6e-24
BM092850 106 2e-23
AW756548 84 1e-16
TC228098 76 4e-14
BM085708 70 3e-12
BM086209 57 1e-08
BU761290 50 2e-06
AW703741 44 2e-04
TC212166 39 0.005
AI901159 35 0.052
BI425174 30 1.7
TC213647 similar to UP|Q8YRJ8 (Q8YRJ8) Alr3448 protein, partial ... 30 1.7
TC219686 30 2.2
TC203337 similar to UP|Q9S728 (Q9S728) En/Spm-like transposon pr... 29 3.8
CO984605 29 4.9
TC208305 similar to UP|Q9FJZ3 (Q9FJZ3) Mannan endo-1,4-beta-mann... 28 6.4
TC225994 similar to UP|Q7XUF4 (Q7XUF4) OJ991113_30.14 protein, p... 28 8.4
>TC203512 weakly similar to PIR|C86287|C86287 F9L1.24 protein - Arabidopsis
thaliana {Arabidopsis thaliana;} , partial (9%)
Length = 988
Score = 114 bits (284), Expect = 1e-25
Identities = 72/257 (28%), Positives = 126/257 (49%), Gaps = 3/257 (1%)
Frame = +3
Query: 194 VSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKKILSFNLIAD---HKGETIGIALENCMK 250
V LT D WTS Q++ Y+ +T H++D +W L+++IL N++ + + + A+ C+
Sbjct: 42 VCLTLDVWTSSQSVGYVFITGHFVDSDWKLQRRIL--NVVMEPYPNSDSALSHAVAVCIS 215
Query: 251 DWGIRSICCVTVDNASANNLAINYLNRGMRVWNGRTLFNGEYLHMRCCAHILNLIVLDGL 310
DW S + +A+ L + V N L NG+ L C A L+ + D L
Sbjct: 216 DWNFEGKLFSITCGPSLSEVALGNLRPLLFVKNPLIL-NGQLLIGNCIAQTLSSVANDLL 392
Query: 311 KDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIA 370
+ +V++IR + K+VK+S S F + V S+ + +D T+WN+TY ML A
Sbjct: 393 SSVHLTVKKIRDSVKYVKTSESHEEKFLDLKQQLQVPSERNLFIDDQTKWNTTYQMLVAA 572
Query: 371 EKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSFSGSLHV 430
+ + VF L+ +D + G P DW+ +LK +DA + + H
Sbjct: 573 SELQEVFSCLDTSDP--------DYKGAPSMQDWKLVETLCTYLKPLFDAANILTTATHP 728
Query: 431 SANSFFKQLMEIQKTLN 447
+ +FF ++ ++Q L+
Sbjct: 729 TVITFFHEVWKLQLDLS 779
>AW201249
Length = 405
Score = 108 bits (269), Expect = 6e-24
Identities = 52/84 (61%), Positives = 66/84 (77%)
Frame = +1
Query: 217 IDDEWVLKKKILSFNLIADHKGETIGIALENCMKDWGIRSICCVTVDNASANNLAINYLN 276
ID+ + L KKIL F LI DHKGET IALENC+K+ GI+ ICC+TVDNASAN++AI+YL+
Sbjct: 7 IDEVFRLHKKILKFCLIGDHKGETTWIALENCLKE*GIQKICCITVDNASANSVAISYLS 186
Query: 277 RGMRVWNGRTLFNGEYLHMRCCAH 300
RG+ V N TL N E++HM+ AH
Sbjct: 187 RGLSV*NDHTLLNEEFIHMQRSAH 258
>BM092850
Length = 421
Score = 106 bits (264), Expect = 2e-23
Identities = 50/140 (35%), Positives = 86/140 (60%), Gaps = 1/140 (0%)
Frame = +1
Query: 114 VSFKLVDFSQEKTRLALAKMIIIDELPFKHVENEGFKMFMGEAQPKFKIPSRVSVARDCL 173
++F F QE+++L LA+MII+ P VE GFK+F+ QP F+ SV C+
Sbjct: 1 INFGSSKFDQERSQLDLARMIILHGYPLSLVEQVGFKVFVKNLQPLFEFTPNSSVEISCI 180
Query: 174 RLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKKILSF-NL 232
+Y EKE + +++ + ++L+ +TW+S +N Y+C++AHYID+EW L+KK+L+F L
Sbjct: 181 DIYRREKEKVFDMINRLHGRINLSIETWSSTENSLYLCLSAHYIDEEWTLQKKLLNFVTL 360
Query: 233 IADHKGETIGIALENCMKDW 252
+ H + + + C+ +W
Sbjct: 361 DSSHTEDLLPEVIIKCLNEW 420
>AW756548
Length = 445
Score = 84.0 bits (206), Expect = 1e-16
Identities = 42/143 (29%), Positives = 83/143 (57%), Gaps = 4/143 (2%)
Frame = +3
Query: 168 VARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKKI 227
V DC+ +Y E++ + +L +SL+ D W ++ + Y+C+T++YID+ W L+++I
Sbjct: 18 VEADCIEIYERERKKVNEMLDKLPGKISLSADVWNAVDDAEYLCLTSNYIDESWQLRRRI 197
Query: 228 LSF-NLIADHKGETIGIALENCMKDWGI-RSICCVTVDNAS-ANNLAINYLNRGMRVWNG 284
L+F + H + + A+ C+ DW I R + + +D+ S ++ +A+ G R+
Sbjct: 198 LNFIRIDPSHTEDMVSEAIMTCLMDWDIDRKLFSMILDSCSTSDKIAVRI---GERLLQN 368
Query: 285 RTLF-NGEYLHMRCCAHILNLIV 306
R L+ NG+ +RC A+++N +V
Sbjct: 369 RFLYCNGQLFDIRCAANVINTMV 437
>TC228098
Length = 720
Score = 75.9 bits (185), Expect = 4e-14
Identities = 42/156 (26%), Positives = 77/156 (48%)
Frame = +2
Query: 291 EYLHMRCCAHILNLIVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKA 350
+ L +R AH++N I D + + +Q+IR + ++++SS F + A + ++
Sbjct: 116 QLLDVRSAAHLVNSIAQDAMDALHEVIQKIRESIRYIRSSQVVQGKFNEIAQRARINTQK 295
Query: 351 MVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVF 410
+ LD +W STYLML A +Y F + D ++ SL+ E +WE +
Sbjct: 296 FLFLDFPVQWKSTYLMLETALEYRSAFSLFQEHDPSYSASLTDE--------EWEWASSV 451
Query: 411 VKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQKTL 446
+LK + T FSG+ +AN +F ++ ++ L
Sbjct: 452 TGYLKLLVEITNVFSGNKFPTANIYFPEICDVHMQL 559
>BM085708
Length = 428
Score = 69.7 bits (169), Expect = 3e-12
Identities = 43/139 (30%), Positives = 70/139 (49%)
Frame = +2
Query: 305 IVLDGLKDIDASVQRIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTY 364
+V L + V +IR ++KSS LA F A VG+ S+ + LD ++WNSTY
Sbjct: 2 MVQHALGTVREIVNKIRQTIGYIKSSQIILAKFNVMAKEVGILSQKGLFLDNPSQWNSTY 181
Query: 365 LMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWERSRVFVKFLKTFYDATLSF 424
ML +A +++ V L+ D A+ LS +WER +LK F + F
Sbjct: 182 SMLVVALEFKDVLILLQENDTAYKVCLS--------EVEWERVTAVTSYLKLFVEVINVF 337
Query: 425 SGSLHVSANSFFKQLMEIQ 443
+ S + +AN +F +L +++
Sbjct: 338 TKSKYPTANIYFPELCDVK 394
>BM086209
Length = 389
Score = 57.4 bits (137), Expect = 1e-08
Identities = 28/91 (30%), Positives = 50/91 (54%), Gaps = 2/91 (2%)
Frame = +1
Query: 167 SVARDCLRLYFDEKETLKSVLSANNQMVSLTTDTWTSIQNMNYMCVTAHYIDDEWVLKKK 226
++ DC+ +Y EK+ L +++ V+LT D WTS Q Y+ + H+ID +W L
Sbjct: 19 AIQGDCVTMYLREKQNLLNLIDGIPGRVNLTLDLWTSNQTSAYVFLRGHFIDGDWNLHHP 198
Query: 227 ILSFNLI--ADHKGETIGIALENCMKDWGIR 255
IL+ ++ D G ++ + NC+ DW ++
Sbjct: 199 ILNVFMVPFPDSDG-SLNQTIVNCLSDWHLK 288
>BU761290
Length = 441
Score = 50.4 bits (119), Expect = 2e-06
Identities = 37/128 (28%), Positives = 61/128 (46%), Gaps = 10/128 (7%)
Frame = +2
Query: 45 SKVWRHFNRNK----NLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNPYRVVDKKQKT 100
SK+W+HF + K + CKYC K+ S +GT +L +H ++C++ Y++ + K
Sbjct: 92 SKIWQHFKKIKVNGLDKTECKYC-KKLLGGKSKNGTKHLWQHNEICVQ--YKIFMRGMKG 262
Query: 101 LAIGMGSEDDPNSVSFKLVDFSQE------KTRLALAKMIIIDELPFKHVENEGFKMFMG 154
A ++ K+V QE R LAK II+ E P + GF+ +
Sbjct: 263 QAF----------LTPKVVQGKQELGAGTYDAREQLAKAIIMHEYPLSIGDRLGFRRYSA 412
Query: 155 EAQPKFKI 162
QP F++
Sbjct: 413 ALQPVFQV 436
>AW703741
Length = 412
Score = 43.5 bits (101), Expect = 2e-04
Identities = 25/98 (25%), Positives = 46/98 (46%)
Frame = +3
Query: 346 VCSKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFVTSLSGELGGCPDHDDWE 405
V S+ + +D WN +Y ML A + + VF L+ +D + G P DW+
Sbjct: 33 VPSERSLFIDDQIHWNRSYQMLVAASELKEVFSCLDTSDPDYK-------GAPPSMQDWK 191
Query: 406 RSRVFVKFLKTFYDATLSFSGSLHVSANSFFKQLMEIQ 443
+ +LK +DA + + H + +FF ++ ++Q
Sbjct: 192 LVEILCSYLKPLFDAANILTTTTHPTTIAFFHEVWKLQ 305
>TC212166
Length = 653
Score = 38.9 bits (89), Expect = 0.005
Identities = 20/56 (35%), Positives = 30/56 (52%), Gaps = 4/56 (7%)
Frame = +1
Query: 39 KKPQTCSKVWRHFNR----NKNLAVCKYCGKEYAANSSSHGTTNLGKHLKVCLKNP 90
K+P+ SKVW R + +C++CGK N GT++L +HL +C K P
Sbjct: 310 KRPKMTSKVWEEMQRIQTTEGSKVLCRHCGKLLQDNC---GTSHLKRHLIICPKRP 468
>AI901159
Length = 388
Score = 35.4 bits (80), Expect = 0.052
Identities = 14/51 (27%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Frame = +1
Query: 205 QNMNYMCVTAHYIDDEWVLKKKILSFNLIAD---HKGETIGIALENCMKDW 252
Q++ Y+ +T H++D +W L+++IL N++ + + + A+ C+ DW
Sbjct: 190 QSVGYVFITGHFVDSDWKLQRRIL--NVVMEPYPNSDSALSHAVAVCISDW 336
>BI425174
Length = 336
Score = 30.4 bits (67), Expect = 1.7
Identities = 23/58 (39%), Positives = 31/58 (52%)
Frame = -2
Query: 319 RIRAACKFVKSSPSRLATFKKCADSVGVCSKAMVTLDVVTRWNSTYLMLNIAEKYEHV 376
R+R A KF+ + TF++ V V K + L TRWNSTY ML +A Y+ V
Sbjct: 326 RLRVA-KFITRTNK---TFRERQIKVSVTKKLI--LQCKTRWNSTYHMLVVALAYKGV 171
>TC213647 similar to UP|Q8YRJ8 (Q8YRJ8) Alr3448 protein, partial (6%)
Length = 663
Score = 30.4 bits (67), Expect = 1.7
Identities = 23/81 (28%), Positives = 34/81 (41%), Gaps = 8/81 (9%)
Frame = +2
Query: 31 ATAVSKQKKKPQTCSKVWR--------HFNRNKNLAVCKYCGKEYAANSSSHGTTNLGKH 82
A+ SK KKK Q + R H+ R + A + C K+Y + T +
Sbjct: 203 ASKASKYKKKTQIECNILRSQQSSLNHHYKRIEGGATYQDCDKKYVVKAIPEPTFDS--- 373
Query: 83 LKVCLKNPYRVVDKKQKTLAI 103
+ C NP VVD +K L +
Sbjct: 374 -EPCASNPENVVDSAKKILDV 433
>TC219686
Length = 819
Score = 30.0 bits (66), Expect = 2.2
Identities = 18/61 (29%), Positives = 33/61 (53%), Gaps = 1/61 (1%)
Frame = +3
Query: 84 KVCLKNPYRVVDKKQKTLAIGMGSEDDPNSVSFKLVDFSQEKTRLALAKMIIIDE-LPFK 142
K C+K+ + D Q +G+ +DD ++SF+L S++K+ A ++ + E LP
Sbjct: 318 KKCIKSQFETADVHQFFKTVGL-EDDDHMAISFELSAISKQKSESAYLELEEVPEWLPEV 494
Query: 143 H 143
H
Sbjct: 495 H 497
>TC203337 similar to UP|Q9S728 (Q9S728) En/Spm-like transposon protein
(Protodermal factor 1), partial (46%)
Length = 1410
Score = 29.3 bits (64), Expect = 3.8
Identities = 10/33 (30%), Positives = 19/33 (57%)
Frame = +2
Query: 50 HFNRNKNLAVCKYCGKEYAANSSSHGTTNLGKH 82
H R+ + ++C YC ++ + SS HG++ H
Sbjct: 20 HAGRDLSNSLCSYCSPDHGSGSSGHGSSPHHSH 118
>CO984605
Length = 802
Score = 28.9 bits (63), Expect = 4.9
Identities = 17/41 (41%), Positives = 24/41 (58%)
Frame = -2
Query: 348 SKAMVTLDVVTRWNSTYLMLNIAEKYEHVFYRLEHADAAFV 388
SK++V+ RWNST LML A K + F RLE + ++
Sbjct: 648 SKSLVSRGARIRWNST-LMLESAFKLQTTF*RLEDTNGNYL 529
>TC208305 similar to UP|Q9FJZ3 (Q9FJZ3) Mannan endo-1,4-beta-mannosidase
(At5g66460), partial (51%)
Length = 1184
Score = 28.5 bits (62), Expect = 6.4
Identities = 32/142 (22%), Positives = 61/142 (42%), Gaps = 21/142 (14%)
Frame = +1
Query: 108 EDDPNSVSFKLV-------DFSQEKTRLALAKMI-IIDELPFKHVENEGFKMFMGEAQPK 159
+DDP ++++L+ D S + + +M + + H+ G + F G++ P+
Sbjct: 1 KDDPTIMAWELMNEPRCTSDPSGRTIQAWITEMASFVKSIDRNHLLEAGLEGFYGQSTPQ 180
Query: 160 FKIPSRVSVARDCLRLYFDEKETLKSVLSANNQMVSLT-------TDTWTSIQNMNYMC- 211
R L FD + + ANN++ ++ D W S N+ Y
Sbjct: 181 ----------RKRLNPGFD----IGTDFIANNRIPAIDFATVHCYPDQWVSSSNIQYQLS 318
Query: 212 -----VTAHYIDDEWVLKKKIL 228
++AH+ID ++ +KK IL
Sbjct: 319 FLNNWLSAHFIDAQYRIKKPIL 384
>TC225994 similar to UP|Q7XUF4 (Q7XUF4) OJ991113_30.14 protein, partial (43%)
Length = 2770
Score = 28.1 bits (61), Expect = 8.4
Identities = 9/24 (37%), Positives = 16/24 (66%)
Frame = +2
Query: 216 YIDDEWVLKKKILSFNLIADHKGE 239
Y+ D WV+ + + S N++ +H GE
Sbjct: 989 YLHDNWVIHRDLKSSNILLNHDGE 1060
Database: GMGI
Posted date: Oct 22, 2004 4:58 PM
Number of letters in database: 37,918,896
Number of sequences in database: 63,676
Lambda K H
0.320 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,123,122
Number of Sequences: 63676
Number of extensions: 284326
Number of successful extensions: 1362
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 1354
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1357
length of query: 447
length of database: 12,639,632
effective HSP length: 100
effective length of query: 347
effective length of database: 6,272,032
effective search space: 2176395104
effective search space used: 2176395104
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC119416.7


