

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0340.11
(420 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
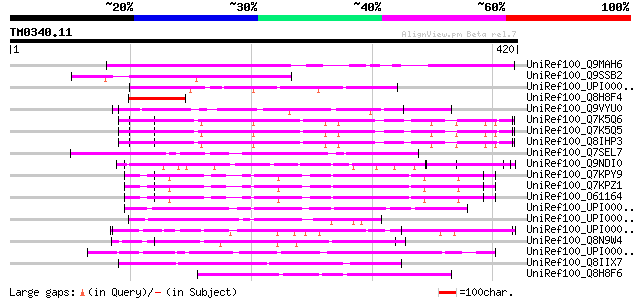
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9MAH6 F12M16.15 [Arabidopsis thaliana] 201 2e-50
UniRef100_Q9SSB2 T18A20.4 protein [Arabidopsis thaliana] 98 5e-19
UniRef100_UPI000034A703 UPI000034A703 UniRef100 entry 65 4e-09
UniRef100_Q8H8F4 Hypothetical protein OSJNBa0041L14.18 [Oryza sa... 65 5e-09
UniRef100_Q9VYU0 CG32662-PA [Drosophila melanogaster] 63 2e-08
UniRef100_Q7K5Q6 Erythrocyte binding protein 3 [Plasmodium falci... 62 3e-08
UniRef100_Q7K5Q5 Erythrocyte binding protein 2 [Plasmodium falci... 62 3e-08
UniRef100_Q8IHP3 MAEBL, putative [Plasmodium falciparum] 62 3e-08
UniRef100_Q7SEL7 Hypothetical protein [Neurospora crassa] 62 3e-08
UniRef100_Q9NDI0 200 kDa antigen p200 [Babesia bigemina] 61 7e-08
UniRef100_Q7KPY9 Erythrocyte binding protein 4 [Plasmodium yoeli... 60 9e-08
UniRef100_Q7KPZ1 Erythrocyte binding protein 3 [Plasmodium yoeli... 60 9e-08
UniRef100_O61164 Erythrocyte binding protein 1 [Plasmodium yoeli... 60 9e-08
UniRef100_UPI000046B0E5 UPI000046B0E5 UniRef100 entry 60 1e-07
UniRef100_UPI0000335A68 UPI0000335A68 UniRef100 entry 60 2e-07
UniRef100_UPI0000249BD4 UPI0000249BD4 UniRef100 entry 59 2e-07
UniRef100_Q8N9W4 Hypothetical protein FLJ36144 [Homo sapiens] 59 3e-07
UniRef100_UPI0000499259 UPI0000499259 UniRef100 entry 58 6e-07
UniRef100_Q8IIX7 Hypothetical protein [Plasmodium falciparum] 58 6e-07
UniRef100_Q8H8F6 Hypothetical protein OSJNBa0041L14.16 [Oryza sa... 57 1e-06
>UniRef100_Q9MAH6 F12M16.15 [Arabidopsis thaliana]
Length = 363
Score = 201 bits (512), Expect = 2e-50
Identities = 129/338 (38%), Positives = 181/338 (53%), Gaps = 66/338 (19%)
Query: 81 VVGLSDSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALR 140
V ++ E +S S + +KE RR KIGLANKG+VPWNKGRKHS +TR RIK+RT EAL
Sbjct: 75 VKAMNKDTEADSDSDRKIKEEERRRKIGLANKGKVPWNKGRKHSEDTRRRIKQRTIEALT 134
Query: 141 EPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGG 200
PKVRKKM++H + HS++ KEKI S+++VW ER +SKR +E+ SW + IA AA+KGG
Sbjct: 135 NPKVRKKMSDHQQPHSNETKEKIRASVKQVWAERSRSKRLKEKFMSSWSENIAEAARKGG 194
Query: 201 CGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGS 260
G+ ELDWDSY KIK+ +QL AEEK + K ++ +AKEA K +
Sbjct: 195 SGEAELDWDSYEKIKQDFSSEQLQLAEEKARAK------------EQTKMIAKEAAKART 242
Query: 261 GEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKD 320
++RR EK K +E K +E +I+ K K
Sbjct: 243 --------------------EKMRRAAEKKKEREE---KDRREGKIR---------KPKQ 270
Query: 321 LRERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSN 380
RE I + KL +LTKI+ K ++A
Sbjct: 271 EREN----------------------PTIASRSKLKKRLTKIHKKKTSLGKIAIGTDRVV 308
Query: 381 SIYPTYNKLDLEVIKREKLRKEASFADRIQAARLKKGN 418
S+ KLDL++I++E+ R + S AD+IQAA+ ++G+
Sbjct: 309 SVAAKLEKLDLDLIRKERTRGDISLADQIQAAKNQRGS 346
>UniRef100_Q9SSB2 T18A20.4 protein [Arabidopsis thaliana]
Length = 603
Score = 97.8 bits (242), Expect = 5e-19
Identities = 63/219 (28%), Positives = 98/219 (43%), Gaps = 49/219 (22%)
Query: 52 ELQINVDDDSDNQSETSVDSSDYLNGG--DGVVGLSDSLEIESLSRKAVKERMRRFKIGL 109
E + +++ S S SS NG DG + D +E++RR +I
Sbjct: 67 ETKYPAQKENERSSSLSSASSKSSNGSADDGEEQVDD------------REKLRRMRISK 114
Query: 110 ANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPR---------------- 153
AN+G PWNKGRKHS ET ++I+ RT A+++PKV E
Sbjct: 115 ANRGNTPWNKGRKHSPETLQKIRERTKIAMQDPKVMSYQDEISESWARTKVSTGFSFAFK 174
Query: 154 -------------------FHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIAN 194
F+S + + KI +R W R + ++ +E W+ +A
Sbjct: 175 PVGAFGFCYHNTIFPNLHDFYSKETRMKIGEGVRMRWARRKERRKVQETCHFEWQNLLAE 234
Query: 195 AAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEK 233
AAK+G +EEL WDSYN + +Q +L+ L E+++ K
Sbjct: 235 AAKQGYTDEEELQWDSYNILDQQNQLEWLESVEQRKAIK 273
>UniRef100_UPI000034A703 UPI000034A703 UniRef100 entry
Length = 235
Score = 65.1 bits (157), Expect = 4e-09
Identities = 61/235 (25%), Positives = 113/235 (47%), Gaps = 20/235 (8%)
Query: 100 ERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKM--------AEH 151
E++RR +IG AN G+VPWNKG KHS ET E+I+ T + +++PK R+++ A H
Sbjct: 7 EQLRRKRIGSANAGKVPWNKGGKHSKETIEKIRATTLKHMQDPKYRERLKQSYNGSNARH 66
Query: 152 PRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGG--CGQEELDWD 209
F KIK S+ R ++++ K E + + + N A G +
Sbjct: 67 SAFTRVKIKRA---SVERARVKKIE-KSTDESVRVWGPKRGRNGAASSGMFARRNSAVMT 122
Query: 210 SYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKE---AKKGGSGEKELD 266
Y + ++ + L+ +KE+EK+ + E ++ S +AK +KK ++
Sbjct: 123 VYFGVNGSADIAR-LRKRQKEEEKVRQKEQRELKKKTISTLVAKRRAMSKKKPKKPQKRS 181
Query: 267 WDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDL 321
+ +KI + ++N+ R AK + ++ A + + + K+ ++ KK L
Sbjct: 182 SEHRKKISQA--IVNKWRDPEYVAKMRSPRKSPATRPTQRRQAKRSSIDPAKKKL 234
>UniRef100_Q8H8F4 Hypothetical protein OSJNBa0041L14.18 [Oryza sativa]
Length = 162
Score = 64.7 bits (156), Expect = 5e-09
Identities = 27/47 (57%), Positives = 37/47 (78%)
Query: 99 KERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVR 145
+ER+RR +I ANKG PWNKGRKH+ ET +RI+ RT A+++PKV+
Sbjct: 95 RERLRRMRISKANKGNTPWNKGRKHTPETLQRIRERTRIAMQDPKVK 141
>UniRef100_Q9VYU0 CG32662-PA [Drosophila melanogaster]
Length = 1168
Score = 62.8 bits (151), Expect = 2e-08
Identities = 66/240 (27%), Positives = 116/240 (47%), Gaps = 27/240 (11%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIK-RRTSEALREPKVRKKMA 149
E L + +KE+ R K+ K + + R E E+ K ++ E LRE K+++K
Sbjct: 516 EKLKEEKIKEKQREEKLK-EEKLKEKEREERMKEKEREEKAKEKQREEKLREEKIKEKER 574
Query: 150 EHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWD 209
E +K+KEK+ +E++K K E++ E+ + ++ ++E
Sbjct: 575 E------EKLKEKLR-------EEKIKEKEKEEKLRKEREEKMREKEREEKIKEKE---- 617
Query: 210 SYNKIKEQLELQQLL-QAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWD 268
KIKE+ ++L + EEK KEK + + EK + E L ++ ++ EKE +
Sbjct: 618 RVEKIKEKEREEKLKKEKEEKLKEKEELLKKKEKEEKEREEKLKEKERQEKLKEKERE-- 675
Query: 269 SYEKIQEEMFLLNQIRRTTEKAKAKERAR--TKAEKEARIKAIKKVMLNQKKKDLRERIK 326
EK++ E + + EK K KERA EKE ++K K+ L +K+K+L+ + K
Sbjct: 676 --EKLKRETEERQREKEREEKLKEKERAEKLKDLEKEVKLKE-KEEQLKEKEKELKLKEK 732
Score = 52.0 bits (123), Expect = 3e-05
Identities = 70/293 (23%), Positives = 128/293 (42%), Gaps = 29/293 (9%)
Query: 86 DSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVR 145
+ L+ E + K +E+++ K+ + K R+ A+ ++R ++ E ++E +
Sbjct: 516 EKLKEEKIKEKQREEKLKEEKLKEKEREERMKEKEREEKAKEKQREEKLREEKIKEKERE 575
Query: 146 KKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEE 205
+K+ E R K KEK LR+ +E+++ K E+I ++ + +K +E+
Sbjct: 576 EKLKEKLREEKIKEKEK-EEKLRKEREEKMREKEREEKI--KEKERVEKIKEKER--EEK 630
Query: 206 LDWDSYNKIKEQLELQQLLQAEEKE---------K-EKLMAIARAEKFIQSWSECLAKEA 255
L + K+KE+ EL + + EEKE + EKL R EK + E ++
Sbjct: 631 LKKEKEEKLKEKEELLKKKEKEEKEREEKLKEKERQEKLKEKEREEKLKRETEERQREKE 690
Query: 256 KKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIK-KVML 314
++ EKE + + +++E+ K K KE + EKE ++K K K +
Sbjct: 691 REEKLKEKERA-EKLKDLEKEV-----------KLKEKEEQLKEKEKELKLKEKKEKDKV 738
Query: 315 NQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQF-KLDTKLTKINISK 366
+K+K L S + ED L Q + L K TK + K
Sbjct: 739 KEKEKSLESEKLLISATVSNPWRRVVEDTPPKLPAVQDYPSLGKKPTKASPEK 791
Score = 46.2 bits (108), Expect = 0.002
Identities = 84/387 (21%), Positives = 162/387 (41%), Gaps = 57/387 (14%)
Query: 45 QRVLSIEELQINVDDDSDNQSETSVDSSDYLNGGDGVVGLSD------SLEIESLSRKAV 98
+R + + + IN + DSD +S+ + DS V +S SL +E + A
Sbjct: 360 ERAVPMVKKTINKEQDSDAESDHA-DSLLANKSSIAAVMVSSASAQGLSLHVEMSAADAE 418
Query: 99 KERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDK 158
+ + GL + +K K + + + T + +E K++ + K
Sbjct: 419 QGEDEEIE-GLDEEPPKTMSKDNKKKQKPGDAVATMTIDKEKEKAKEKELKLKEKEREAK 477
Query: 159 IKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQL 218
++EK +E+LK K E + + E+ + +EE KIKE+
Sbjct: 478 LQEKEK-------EEKLKLKEREESLRMEREEKL----------KEE-------KIKEKE 513
Query: 219 ELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMF 278
++L + + KEK+ R EK + E L ++ ++ EKE + + EK +EE
Sbjct: 514 REEKLKEEKIKEKQ------REEKLKE---EKLKEKEREERMKEKEREEKAKEKQREEKL 564
Query: 279 LLNQIRRTTEKAKAKERARTK--AEKEARIKAIKKVMLNQKKKDLRERIKGRGNI----- 331
+I+ + K KE+ R + EKE K K+ ++K+ E+IK + +
Sbjct: 565 REEKIKEKEREEKLKEKLREEKIKEKEKEEKLRKEREEKMREKEREEKIKEKERVEKIKE 624
Query: 332 ---KSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNK 388
+ +L K E + E+ ++ + + K + + + E +E + K
Sbjct: 625 KEREEKLKKEKEEKLKEKEELLKKKEKEEKEREEKLKEKERQEKLKEKEREEKL-----K 679
Query: 389 LDLEVIKREKLRKEASFADRIQAARLK 415
+ E +REK R+E ++ +A +LK
Sbjct: 680 RETEERQREKEREE-KLKEKERAEKLK 705
>UniRef100_Q7K5Q6 Erythrocyte binding protein 3 [Plasmodium falciparum]
Length = 2006
Score = 62.0 bits (149), Expect = 3e-08
Identities = 66/326 (20%), Positives = 151/326 (46%), Gaps = 24/326 (7%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAE 150
+ L +KA +++ A + + N + + + +K++ E + +++KK E
Sbjct: 1415 DELKKKAEEKKKADELKKKAEEKKKAENLKKAEEKKKADELKKKAEEKKKADELKKKAEE 1474
Query: 151 HPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDS 210
+ +D++K+K + ++ + K+ +++ + E+ A+ KK ++ +
Sbjct: 1475 KKK--ADELKKKAEEKKKADELKKAEEKKKADELKKAEEKKKADELKKAEEKKKADELKK 1532
Query: 211 YNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSY 270
++K+ E +++ Q + +E+ + MA+ RAE ++ + +E K EK++ +
Sbjct: 1533 AEELKKAEEKKKVEQKKREEERRNMALRRAE-ILKQIEKKRIEEVMKLYEEEKKMKAEQL 1591
Query: 271 EKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGN 330
+K +EE Q+++ E+ K E+ + K E+E KK KK++ +IK
Sbjct: 1592 KKEEEEKIKAEQLKKEEEEKKKVEQLKKKEEEE------KKKAEQLKKEEEENKIKA--- 1642
Query: 331 IKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLD 390
QL K E+ + A E+ ++ + + K + ++ +E + K +
Sbjct: 1643 --EQLKKKEEEEKKKAEELKKEEEEEKKKAE---------QLKKEEEEKKKVEQLKKKEE 1691
Query: 391 LEVIKREKLRKEASFADRIQAARLKK 416
E K E+L+KE ++I+ +LKK
Sbjct: 1692 EEKKKAEQLKKEEE-ENKIKVEQLKK 1716
Score = 60.1 bits (144), Expect = 1e-07
Identities = 71/327 (21%), Positives = 148/327 (44%), Gaps = 29/327 (8%)
Query: 121 RKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKE-KISYSLRRVW-QERLKSK 178
+K + E ++ +++ ++ L++ + KK E + K +E + + +LRR ++++ K
Sbjct: 1512 KKKADELKKAEEKKKADELKKAEELKKAEEKKKVEQKKREEERRNMALRRAEILKQIEKK 1571
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLE-LQQLLQAEEKEKEKLMAI 237
R E + L E+ A + +E++ + K +E+ + ++QL + EE+EK+K +
Sbjct: 1572 RIEEVMKLYEEEKKMKAEQLKKEEEEKIKAEQLKKEEEEKKKVEQLKKKEEEEKKKAEQL 1631
Query: 238 ARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYE---------KIQEEMFLLNQIRRTTE 288
+ E+ + +E L K+ ++ +EL + E K +EE + Q+++ E
Sbjct: 1632 KKEEEENKIKAEQLKKKEEEEKKKAEELKKEEEEEKKKAEQLKKEEEEKKKVEQLKKKEE 1691
Query: 289 KAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALE 348
+ K K K E+E +IK + +++K E +K K ++ + E+ + A E
Sbjct: 1692 EEKKKAEQLKKEEEENKIKVEQLKKEEEEEKKKAEELKKEEEEKKKVQQLKKEEEKKAEE 1751
Query: 349 --------ITQQFKLDTKLTKINISKNI-----NSEVAREGGFSNSIYPTYNKLDLE--- 392
I ++ K + + ++ + K I N E +EG N+ Y D E
Sbjct: 1752 IRKEKEAVIEEELKKEDEKRRMEVEKKIKDTKDNFENIQEGNNKNTPYINKEMFDSEIKE 1811
Query: 393 -VIKREKLRKEASFADRIQAARLKKGN 418
VI + EA ++ + K N
Sbjct: 1812 VVITKNMQLNEADAFEKHNSENSKSSN 1838
Score = 51.6 bits (122), Expect = 4e-05
Identities = 77/337 (22%), Positives = 152/337 (44%), Gaps = 29/337 (8%)
Query: 100 ERMRRFKIGLANKGRVPWNKGRKHSAETR-ERIKRRTSEALREPKVRKKMAEHPRFHSD- 157
E RR++ N+ R+ + R++ E R E +KR +E +R+ + K AE R + +
Sbjct: 1186 EAARRYE----NERRI--EEARRYEDEKRIEAVKR--AEEVRKDEEEAKRAEKERNNEEI 1237
Query: 158 -KIKE-KISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGG----------CGQEE 205
K +E ++++ RR + + KR +++ + E+ A+ KK +E+
Sbjct: 1238 RKFEEARMAHFARRQAAIKAEEKRKADELKKAEEKKKADELKKSEEKKKADELKKKAEEK 1297
Query: 206 LDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGS---GE 262
D K E+ + L+ + +EK+K + +AE+ ++ ++E KK E
Sbjct: 1298 KKADELKKKAEEKKKADELKKKAEEKKKADEVKKAEEKKKADELKKSEEKKKADELKKSE 1357
Query: 263 KELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLR 322
++ D +K EE +++++ E+ K + + KAE++ + +KK +KK D
Sbjct: 1358 EKKKADELKKKAEEKKKADELKKKAEEKKKADELKKKAEEKKKADELKKKAEEKKKADEL 1417
Query: 323 ERIKGRGNIKSQLCKNANEDGEAA-LEITQQFKLDTKLTKINISKNINSEVAREGGFSNS 381
++ +L K A E +A L+ ++ K +L K K E+ ++
Sbjct: 1418 KKKAEEKKKADELKKKAEEKKKAENLKKAEEKKKADELKKKAEEKKKADELKKKAEEKKK 1477
Query: 382 IYPTYNKLDLEVIKREKLRK--EASFADRIQAARLKK 416
K + E K ++L+K E AD ++ A KK
Sbjct: 1478 ADELKKKAE-EKKKADELKKAEEKKKADELKKAEEKK 1513
>UniRef100_Q7K5Q5 Erythrocyte binding protein 2 [Plasmodium falciparum]
Length = 2019
Score = 62.0 bits (149), Expect = 3e-08
Identities = 66/326 (20%), Positives = 151/326 (46%), Gaps = 24/326 (7%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAE 150
+ L +KA +++ A + + N + + + +K++ E + +++KK E
Sbjct: 1415 DELKKKAEEKKKADELKKKAEEKKKAENLKKAEEKKKADELKKKAEEKKKADELKKKAEE 1474
Query: 151 HPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDS 210
+ +D++K+K + ++ + K+ +++ + E+ A+ KK ++ +
Sbjct: 1475 KKK--ADELKKKAEEKKKADELKKAEEKKKADELKKAEEKKKADELKKAEEKKKADELKK 1532
Query: 211 YNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSY 270
++K+ E +++ Q + +E+ + MA+ RAE ++ + +E K EK++ +
Sbjct: 1533 AEELKKAEEKKKVEQKKREEERRNMALRRAE-ILKQIEKKRIEEVMKLYEEEKKMKAEQL 1591
Query: 271 EKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGN 330
+K +EE Q+++ E+ K E+ + K E+E KK KK++ +IK
Sbjct: 1592 KKEEEEKIKAEQLKKEEEEKKKVEQLKKKEEEE------KKKAEQLKKEEEENKIKA--- 1642
Query: 331 IKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLD 390
QL K E+ + A E+ ++ + + K + ++ +E + K +
Sbjct: 1643 --EQLKKKEEEEKKKAEELKKEEEEEKKKAE---------QLKKEEEEKKKVEQLKKKEE 1691
Query: 391 LEVIKREKLRKEASFADRIQAARLKK 416
E K E+L+KE ++I+ +LKK
Sbjct: 1692 EEKKKAEQLKKEEE-ENKIKVEQLKK 1716
Score = 60.1 bits (144), Expect = 1e-07
Identities = 71/327 (21%), Positives = 148/327 (44%), Gaps = 29/327 (8%)
Query: 121 RKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKE-KISYSLRRVW-QERLKSK 178
+K + E ++ +++ ++ L++ + KK E + K +E + + +LRR ++++ K
Sbjct: 1512 KKKADELKKAEEKKKADELKKAEELKKAEEKKKVEQKKREEERRNMALRRAEILKQIEKK 1571
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLE-LQQLLQAEEKEKEKLMAI 237
R E + L E+ A + +E++ + K +E+ + ++QL + EE+EK+K +
Sbjct: 1572 RIEEVMKLYEEEKKMKAEQLKKEEEEKIKAEQLKKEEEEKKKVEQLKKKEEEEKKKAEQL 1631
Query: 238 ARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYE---------KIQEEMFLLNQIRRTTE 288
+ E+ + +E L K+ ++ +EL + E K +EE + Q+++ E
Sbjct: 1632 KKEEEENKIKAEQLKKKEEEEKKKAEELKKEEEEEKKKAEQLKKEEEEKKKVEQLKKKEE 1691
Query: 289 KAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALE 348
+ K K K E+E +IK + +++K E +K K ++ + E+ + A E
Sbjct: 1692 EEKKKAEQLKKEEEENKIKVEQLKKEEEEEKKKAEELKKEEEEKKKVQQLKKEEEKKAEE 1751
Query: 349 --------ITQQFKLDTKLTKINISKNI-----NSEVAREGGFSNSIYPTYNKLDLE--- 392
I ++ K + + ++ + K I N E +EG N+ Y D E
Sbjct: 1752 IRKEKEAVIEEELKKEDEKRRMEVEKKIKDTKDNFENIQEGNNKNTPYINKEMFDSEIKE 1811
Query: 393 -VIKREKLRKEASFADRIQAARLKKGN 418
VI + EA ++ + K N
Sbjct: 1812 VVITKNMQLNEADAFEKHNSENSKSSN 1838
Score = 51.6 bits (122), Expect = 4e-05
Identities = 77/337 (22%), Positives = 152/337 (44%), Gaps = 29/337 (8%)
Query: 100 ERMRRFKIGLANKGRVPWNKGRKHSAETR-ERIKRRTSEALREPKVRKKMAEHPRFHSD- 157
E RR++ N+ R+ + R++ E R E +KR +E +R+ + K AE R + +
Sbjct: 1186 EAARRYE----NERRI--EEARRYEDEKRIEAVKR--AEEVRKDEEEAKRAEKERNNEEI 1237
Query: 158 -KIKE-KISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGG----------CGQEE 205
K +E ++++ RR + + KR +++ + E+ A+ KK +E+
Sbjct: 1238 RKFEEARMAHFARRQAAIKAEEKRKADELKKAEEKKKADELKKSEEKKKADELKKKAEEK 1297
Query: 206 LDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGS---GE 262
D K E+ + L+ + +EK+K + +AE+ ++ ++E KK E
Sbjct: 1298 KKADELKKKAEEKKKADELKKKAEEKKKADEVKKAEEKKKADELKKSEEKKKADELKKSE 1357
Query: 263 KELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLR 322
++ D +K EE +++++ E+ K + + KAE++ + +KK +KK D
Sbjct: 1358 EKKKADELKKKAEEKKKADELKKKAEEKKKADELKKKAEEKKKADELKKKAEEKKKADEL 1417
Query: 323 ERIKGRGNIKSQLCKNANEDGEAA-LEITQQFKLDTKLTKINISKNINSEVAREGGFSNS 381
++ +L K A E +A L+ ++ K +L K K E+ ++
Sbjct: 1418 KKKAEEKKKADELKKKAEEKKKAENLKKAEEKKKADELKKKAEEKKKADELKKKAEEKKK 1477
Query: 382 IYPTYNKLDLEVIKREKLRK--EASFADRIQAARLKK 416
K + E K ++L+K E AD ++ A KK
Sbjct: 1478 ADELKKKAE-EKKKADELKKAEEKKKADELKKAEEKK 1513
>UniRef100_Q8IHP3 MAEBL, putative [Plasmodium falciparum]
Length = 2055
Score = 62.0 bits (149), Expect = 3e-08
Identities = 66/326 (20%), Positives = 151/326 (46%), Gaps = 24/326 (7%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAE 150
+ L +KA +++ A + + N + + + +K++ E + +++KK E
Sbjct: 1415 DELKKKAEEKKKADELKKKAEEKKKAENLKKAEEKKKADELKKKAEEKKKADELKKKAEE 1474
Query: 151 HPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDS 210
+ +D++K+K + ++ + K+ +++ + E+ A+ KK ++ +
Sbjct: 1475 KKK--ADELKKKAEEKKKADELKKAEEKKKADELKKAEEKKKADELKKAEEKKKADELKK 1532
Query: 211 YNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSY 270
++K+ E +++ Q + +E+ + MA+ RAE ++ + +E K EK++ +
Sbjct: 1533 AEELKKAEEKKKVEQKKREEERRNMALRRAE-ILKQIEKKRIEEVMKLYEEEKKMKAEQL 1591
Query: 271 EKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGN 330
+K +EE Q+++ E+ K E+ + K E+E KK KK++ +IK
Sbjct: 1592 KKEEEEKIKAEQLKKEEEEKKKVEQLKKKEEEE------KKKAEQLKKEEEENKIKA--- 1642
Query: 331 IKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLD 390
QL K E+ + A E+ ++ + + K + ++ +E + K +
Sbjct: 1643 --EQLKKKEEEEKKKAEELKKEEEEEKKKAE---------QLKKEEEEKKKVEQLKKKEE 1691
Query: 391 LEVIKREKLRKEASFADRIQAARLKK 416
E K E+L+KE ++I+ +LKK
Sbjct: 1692 EEKKKAEQLKKEEE-ENKIKVEQLKK 1716
Score = 60.1 bits (144), Expect = 1e-07
Identities = 71/327 (21%), Positives = 148/327 (44%), Gaps = 29/327 (8%)
Query: 121 RKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKE-KISYSLRRVW-QERLKSK 178
+K + E ++ +++ ++ L++ + KK E + K +E + + +LRR ++++ K
Sbjct: 1512 KKKADELKKAEEKKKADELKKAEELKKAEEKKKVEQKKREEERRNMALRRAEILKQIEKK 1571
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLE-LQQLLQAEEKEKEKLMAI 237
R E + L E+ A + +E++ + K +E+ + ++QL + EE+EK+K +
Sbjct: 1572 RIEEVMKLYEEEKKMKAEQLKKEEEEKIKAEQLKKEEEEKKKVEQLKKKEEEEKKKAEQL 1631
Query: 238 ARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYE---------KIQEEMFLLNQIRRTTE 288
+ E+ + +E L K+ ++ +EL + E K +EE + Q+++ E
Sbjct: 1632 KKEEEENKIKAEQLKKKEEEEKKKAEELKKEEEEEKKKAEQLKKEEEEKKKVEQLKKKEE 1691
Query: 289 KAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALE 348
+ K K K E+E +IK + +++K E +K K ++ + E+ + A E
Sbjct: 1692 EEKKKAEQLKKEEEENKIKVEQLKKEEEEEKKKAEELKKEEEEKKKVQQLKKEEEKKAEE 1751
Query: 349 --------ITQQFKLDTKLTKINISKNI-----NSEVAREGGFSNSIYPTYNKLDLE--- 392
I ++ K + + ++ + K I N E +EG N+ Y D E
Sbjct: 1752 IRKEKEAVIEEELKKEDEKRRMEVEKKIKDTKDNFENIQEGNNKNTPYINKEMFDSEIKE 1811
Query: 393 -VIKREKLRKEASFADRIQAARLKKGN 418
VI + EA ++ + K N
Sbjct: 1812 VVITKNMQLNEADAFEKHNSENSKSSN 1838
Score = 51.6 bits (122), Expect = 4e-05
Identities = 77/337 (22%), Positives = 152/337 (44%), Gaps = 29/337 (8%)
Query: 100 ERMRRFKIGLANKGRVPWNKGRKHSAETR-ERIKRRTSEALREPKVRKKMAEHPRFHSD- 157
E RR++ N+ R+ + R++ E R E +KR +E +R+ + K AE R + +
Sbjct: 1186 EAARRYE----NERRI--EEARRYEDEKRIEAVKR--AEEVRKDEEEAKRAEKERNNEEI 1237
Query: 158 -KIKE-KISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGG----------CGQEE 205
K +E ++++ RR + + KR +++ + E+ A+ KK +E+
Sbjct: 1238 RKFEEARMAHFARRQAAIKAEEKRKADELKKAEEKKKADELKKSEEKKKADELKKKAEEK 1297
Query: 206 LDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGS---GE 262
D K E+ + L+ + +EK+K + +AE+ ++ ++E KK E
Sbjct: 1298 KKADELKKKAEEKKKADELKKKAEEKKKADEVKKAEEKKKADELKKSEEKKKADELKKSE 1357
Query: 263 KELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLR 322
++ D +K EE +++++ E+ K + + KAE++ + +KK +KK D
Sbjct: 1358 EKKKADELKKKAEEKKKADELKKKAEEKKKADELKKKAEEKKKADELKKKAEEKKKADEL 1417
Query: 323 ERIKGRGNIKSQLCKNANEDGEAA-LEITQQFKLDTKLTKINISKNINSEVAREGGFSNS 381
++ +L K A E +A L+ ++ K +L K K E+ ++
Sbjct: 1418 KKKAEEKKKADELKKKAEEKKKAENLKKAEEKKKADELKKKAEEKKKADELKKKAEEKKK 1477
Query: 382 IYPTYNKLDLEVIKREKLRK--EASFADRIQAARLKK 416
K + E K ++L+K E AD ++ A KK
Sbjct: 1478 ADELKKKAE-EKKKADELKKAEEKKKADELKKAEEKK 1513
>UniRef100_Q7SEL7 Hypothetical protein [Neurospora crassa]
Length = 1758
Score = 62.0 bits (149), Expect = 3e-08
Identities = 65/289 (22%), Positives = 125/289 (42%), Gaps = 24/289 (8%)
Query: 51 EELQINVDDDSDNQSETSVDSSDYLNGGDGVVGLSDSLEIESLSRKAVKERMRRFKIGLA 110
E+ Q NVD+ ++ ++ +S +Y + +I + RK K+R ++ +
Sbjct: 1008 EDGQANVDEKDKDEMDSEENSDEYNTAAEEQSPQPKKSKINNAKRKRRKQRKQKAEQAAK 1067
Query: 111 NKG-RVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRR 169
K R W K ++ + E +ER K+R EA + RKK A + +EK + R
Sbjct: 1068 EKAEREAWEKKKQEAKERKER-KKREEEA--QKAARKKEAREKETREKEAREK---AARE 1121
Query: 170 VWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEK 229
++ ++ K RE A KK +E + ++ K + E ++ E++
Sbjct: 1122 KEEKAMREKAARE----------IAARKKEAREKETREKEAREKAAREKEAREKTAREKE 1171
Query: 230 EKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEK 289
+EK + A K + K+A++ + E+E EK+ E + R E+
Sbjct: 1172 AREKEVREKEARKKEAREKQVREKDAREKAAKERE------EKVAREK-EAQKARERQEQ 1224
Query: 290 AKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKN 338
K ++AR + E+EAR + ++ ++K + IK +K + N
Sbjct: 1225 EKEAQKAREQQEQEARERKKLDEVIVVEEKVNEDDIKQEDEVKEEADNN 1273
Score = 36.2 bits (82), Expect = 1.8
Identities = 43/200 (21%), Positives = 83/200 (41%), Gaps = 12/200 (6%)
Query: 215 KEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKEL-DWDSYEKI 273
K++ E +AE + EK A+ K + E K A+K + EKE + ++ EK
Sbjct: 1059 KQKAEQAAKEKAEREAWEKKKQEAKERKERKKREEEAQKAARKKEAREKETREKEAREKA 1118
Query: 274 ---QEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQK--------KKDLR 322
+EE + + R K + R + EKEAR KA ++ +K +K++R
Sbjct: 1119 AREKEEKAMREKAAREIAARKKEAREKETREKEAREKAAREKEAREKTAREKEAREKEVR 1178
Query: 323 ERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSI 382
E+ + + + + + +AA E ++ + + K + E +
Sbjct: 1179 EKEARKKEAREKQVREKDAREKAAKEREEKVAREKEAQKARERQEQEKEAQKAREQQEQE 1238
Query: 383 YPTYNKLDLEVIKREKLRKE 402
KLD ++ EK+ ++
Sbjct: 1239 ARERKKLDEVIVVEEKVNED 1258
>UniRef100_Q9NDI0 200 kDa antigen p200 [Babesia bigemina]
Length = 1108
Score = 60.8 bits (146), Expect = 7e-08
Identities = 79/330 (23%), Positives = 143/330 (42%), Gaps = 29/330 (8%)
Query: 96 KAVKERMRRFKIGLANKGRVPWNKGRKHSAE--TRERIKRRTSEALREPKVRKKMAEHPR 153
KA +E+ R K L K + + + AE +E+ +R E + + ++ AE +
Sbjct: 353 KAEREQREREKAELEAKEKAEREQREREKAEREAKEKAEREQREREKAEREAREKAEREQ 412
Query: 154 FHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNK 213
+K + + R +ER K++R L+ E+ A +K Q E +
Sbjct: 413 REREKAEREAREKAEREQREREKAER------LAREKAEREAREKAEREQRERE------ 460
Query: 214 IKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKI 273
K + E ++ + E++E+EK +AR EK + E +E ++ E+E + E+
Sbjct: 461 -KAEREAREKAEREQREREKAERLAR-EKAEREAREKAEREQREREKAEREAR-EKAERE 517
Query: 274 QEEMFLLNQI------RRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKG 327
Q E ++ R EKA+ ++R R KAE+EAR KA ++ Q++++ ER+
Sbjct: 518 QREREKAERLAREKAEREAREKAEREQREREKAEREAREKAERE----QREREKAERLAR 573
Query: 328 RGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVARE-GGFSNSIYPTY 386
+ K E E A ++ + +L + + + RE +
Sbjct: 574 EKAEREAREKAEREAREKAEREQREREKAERLAREKAEREAREKAEREQREREKAEREAK 633
Query: 387 NKLDLEVIKREKLRKEA-SFADRIQAARLK 415
K + E +REK +EA A+R Q R K
Sbjct: 634 EKAEREQREREKAEREAKEKAEREQREREK 663
Score = 55.8 bits (133), Expect = 2e-06
Identities = 86/346 (24%), Positives = 147/346 (41%), Gaps = 35/346 (10%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVR--- 145
E E R+A KE+ R A + + K + + E ER +R +A RE K +
Sbjct: 246 EREKAEREA-KEKAEREAKEKAEREQREREKAEREAKEKAEREQREREKAEREAKEKAER 304
Query: 146 --KKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQ 203
K+ AE + +K + + R +ER K++R + E+ A +K Q
Sbjct: 305 EAKEKAEREQREREKAELEAKEKAEREQREREKAEREAK------EKAEREAKEKAEREQ 358
Query: 204 EELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEK 263
E + K +LE ++ + E++E+EK A+ EK + E ++A++ +
Sbjct: 359 RERE-------KAELEAKEKAEREQREREKAEREAK-EKAEREQRE--REKAEREAREKA 408
Query: 264 ELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQK------ 317
E + EK + E + R E+ KA+ AR KAE+EAR KA ++ +K
Sbjct: 409 EREQREREKAEREAREKAE-REQREREKAERLAREKAEREAREKAEREQREREKAEREAR 467
Query: 318 ---KKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAR 374
+++ RER K + + + A E E E ++ K + + + + E A
Sbjct: 468 EKAEREQREREKAERLAREKAEREAREKAER--EQREREKAEREAREKAEREQREREKAE 525
Query: 375 EGGFSNSIYPTYNKLDLEVIKREKLRKEA-SFADRIQAARLKKGNL 419
+ K + E +REK +EA A+R Q R K L
Sbjct: 526 RLAREKAEREAREKAEREQREREKAEREAREKAEREQREREKAERL 571
Score = 55.8 bits (133), Expect = 2e-06
Identities = 62/258 (24%), Positives = 117/258 (45%), Gaps = 16/258 (6%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKM 148
E E R+A +E+ R + R+ K + + E ER +R +A RE + + +
Sbjct: 502 EREKAEREA-REKAEREQREREKAERLAREKAEREAREKAEREQREREKAEREAREKAER 560
Query: 149 AEHPRFHSDKI-KEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELD 207
+ R ++++ +EK R + + K REQ + +A K +E+ +
Sbjct: 561 EQREREKAERLAREKAEREAREKAEREAREKAEREQREREKAERLARE-KAEREAREKAE 619
Query: 208 WDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDW 267
+ + K + E ++ + E++E+EK A+ EK + E ++A++ + E +
Sbjct: 620 REQREREKAEREAKEKAEREQREREKAEREAK-EKAEREQRE--REKAEREAKEKAEREQ 676
Query: 268 DSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKG 327
+EK + E + R EKA+ ++R R KAE+EA+ KA +++ RER K
Sbjct: 677 REHEKAEREAREKAE-REAREKAEREQREREKAEREAKEKA---------EREQREREKA 726
Query: 328 RGNIKSQLCKNANEDGEA 345
K + + E EA
Sbjct: 727 EREAKEKAEREQREREEA 744
Score = 53.9 bits (128), Expect = 8e-06
Identities = 63/262 (24%), Positives = 123/262 (46%), Gaps = 16/262 (6%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKM 148
E E R+A +E+ R + R+ K + + E ER +R +A RE + + +
Sbjct: 458 EREKAEREA-REKAEREQREREKAERLAREKAEREAREKAEREQREREKAEREAREKAER 516
Query: 149 AEHPRFHSDKI-KEKISYSLR----RVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQ 203
+ R ++++ +EK R R +ER K++R + E+ K +
Sbjct: 517 EQREREKAERLAREKAEREAREKAEREQREREKAEREARE---KAEREQREREKAERLAR 573
Query: 204 EELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEK 263
E+ + ++ K + E ++ + E++E+EK +AR EK + E +E ++ E+
Sbjct: 574 EKAEREAREKAER--EAREKAEREQREREKAERLAR-EKAEREAREKAEREQREREKAER 630
Query: 264 ELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQK-KKDLR 322
E + E+ Q E R EKA+ ++R R KAE+EA+ KA ++ ++K +++ R
Sbjct: 631 EAK-EKAEREQREREKAE--REAKEKAEREQREREKAEREAKEKAEREQREHEKAEREAR 687
Query: 323 ERIKGRGNIKSQLCKNANEDGE 344
E+ + K++ + E E
Sbjct: 688 EKAEREAREKAEREQREREKAE 709
Score = 49.3 bits (116), Expect = 2e-04
Identities = 72/323 (22%), Positives = 134/323 (41%), Gaps = 43/323 (13%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKM 148
E + + +E+ R + R+ K + + E ER +R +A RE K +
Sbjct: 579 EAREKAEREAREKAEREQREREKAERLAREKAEREAREKAEREQREREKAEREAKEK--- 635
Query: 149 AEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDW 208
AE + +K + + R +ER K++R A +K Q E +
Sbjct: 636 AEREQREREKAEREAKEKAEREQREREKAER--------------EAKEKAEREQREHE- 680
Query: 209 DSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKE-LDW 267
+ + +E+ E + +AE +++E+ +AE+ + +E +E +K KE +
Sbjct: 681 KAEREAREKAEREAREKAEREQRER----EKAEREAKEKAEREQREREKAEREAKEKAER 736
Query: 268 DSYEKIQEEMFLLNQIRRTT-EKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIK 326
+ E+ + E Q R E+ +A+ AR +AE+EAR KA +++ RER +
Sbjct: 737 EQREREEAERLAREQAEREQREREEAERLAREQAEREAREKA---------EREQREREE 787
Query: 327 GRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTY 386
+ Q + A E EA +Q + + + K +ARE
Sbjct: 788 AERLAREQADREAREKEEAERLAREQEEREAR------EKEEAERLARE----QKEREAR 837
Query: 387 NKLDLEVIKREKLRKEASFADRI 409
K + E + +E+ +EA A R+
Sbjct: 838 EKEEAERLAQEQAEREAEEARRL 860
Score = 48.1 bits (113), Expect = 5e-04
Identities = 70/306 (22%), Positives = 129/306 (41%), Gaps = 42/306 (13%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEA---------- 138
E E L+R+ + R + R+ + + + E ER +R EA
Sbjct: 743 EAERLAREQAEREQRERE----EAERLAREQAEREAREKAEREQREREEAERLAREQADR 798
Query: 139 -LREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAK 197
RE + +++A + KE+ R + + K E+ L+ EQ A +
Sbjct: 799 EAREKEEAERLAREQEEREAREKEEAERLAREQKEREAREKEEAER--LAQEQAEREAEE 856
Query: 198 KGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKK 257
QE+ D ++ K + + ++ + E +EKE+ AE+ Q +E A+EA++
Sbjct: 857 ARRLAQEQADREALEKEEAERLAREQEEREAREKEE------AERLAQEQAEREAREAEE 910
Query: 258 GGSGEKELDWDSYEKIQEEMFLLNQI---RRTTEKAKAKERARTKAEK---EARIKAIKK 311
+E E+ EE L Q R EK +A RAR +AE+ EAR K ++
Sbjct: 911 ADRLARE----QAEREAEEARRLAQEQEEREAREKEEADRRAREQAEREAEEARQKEAER 966
Query: 312 VMLNQKKKDLRERIKGRGNIKSQ-------LCKNANEDGEAALEITQQFKLDTKLTKINI 364
+ ++ + RE+ G + + + NE EA+LE ++ + K ++ +
Sbjct: 967 LEHEHEEPEAREQQDGESISPEEAHPTPYTMVRELNE--EASLEDEEKQAHEQKDSEYDE 1024
Query: 365 SKNINS 370
+N N+
Sbjct: 1025 KQNENN 1030
Score = 35.4 bits (80), Expect = 3.1
Identities = 38/142 (26%), Positives = 61/142 (42%), Gaps = 12/142 (8%)
Query: 284 RRTTEKAKAKERARTKAEKEARIKAIKKVMLNQK---------KKDLRERIKGRGNIKSQ 334
R EKA+ ++R R KAE+EA+ KA ++ +K +++ RER K K +
Sbjct: 198 REAKEKAEREQREREKAEREAKEKAEREQREREKAEREAKEKAEREQREREKAEREAKEK 257
Query: 335 LCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLEVI 394
+ A E E E ++ K + + + + E A + K + E
Sbjct: 258 AEREAKEKAER--EQREREKAEREAKEKAEREQREREKAEREAKEKAEREAKEKAEREQR 315
Query: 395 KREKLRKEA-SFADRIQAARLK 415
+REK EA A+R Q R K
Sbjct: 316 EREKAELEAKEKAEREQREREK 337
>UniRef100_Q7KPY9 Erythrocyte binding protein 4 [Plasmodium yoelii yoelii]
Length = 1680
Score = 60.5 bits (145), Expect = 9e-08
Identities = 66/300 (22%), Positives = 132/300 (44%), Gaps = 36/300 (12%)
Query: 96 KAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFH 155
K +E +R K+ R P K ++ S + R K++ + R+ K +KK + R
Sbjct: 1296 KKAEEEKKRLKLQ-----RKPERKKKRRSGKESRRRKKKRLKLQRKQKKKKKRLKRQRKQ 1350
Query: 156 SDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIK 215
K K L+R ++R K K+ A AAKK +E ++ K +
Sbjct: 1351 KKKKKR-----LKRQRKQRRKEKK-------------AEAAKKAE--EERKRIEAEKKAE 1390
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQE 275
E+ + + + E+E++++ A +AE+ E EA+K E++ ++ +K +E
Sbjct: 1391 EERKKAEAAKKAEEERKRIEAEKKAEE------ERKRIEAEKKAEEERKRI-EAEKKAEE 1443
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
E + +++ E+ K E + E+ RI+A+KK +K+ + ++ + I +
Sbjct: 1444 ERKRIEAVKKAEEERKRIEAEKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKI-IEA 1502
Query: 336 CKNANED---GEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLE 392
K A E+ EA + + K ++ L++ IS N + + F Y +++
Sbjct: 1503 AKKAEEERIKAEAVKKEEEVIKKNSNLSETKISNNYETRNIDDNSFKKLDEEEYKSRNID 1562
Score = 51.2 bits (121), Expect = 5e-05
Identities = 68/296 (22%), Positives = 126/296 (41%), Gaps = 27/296 (9%)
Query: 121 RKHSAETRERI--KRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSK 178
RK AE ++ +++ SEA ++ RKK AE + +K K + + ++ +K
Sbjct: 1237 RKKKAEAAKKALERKKKSEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKAEAAK 1296
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQ-------QLLQAEEKEK 231
+ E+ Q KK G+E S + K++L+LQ + L+ + K+K
Sbjct: 1297 KAEEEKKRLKLQRKPERKKKRRSGKE-----SRRRKKKRLKLQRKQKKKKKRLKRQRKQK 1351
Query: 232 EKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAK 291
+K + R K Q E A+ AKK K + ++ +K +EE ++ E+ K
Sbjct: 1352 KKKKRLKRQRK--QRRKEKKAEAAKKAEEERKRI--EAEKKAEEERKKAEAAKKAEEERK 1407
Query: 292 AKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQ 351
E + E+ RI+A KK +K+ + ++ + + + K A E+ + I
Sbjct: 1408 RIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERK-RIEAVKKAEEERK---RIEA 1463
Query: 352 QFKLDTKLTKINISKNINS-----EVAREGGFSNSIYPTYNKLDLEVIKREKLRKE 402
+ K + + +I K E ++ I K + E IK E ++KE
Sbjct: 1464 EKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKIIEAAKKAEEERIKAEAVKKE 1519
Score = 45.8 bits (107), Expect = 0.002
Identities = 47/211 (22%), Positives = 98/211 (46%), Gaps = 23/211 (10%)
Query: 114 RVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQE 173
+ W K + + E +K+ E R +K E R ++K E+ ++
Sbjct: 1110 KAAWAKKAEEERKKAEAVKKAEEERKRIEAEKKAEEERKRIEAEKKAEEE--------RK 1161
Query: 174 RLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEK 233
R+++++ E+ E+ I AAKK +E + K +E+ + + + E+E++K
Sbjct: 1162 RIEAEKKAEE-----ERKIIEAAKKAE--EERKRIEEAKKAEEERKKIEAAKKAEEERKK 1214
Query: 234 LMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAK 293
A+ +AE+ ++ A+ AKK +K+ ++ +K E ++ E+ K
Sbjct: 1215 AEAVKKAEE-----AKKKAEAAKKAEERKKKA--EAAKKALERKKKSEAAKKALERKKKA 1267
Query: 294 ERARTKAEKEARIKAIKKVMLNQKKKDLRER 324
E A+ KAE++ + +A KK +KK + ++
Sbjct: 1268 EAAK-KAEEKKKAEAAKKAEEEKKKAEAAKK 1297
Score = 41.6 bits (96), Expect = 0.043
Identities = 51/236 (21%), Positives = 106/236 (44%), Gaps = 14/236 (5%)
Query: 110 ANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRK----KMAEHPRFHSDKIKEKISY 165
++ R +N ++S T R E R K+ + E+ SD +
Sbjct: 1024 SSHNRAKYNTPIENSESTIVRKHNSAPEHFRSLKINSYTPNRRGENFAKESDSTRNTDES 1083
Query: 166 SLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQ 225
+ V ++R ++ + E I E A AKK +E ++ K +E+ + + +
Sbjct: 1084 KMDEVIRKREEAAKNAEIIRKFEEAQKAAWAKKAE--EERKKAEAVKKAEEERKRIEAEK 1141
Query: 226 AEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRR 285
E+E++++ A +AE+ E EA+K E+++ ++ +K +EE + + ++
Sbjct: 1142 KAEEERKRIEAEKKAEE------ERKRIEAEKKAEEERKII-EAAKKAEEERKRIEEAKK 1194
Query: 286 TTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANE 341
E+ K E A+ E+ + +A+KK +KK + ++ + R K++ K A E
Sbjct: 1195 AEEERKKIEAAKKAEEERKKAEAVKKAEEAKKKAEAAKKAEERKK-KAEAAKKALE 1249
Score = 40.4 bits (93), Expect = 0.095
Identities = 69/319 (21%), Positives = 129/319 (39%), Gaps = 34/319 (10%)
Query: 122 KHSAETRERI---KRRTSEALREPKVRKKMAEHPRFHSDKIKE---KISYSLRRVWQERL 175
K + E R+RI K+ E R +K E R ++K E KI + ++ +ER
Sbjct: 1128 KKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKIIEAAKKAEEER- 1186
Query: 176 KSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLM 235
KR E E+ AAKK +E ++ K +E + + + E+ K+K
Sbjct: 1187 --KRIEEAKKAEEERKKIEAAKKAE--EERKKAEAVKKAEEAKKKAEAAKKAEERKKKAE 1242
Query: 236 AIARA---EKFIQSWSECL-----------------AKEAKKGGSGEKELDWDSYEKIQE 275
A +A +K ++ + L A+ AKK +K+ ++ +K +E
Sbjct: 1243 AAKKALERKKKSEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKA--EAAKKAEE 1300
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKK-KDLRERIKGRGNIKSQ 334
E L R+ K K + ++ K+ R+K +K +K+ K R++ K + +K Q
Sbjct: 1301 EKKRLKLQRKPERKKKRRSGKESRRRKKKRLKLQRKQKKKKKRLKRQRKQKKKKKRLKRQ 1360
Query: 335 LCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLEVI 394
+ E A + ++ + + K + +E A++ K + E
Sbjct: 1361 RKQRRKEKKAEAAKKAEEERKRIEAEKKAEEERKKAEAAKKAEEERKRIEAEKKAEEERK 1420
Query: 395 KREKLRKEASFADRIQAAR 413
+ E +K RI+A +
Sbjct: 1421 RIEAEKKAEEERKRIEAEK 1439
>UniRef100_Q7KPZ1 Erythrocyte binding protein 3 [Plasmodium yoelii yoelii]
Length = 1652
Score = 60.5 bits (145), Expect = 9e-08
Identities = 66/300 (22%), Positives = 132/300 (44%), Gaps = 36/300 (12%)
Query: 96 KAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFH 155
K +E +R K+ R P K ++ S + R K++ + R+ K +KK + R
Sbjct: 1296 KKAEEEKKRLKLQ-----RKPERKKKRRSGKESRRRKKKRLKLQRKQKKKKKRLKRQRKQ 1350
Query: 156 SDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIK 215
K K L+R ++R K K+ A AAKK +E ++ K +
Sbjct: 1351 KKKKKR-----LKRQRKQRRKEKK-------------AEAAKKAE--EERKRIEAEKKAE 1390
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQE 275
E+ + + + E+E++++ A +AE+ E EA+K E++ ++ +K +E
Sbjct: 1391 EERKKAEAAKKAEEERKRIEAEKKAEE------ERKRIEAEKKAEEERKRI-EAEKKAEE 1443
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
E + +++ E+ K E + E+ RI+A+KK +K+ + ++ + I +
Sbjct: 1444 ERKRIEAVKKAEEERKRIEAEKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKI-IEA 1502
Query: 336 CKNANED---GEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLE 392
K A E+ EA + + K ++ L++ IS N + + F Y +++
Sbjct: 1503 AKKAEEERIKAEAVKKEEEVIKKNSNLSETKISNNYETRNIDDNSFKKLDEEEYKSRNID 1562
Score = 51.2 bits (121), Expect = 5e-05
Identities = 68/296 (22%), Positives = 126/296 (41%), Gaps = 27/296 (9%)
Query: 121 RKHSAETRERI--KRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSK 178
RK AE ++ +++ SEA ++ RKK AE + +K K + + ++ +K
Sbjct: 1237 RKKKAEAAKKALERKKKSEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKAEAAK 1296
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQ-------QLLQAEEKEK 231
+ E+ Q KK G+E S + K++L+LQ + L+ + K+K
Sbjct: 1297 KAEEEKKRLKLQRKPERKKKRRSGKE-----SRRRKKKRLKLQRKQKKKKKRLKRQRKQK 1351
Query: 232 EKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAK 291
+K + R K Q E A+ AKK K + ++ +K +EE ++ E+ K
Sbjct: 1352 KKKKRLKRQRK--QRRKEKKAEAAKKAEEERKRI--EAEKKAEEERKKAEAAKKAEEERK 1407
Query: 292 AKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQ 351
E + E+ RI+A KK +K+ + ++ + + + K A E+ + I
Sbjct: 1408 RIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERK-RIEAVKKAEEERK---RIEA 1463
Query: 352 QFKLDTKLTKINISKNINS-----EVAREGGFSNSIYPTYNKLDLEVIKREKLRKE 402
+ K + + +I K E ++ I K + E IK E ++KE
Sbjct: 1464 EKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKIIEAAKKAEEERIKAEAVKKE 1519
Score = 45.8 bits (107), Expect = 0.002
Identities = 47/211 (22%), Positives = 98/211 (46%), Gaps = 23/211 (10%)
Query: 114 RVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQE 173
+ W K + + E +K+ E R +K E R ++K E+ ++
Sbjct: 1110 KAAWAKKAEEERKKAEAVKKAEEERKRIEAEKKAEEERKRIEAEKKAEEE--------RK 1161
Query: 174 RLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEK 233
R+++++ E+ E+ I AAKK +E + K +E+ + + + E+E++K
Sbjct: 1162 RIEAEKKAEE-----ERKIIEAAKKAE--EERKRIEEAKKAEEERKKIEAAKKAEEERKK 1214
Query: 234 LMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAK 293
A+ +AE+ ++ A+ AKK +K+ ++ +K E ++ E+ K
Sbjct: 1215 AEAVKKAEE-----AKKKAEAAKKAEERKKKA--EAAKKALERKKKSEAAKKALERKKKA 1267
Query: 294 ERARTKAEKEARIKAIKKVMLNQKKKDLRER 324
E A+ KAE++ + +A KK +KK + ++
Sbjct: 1268 EAAK-KAEEKKKAEAAKKAEEEKKKAEAAKK 1297
Score = 41.6 bits (96), Expect = 0.043
Identities = 51/236 (21%), Positives = 106/236 (44%), Gaps = 14/236 (5%)
Query: 110 ANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRK----KMAEHPRFHSDKIKEKISY 165
++ R +N ++S T R E R K+ + E+ SD +
Sbjct: 1024 SSHNRAKYNTPIENSESTIVRKHNSAPEHFRSLKINSYTPNRRGENFAKESDSTRNTDES 1083
Query: 166 SLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQ 225
+ V ++R ++ + E I E A AKK +E ++ K +E+ + + +
Sbjct: 1084 KMDEVIRKREEAAKNAEIIRKFEEAQKAAWAKKAE--EERKKAEAVKKAEEERKRIEAEK 1141
Query: 226 AEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRR 285
E+E++++ A +AE+ E EA+K E+++ ++ +K +EE + + ++
Sbjct: 1142 KAEEERKRIEAEKKAEE------ERKRIEAEKKAEEERKII-EAAKKAEEERKRIEEAKK 1194
Query: 286 TTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANE 341
E+ K E A+ E+ + +A+KK +KK + ++ + R K++ K A E
Sbjct: 1195 AEEERKKIEAAKKAEEERKKAEAVKKAEEAKKKAEAAKKAEERKK-KAEAAKKALE 1249
Score = 40.4 bits (93), Expect = 0.095
Identities = 69/319 (21%), Positives = 129/319 (39%), Gaps = 34/319 (10%)
Query: 122 KHSAETRERI---KRRTSEALREPKVRKKMAEHPRFHSDKIKE---KISYSLRRVWQERL 175
K + E R+RI K+ E R +K E R ++K E KI + ++ +ER
Sbjct: 1128 KKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKIIEAAKKAEEER- 1186
Query: 176 KSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLM 235
KR E E+ AAKK +E ++ K +E + + + E+ K+K
Sbjct: 1187 --KRIEEAKKAEEERKKIEAAKKAE--EERKKAEAVKKAEEAKKKAEAAKKAEERKKKAE 1242
Query: 236 AIARA---EKFIQSWSECL-----------------AKEAKKGGSGEKELDWDSYEKIQE 275
A +A +K ++ + L A+ AKK +K+ ++ +K +E
Sbjct: 1243 AAKKALERKKKSEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKA--EAAKKAEE 1300
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKK-KDLRERIKGRGNIKSQ 334
E L R+ K K + ++ K+ R+K +K +K+ K R++ K + +K Q
Sbjct: 1301 EKKRLKLQRKPERKKKRRSGKESRRRKKKRLKLQRKQKKKKKRLKRQRKQKKKKKRLKRQ 1360
Query: 335 LCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLEVI 394
+ E A + ++ + + K + +E A++ K + E
Sbjct: 1361 RKQRRKEKKAEAAKKAEEERKRIEAEKKAEEERKKAEAAKKAEEERKRIEAEKKAEEERK 1420
Query: 395 KREKLRKEASFADRIQAAR 413
+ E +K RI+A +
Sbjct: 1421 RIEAEKKAEEERKRIEAEK 1439
>UniRef100_O61164 Erythrocyte binding protein 1 [Plasmodium yoelii yoelii]
Length = 1701
Score = 60.5 bits (145), Expect = 9e-08
Identities = 66/300 (22%), Positives = 132/300 (44%), Gaps = 36/300 (12%)
Query: 96 KAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFH 155
K +E +R K+ R P K ++ S + R K++ + R+ K +KK + R
Sbjct: 1296 KKAEEEKKRLKLQ-----RKPERKKKRRSGKESRRRKKKRLKLQRKQKKKKKRLKRQRKQ 1350
Query: 156 SDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIK 215
K K L+R ++R K K+ A AAKK +E ++ K +
Sbjct: 1351 KKKKKR-----LKRQRKQRRKEKK-------------AEAAKKAE--EERKRIEAEKKAE 1390
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQE 275
E+ + + + E+E++++ A +AE+ E EA+K E++ ++ +K +E
Sbjct: 1391 EERKKAEAAKKAEEERKRIEAEKKAEE------ERKRIEAEKKAEEERKRI-EAEKKAEE 1443
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
E + +++ E+ K E + E+ RI+A+KK +K+ + ++ + I +
Sbjct: 1444 ERKRIEAVKKAEEERKRIEAEKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKI-IEA 1502
Query: 336 CKNANED---GEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLE 392
K A E+ EA + + K ++ L++ IS N + + F Y +++
Sbjct: 1503 AKKAEEERIKAEAVKKEEEVIKKNSNLSETKISNNYETRNIDDNSFKKLDEEEYKSRNID 1562
Score = 51.2 bits (121), Expect = 5e-05
Identities = 68/296 (22%), Positives = 126/296 (41%), Gaps = 27/296 (9%)
Query: 121 RKHSAETRERI--KRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSK 178
RK AE ++ +++ SEA ++ RKK AE + +K K + + ++ +K
Sbjct: 1237 RKKKAEAAKKALERKKKSEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKAEAAK 1296
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQ-------QLLQAEEKEK 231
+ E+ Q KK G+E S + K++L+LQ + L+ + K+K
Sbjct: 1297 KAEEEKKRLKLQRKPERKKKRRSGKE-----SRRRKKKRLKLQRKQKKKKKRLKRQRKQK 1351
Query: 232 EKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAK 291
+K + R K Q E A+ AKK K + ++ +K +EE ++ E+ K
Sbjct: 1352 KKKKRLKRQRK--QRRKEKKAEAAKKAEEERKRI--EAEKKAEEERKKAEAAKKAEEERK 1407
Query: 292 AKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQ 351
E + E+ RI+A KK +K+ + ++ + + + K A E+ + I
Sbjct: 1408 RIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERK-RIEAVKKAEEERK---RIEA 1463
Query: 352 QFKLDTKLTKINISKNINS-----EVAREGGFSNSIYPTYNKLDLEVIKREKLRKE 402
+ K + + +I K E ++ I K + E IK E ++KE
Sbjct: 1464 EKKAEEERKRIEAVKKAEEERKRIEAEKKAEEERKIIEAAKKAEEERIKAEAVKKE 1519
Score = 45.8 bits (107), Expect = 0.002
Identities = 47/211 (22%), Positives = 98/211 (46%), Gaps = 23/211 (10%)
Query: 114 RVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQE 173
+ W K + + E +K+ E R +K E R ++K E+ ++
Sbjct: 1110 KAAWAKKAEEERKKAEAVKKAEEERKRIEAEKKAEEERKRIEAEKKAEEE--------RK 1161
Query: 174 RLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEK 233
R+++++ E+ E+ I AAKK +E + K +E+ + + + E+E++K
Sbjct: 1162 RIEAEKKAEE-----ERKIIEAAKKAE--EERKRIEEAKKAEEERKKIEAAKKAEEERKK 1214
Query: 234 LMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAK 293
A+ +AE+ ++ A+ AKK +K+ ++ +K E ++ E+ K
Sbjct: 1215 AEAVKKAEE-----AKKKAEAAKKAEERKKKA--EAAKKALERKKKSEAAKKALERKKKA 1267
Query: 294 ERARTKAEKEARIKAIKKVMLNQKKKDLRER 324
E A+ KAE++ + +A KK +KK + ++
Sbjct: 1268 EAAK-KAEEKKKAEAAKKAEEEKKKAEAAKK 1297
Score = 41.6 bits (96), Expect = 0.043
Identities = 51/236 (21%), Positives = 106/236 (44%), Gaps = 14/236 (5%)
Query: 110 ANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRK----KMAEHPRFHSDKIKEKISY 165
++ R +N ++S T R E R K+ + E+ SD +
Sbjct: 1024 SSHNRAKYNTPIENSESTIVRKHNSAPEHFRSLKINSYTPNRRGENFAKESDSTRNTDES 1083
Query: 166 SLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQ 225
+ V ++R ++ + E I E A AKK +E ++ K +E+ + + +
Sbjct: 1084 KMDEVIRKREEAAKNAEIIRKFEEAQKAAWAKKAE--EERKKAEAVKKAEEERKRIEAEK 1141
Query: 226 AEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRR 285
E+E++++ A +AE+ E EA+K E+++ ++ +K +EE + + ++
Sbjct: 1142 KAEEERKRIEAEKKAEE------ERKRIEAEKKAEEERKII-EAAKKAEEERKRIEEAKK 1194
Query: 286 TTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANE 341
E+ K E A+ E+ + +A+KK +KK + ++ + R K++ K A E
Sbjct: 1195 AEEERKKIEAAKKAEEERKKAEAVKKAEEAKKKAEAAKKAEERKK-KAEAAKKALE 1249
Score = 40.4 bits (93), Expect = 0.095
Identities = 69/319 (21%), Positives = 129/319 (39%), Gaps = 34/319 (10%)
Query: 122 KHSAETRERI---KRRTSEALREPKVRKKMAEHPRFHSDKIKE---KISYSLRRVWQERL 175
K + E R+RI K+ E R +K E R ++K E KI + ++ +ER
Sbjct: 1128 KKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKRIEAEKKAEEERKIIEAAKKAEEER- 1186
Query: 176 KSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLM 235
KR E E+ AAKK +E ++ K +E + + + E+ K+K
Sbjct: 1187 --KRIEEAKKAEEERKKIEAAKKAE--EERKKAEAVKKAEEAKKKAEAAKKAEERKKKAE 1242
Query: 236 AIARA---EKFIQSWSECL-----------------AKEAKKGGSGEKELDWDSYEKIQE 275
A +A +K ++ + L A+ AKK +K+ ++ +K +E
Sbjct: 1243 AAKKALERKKKSEAAKKALERKKKAEAAKKAEEKKKAEAAKKAEEEKKKA--EAAKKAEE 1300
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKK-KDLRERIKGRGNIKSQ 334
E L R+ K K + ++ K+ R+K +K +K+ K R++ K + +K Q
Sbjct: 1301 EKKRLKLQRKPERKKKRRSGKESRRRKKKRLKLQRKQKKKKKRLKRQRKQKKKKKRLKRQ 1360
Query: 335 LCKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLEVI 394
+ E A + ++ + + K + +E A++ K + E
Sbjct: 1361 RKQRRKEKKAEAAKKAEEERKRIEAEKKAEEERKKAEAAKKAEEERKRIEAEKKAEEERK 1420
Query: 395 KREKLRKEASFADRIQAAR 413
+ E +K RI+A +
Sbjct: 1421 RIEAEKKAEEERKRIEAEK 1439
>UniRef100_UPI000046B0E5 UPI000046B0E5 UniRef100 entry
Length = 1712
Score = 60.1 bits (144), Expect = 1e-07
Identities = 64/284 (22%), Positives = 125/284 (43%), Gaps = 22/284 (7%)
Query: 96 KAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFH 155
+A K +R K A K K +K A + KR+ EA ++ + ++K AE +
Sbjct: 1282 EAKKVEEKRKKDEAAKKAE---EKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKAEAAKKA 1338
Query: 156 SDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIK 215
K K++ + ++ +K+ E E+ A AKK +E + K++
Sbjct: 1339 ERKEKDEAAKKAEEKRKKDEAAKKVEE------ERKKAEEAKKAE--EERKRIEEAKKVE 1390
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQE 275
E+ + + + E+E++K A +AE+ +EAKK K ++ + +K++E
Sbjct: 1391 EKRKKDEAAKKAEEERKKAEAAKKAEE-----ERKRIEEAKKAEEERKRIE--AAKKVEE 1443
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
E + + ++ E+ K E A+ E+ RI+A KKV +++K + E K K
Sbjct: 1444 ERKRIEEAKKVEEERKRIEEAKKAEEERKRIEAAKKV--EEERKRIEEAKKSEEERKR-- 1499
Query: 336 CKNANEDGEAALEITQQFKLDTKLTKINISKNINSEVAREGGFS 379
+ A + E I K + + +I +K E+ ++ +
Sbjct: 1500 IEEAKKAEEERKRIEAAKKAEEERKRIEEAKKAEEEIKKDSNLA 1543
Score = 46.6 bits (109), Expect = 0.001
Identities = 65/298 (21%), Positives = 125/298 (41%), Gaps = 28/298 (9%)
Query: 119 KGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSK 178
K RK + E E +K+ E R +K E + + K E+ + ++++ K
Sbjct: 1115 KKRKEAGENAEEVKKAEEERKRIEAAKKVEEERKKAEAAKKAEEERKRIEEA--KKVEEK 1172
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIA 238
R +++ AAKK ++ ++ K +E+ + + + E++++K A
Sbjct: 1173 RKKDE-----------AAKKAE--EKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAK 1219
Query: 239 RAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERART 298
+AE+ +EAKK EK ++ +K +E+ ++ EK K E A+
Sbjct: 1220 KAEE-----ERKRIEEAKK--VEEKRKKDEAAKKAEEKRKKDEAAKKAEEKRKKDEAAKK 1272
Query: 299 KAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANE---DGEAALEITQQFKL 355
E+ RI+ KKV +KK + ++ + + K + K A E EAA + ++ K
Sbjct: 1273 VEEERKRIEEAKKVEEKRKKDEAAKKAEEKRK-KDEAAKKAEEKRKKDEAAKKAEEKRKK 1331
Query: 356 DTKLTKINISKNINSEVAREGGFSNSIYPTYNKLDLEVIKREKLRKEASFADRIQAAR 413
K + E A++ K++ E K E+ +K RI+ A+
Sbjct: 1332 AEAAKK--AERKEKDEAAKKAEEKRKKDEAAKKVEEERKKAEEAKKAEEERKRIEEAK 1387
Score = 39.3 bits (90), Expect = 0.21
Identities = 48/206 (23%), Positives = 88/206 (42%), Gaps = 17/206 (8%)
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQE 275
++ + ++++ E E EK I R KF + + K+ K+ G +E+ +K +E
Sbjct: 1081 DRSRMDEVIRVRE-EAEKNAKIIR--KFEELRIADMIKKRKEAGENAEEV-----KKAEE 1132
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
E + ++ E+ K E A+ E+ RI+ KKV +KK + ++ + + K +
Sbjct: 1133 ERKRIEAAKKVEEERKKAEAAKKAEEERKRIEEAKKVEEKRKKDEAAKKAEEKRK-KDEA 1191
Query: 336 CKNANE---DGEAALEITQQFKLDTKLTKI-----NISKNINSEVAREGGFSNSIYPTYN 387
K A E EAA + ++ K D K I + E R+ +
Sbjct: 1192 AKKAEEKRKKDEAAKKAEEKRKKDEAAKKAEEERKRIEEAKKVEEKRKKDEAAKKAEEKR 1251
Query: 388 KLDLEVIKREKLRKEASFADRIQAAR 413
K D K E+ RK+ A +++ R
Sbjct: 1252 KKDEAAKKAEEKRKKDEAAKKVEEER 1277
Score = 35.8 bits (81), Expect = 2.3
Identities = 43/199 (21%), Positives = 78/199 (38%), Gaps = 7/199 (3%)
Query: 96 KAVKERMRRFKIGLANKGRVPWNKGR--KHSAETRERI---KRRTSEALREPKVRKKMAE 150
+A K +R K A K K K + E R+RI K+ E R +K E
Sbjct: 1385 EAKKVEEKRKKDEAAKKAEEERKKAEAAKKAEEERKRIEEAKKAEEERKRIEAAKKVEEE 1444
Query: 151 HPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDS 210
R K E+ + + + KR + E+ AKK +E +
Sbjct: 1445 RKRIEEAKKVEEERKRIEEAKKAEEERKRIEAAKKVEEERKRIEEAKKSE--EERKRIEE 1502
Query: 211 YNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSY 270
K +E+ + + + E+E++++ +AE+ I+ S K+ + +D +S+
Sbjct: 1503 AKKAEEERKRIEAAKKAEEERKRIEEAKKAEEEIKKDSNLAEKKISSSNYETRHIDDNSF 1562
Query: 271 EKIQEEMFLLNQIRRTTEK 289
+K+ E + I T K
Sbjct: 1563 KKLDEAEYKSRNIDSTRNK 1581
>UniRef100_UPI0000335A68 UPI0000335A68 UniRef100 entry
Length = 237
Score = 59.7 bits (143), Expect = 2e-07
Identities = 62/218 (28%), Positives = 105/218 (47%), Gaps = 25/218 (11%)
Query: 99 KERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALR-EPKVRKKMAEHPRFHSD 157
KE KI NK + + K AE R K L+ E + RKK E R ++
Sbjct: 25 KELEDELKITYENK--LKEEEQAKIQAEEEARKKAEEDARLKAEEEARKKAEEEARLKAE 82
Query: 158 KIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQ 217
+ K + +R+ E++ ++ E+ FL E+ A KK +EE + + +++
Sbjct: 83 EEARKKAEEEQRIKNEKIAQQKAEEEAFLKAEE---EARKKA---EEEARLKAEEEARKK 136
Query: 218 LELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKEL--DWDSYEKIQE 275
E + L+AEE+ ++K AR + +EA+K E L + ++ +K +E
Sbjct: 137 AEEEARLKAEEEARKKAEEEARLKA---------EEEARKKAEEEARLKAEEEARKKAEE 187
Query: 276 EMFLLNQI---RRTTEKA--KAKERARTKAEKEARIKA 308
E + N+ ++ E+A KA+E AR KAE+EAR+KA
Sbjct: 188 EQRIKNEKIAQQKAEEEAFLKAEEEARKKAEEEARLKA 225
Score = 42.0 bits (97), Expect = 0.033
Identities = 53/184 (28%), Positives = 86/184 (45%), Gaps = 39/184 (21%)
Query: 203 QEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIAR--AEKFIQSWSECLAK-----EA 255
++EL NK+KE E Q +QAEE+ ++K AR AE+ + +E A+ EA
Sbjct: 28 EDELKITYENKLKE--EEQAKIQAEEEARKKAEEDARLKAEEEARKKAEEEARLKAEEEA 85
Query: 256 KKGGSGEKELDWD--SYEKIQEEMFLL------------------NQIRRTTE---KAKA 292
+K E+ + + + +K +EE FL + R+ E + KA
Sbjct: 86 RKKAEEEQRIKNEKIAQQKAEEEAFLKAEEEARKKAEEEARLKAEEEARKKAEEEARLKA 145
Query: 293 KERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQ 352
+E AR KAE+EAR+KA ++ +KK + R+K + + K A E+ E Q
Sbjct: 146 EEEARKKAEEEARLKAEEEA---RKKAEEEARLKA----EEEARKKAEEEQRIKNEKIAQ 198
Query: 353 FKLD 356
K +
Sbjct: 199 QKAE 202
>UniRef100_UPI0000249BD4 UPI0000249BD4 UniRef100 entry
Length = 414
Score = 59.3 bits (142), Expect = 2e-07
Identities = 69/263 (26%), Positives = 130/263 (49%), Gaps = 42/263 (15%)
Query: 86 DSLEIESLSR-KAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKV 144
+ E E L++ KA KER+ + K+ K R+ K K AE +ER+ + +E + ++
Sbjct: 169 EKAEKERLAKEKAEKERVAKEKV---EKERIEKEKAEKEKAE-KERLAKEKAE---KERL 221
Query: 145 RKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQE 204
K+ AE R +K EK + +V +ER++ ++ + E+ + A+K +E
Sbjct: 222 AKEKAEKERIEKEKA-EKERLAKEKVEKERIEKEKAEK------ERLVKEKAEKERLAKE 274
Query: 205 ELDWDSYNKIKEQLEL-------QQLLQAEEKEKEKL------MAIARAEKF----IQSW 247
+ + + K K + E ++ L+ E+ EKE+L +A +AEK +++
Sbjct: 275 KAEKEKIEKEKAEKERLAKEKAEKERLEREKAEKERLAKEKERLAKEKAEKEKNEKVKAE 334
Query: 248 SECLAKEA------KKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAE 301
E LAK+ +K + ++ L + EK + EM L + ++ EK KA++R + + E
Sbjct: 335 KERLAKKKAEKEKLEKKNTEKERLSKEKPEKERAEMERLTKEKKQVEKEKAEKR-KPQKE 393
Query: 302 KEARIKAIKKVMLNQKKKDLRER 324
KE K K + +KD++E+
Sbjct: 394 KEREEKRASK---GKAEKDVKEK 413
Score = 58.9 bits (141), Expect = 3e-07
Identities = 79/337 (23%), Positives = 146/337 (42%), Gaps = 30/337 (8%)
Query: 84 LSDSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPK 143
+ + +E + +++ A+ E R + + R +K E ++K + +E K
Sbjct: 65 IHEKIEAKKIAKLALAEV--RSVLAQEEEERQATLAAKKAKEEAEAKLKEERFQREQEKK 122
Query: 144 VRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQ 203
+ K+ AE R +K+ ++ + + +ERL ++ E+ + E+ A+K +
Sbjct: 123 LAKEKAERERMEQEKMAKERA---EKAEKERLDEEK-NEKERIEKEKAEKEKAEKERLAK 178
Query: 204 EELDWDSYNKIKEQLELQQLLQAEEKEKEK-LMAIARAEKFIQSWSECLAKEAKKGGSGE 262
E+ + + K K + E + +AE+++ EK +A +AEK E LAKE + E
Sbjct: 179 EKAEKERVAKEKVEKERIEKEKAEKEKAEKERLAKEKAEK------ERLAKEKAEKERIE 232
Query: 263 KELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLR 322
KE K ++E R EK + + + KAEKE +K + K+K +
Sbjct: 233 KE-------KAEKE-------RLAKEKVEKERIEKEKAEKERLVKEKAEKERLAKEKAEK 278
Query: 323 ERIKGRGNIKSQLCKNANEDGEAALEITQQFKL---DTKLTKINISKNINSEVAREGGFS 379
E+I+ K +L K E E ++ +L +L K K N +V E
Sbjct: 279 EKIEKEKAEKERLAKEKAEKERLEREKAEKERLAKEKERLAKEKAEKEKNEKVKAEKERL 338
Query: 380 NSIYPTYNKLDLEVIKREKLRKEASFADRIQAARLKK 416
KL+ + ++E+L KE +R + RL K
Sbjct: 339 AKKKAEKEKLEKKNTEKERLSKEKPEKERAEMERLTK 375
Score = 53.1 bits (126), Expect = 1e-05
Identities = 75/340 (22%), Positives = 145/340 (42%), Gaps = 32/340 (9%)
Query: 86 DSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVR 145
D ++ S++ ++ R+ K L + R+ E+I+ + L +VR
Sbjct: 35 DIVDYNSVAEHPTRDIGRKLKTALKEQLRI-----------IHEKIEAKKIAKLALAEVR 83
Query: 146 KKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFR--EQIFLSWEQCIANAAKKGGCGQ 203
+A+ + K + + +LK +RF+ ++ L+ E+ ++ +
Sbjct: 84 SVLAQEEEERQATLAAK---KAKEEAEAKLKEERFQREQEKKLAKEKAERERMEQEKMAK 140
Query: 204 EELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKE-AKKGGSGE 262
E + ++ E+ ++ ++ E+ EKEK AEK E LAKE A+K +
Sbjct: 141 ERAEKAEKERLDEEKNEKERIEKEKAEKEK------AEK------ERLAKEKAEKERVAK 188
Query: 263 KELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLR 322
++++ + EK + E + R EKA+ + A+ KAEKE K + K+K +
Sbjct: 189 EKVEKERIEKEKAEKEKAEKERLAKEKAEKERLAKEKAEKERIEKEKAEKERLAKEKVEK 248
Query: 323 ERIKGRGNIKSQLCKNANEDGEAALEITQQFKLD-TKLTKINISKN--INSEVAREGGFS 379
ERI+ K +L K E A E ++ K++ K K ++K + RE
Sbjct: 249 ERIEKEKAEKERLVKEKAEKERLAKEKAEKEKIEKEKAEKERLAKEKAEKERLEREKAEK 308
Query: 380 NSIYPTYNKLDLEVIKREKLRKEASFADRIQAARLKKGNL 419
+ +L E ++EK K + +R+ + +K L
Sbjct: 309 ERLAKEKERLAKEKAEKEKNEKVKAEKERLAKKKAEKEKL 348
>UniRef100_Q8N9W4 Hypothetical protein FLJ36144 [Homo sapiens]
Length = 650
Score = 58.9 bits (141), Expect = 3e-07
Identities = 47/215 (21%), Positives = 101/215 (46%), Gaps = 10/215 (4%)
Query: 121 RKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQE----RLK 176
R+ E RE+ K R E + + K + + + EK+ R+W++ R +
Sbjct: 331 REQEKELREQKKLREQEEQMQEQEEKMWEQEEKMREQE--EKMWRQEERLWEQEKQMREQ 388
Query: 177 SKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMA 236
++ R+Q WEQ K+ ++E W+ K++E+ ++Q+ + ++EK+
Sbjct: 389 EQKMRDQEERMWEQDERLREKEERMREQEKMWEQVEKMREEKKMQEQEKKTRDQEEKMQE 448
Query: 237 ---IARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAK 293
I EK ++ E + ++ +K E+ + W+ EK ++ L Q + E+ K +
Sbjct: 449 EERIREREKKMREEEETMREQEEKMQKQEENM-WEQEEKEWQQQRLPEQKEKLWEQEKMQ 507
Query: 294 ERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGR 328
E+ E+E +I+ +++ +KK E++ G+
Sbjct: 508 EQEEKIWEQEEKIRDQEEMWGQEKKMWQEEKMWGQ 542
Score = 54.7 bits (130), Expect = 5e-06
Identities = 52/240 (21%), Positives = 111/240 (45%), Gaps = 11/240 (4%)
Query: 85 SDSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKV 144
S+ EI+ L+ K +K ++ R K L +V N ++ E ++ + + ++ +
Sbjct: 239 SEKSEIQ-LNVKELKRKLERAKFLLP---QVQTNTLQEEMWRQEEELREQEKKIRKQEEK 294
Query: 145 RKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQE 204
+ E R K++E+ R+ + R + K REQ EQ ++ QE
Sbjct: 295 MWRQEERLREQEGKMREQEEKMWRQEKRLREQEKELREQEKELREQKKLREQEEQMQEQE 354
Query: 205 ELDWDSYNKIKEQLEL-----QQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGG 259
E W+ K++EQ E ++L + E++ +E+ + E+ + W + K+
Sbjct: 355 EKMWEQEEKMREQEEKMWRQEERLWEQEKQMREQEQKMRDQEE--RMWEQDERLREKEER 412
Query: 260 SGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKK 319
E+E W+ EK++EE + Q ++T ++ + + E+E +++ ++ M Q++K
Sbjct: 413 MREQEKMWEQVEKMREEKKMQEQEKKTRDQEEKMQEEERIREREKKMREEEETMREQEEK 472
>UniRef100_UPI0000499259 UPI0000499259 UniRef100 entry
Length = 1598
Score = 57.8 bits (138), Expect = 6e-07
Identities = 84/341 (24%), Positives = 156/341 (45%), Gaps = 35/341 (10%)
Query: 65 SETSVDSS-DYLNGGDGVVGLSDSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKH 123
+E+SVDS D + + + + DS+ S +K KE+ KI N K RK
Sbjct: 287 NESSVDSEEDRKDQQNQTIPIIDSIG-SSRIKKTQKEQRGLNKI---NSFDEVVEKKRKE 342
Query: 124 SAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQ 183
E ++ K S+ L E K+ AE + +++ +K+ +R E K K RE+
Sbjct: 343 DLEKLKKAKEELSK-LEEEKIA---AEKEKEEANEKVQKLEEEMREKKIEIEKLKVEREE 398
Query: 184 IFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKF 243
F + + ++ C + ++ + +IKE++E Q EKE E+L+ + +
Sbjct: 399 SF----RLLKGQKEQSLCEIQRVEQEKEKRIKEEVEKAH--QLREKEFEELLKRIKELEA 452
Query: 244 IQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQ-IRRTTEKA-KAKERARTKAE 301
I +E E KK K L+ + ++ +EE+ L+ Q I+ EK K E + K E
Sbjct: 453 INLENEYNKDELKK-----KCLEISNIQEEKEELILIVQEIKEENEKRIKEIENLQEKKE 507
Query: 302 KEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAALEITQQFKLDTKLTK 361
+ ARI K + +++K+L K +++++ + N+ ++T Q ++ K +
Sbjct: 508 ENARIFEEMKKEIEEREKELIIERKQEIELQNEVQRITNKSNNNVEKLTNQIEVLQK--E 565
Query: 362 INISKNINSEVAREGGFSNSIYPTYNKLDLEVIKREKLRKE 402
IN +++ E+ + NKL IK K+++E
Sbjct: 566 INNQRSLLQEIEEK-----------NKLFNNEIKEAKIKEE 595
Score = 40.4 bits (93), Expect = 0.095
Identities = 73/353 (20%), Positives = 151/353 (42%), Gaps = 28/353 (7%)
Query: 51 EELQINVDDDSDNQSETSVDSSDYLNGGDGVVGLSDSLEIESLSRKAVKERMRRFKIGLA 110
E+++ D+ + E + +Y N + D LEI L + E+ + +I +
Sbjct: 731 EQIKEKEDEIIKIKEEMKMAEENYKNNISTIRTKKD-LEIHQLKEE---EQNKEKEIDIL 786
Query: 111 NKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRV 170
+ +K E E +K + +E L+E K +++ + ++++KEK+
Sbjct: 787 TNQKEIISKELITKKEENEILKEKINETLKELKEKEESNNQYQQINEEMKEKL------- 839
Query: 171 WQERLKSKRFREQIFLSWE---QCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAE 227
+E+ + +E+ + E + + N K E+ + N++ +++E Q+
Sbjct: 840 -KEKENEIKIKEEKITNKEKEMEELKNNFNKVENENEKRFKEKINELNKEIEEQKKSIKN 898
Query: 228 EKEKEKLMA-IARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRT 286
KE+E L A I + I+ E + E + + I ++ L Q+ +
Sbjct: 899 IKEEEYLFADIILDNEEIKELKEEMIHSKLNRKMIELIKKKEMEQLINDKKDLEIQLSKH 958
Query: 287 TEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGEAA 346
+ + + + +K + K +KV+LN+ + L+ +I +K + +E+ +
Sbjct: 959 QIENQKIQNQNDELKKINQTKENEKVLLNEINEKLQNKI---NELKEKDKIQEDENNKLQ 1015
Query: 347 LEITQQFKLDTKLT-KINISKNINSEVAREGGFSNSIYPTYNKLDLEVIKREK 398
EIT K T LT KI + K N+++ N I N ++E IK+EK
Sbjct: 1016 SEITNYSKTITSLTEKIELCKTENTKI------ENKIQQKEN--EIEEIKKEK 1060
>UniRef100_Q8IIX7 Hypothetical protein [Plasmodium falciparum]
Length = 324
Score = 57.8 bits (138), Expect = 6e-07
Identities = 57/234 (24%), Positives = 108/234 (45%), Gaps = 7/234 (2%)
Query: 91 ESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEALREPKVRKKMAE 150
E+ RK +++ R+ + NK R K + E RE+ +R+ E RE K RKK E
Sbjct: 73 ENKERKEREKQERKEREERENKERKEREKQERKEREEREKKERKEREE-REKKERKKREE 131
Query: 151 HPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDS 210
+ ++ ++++ ++ +E K + RE+ + K QE+ +
Sbjct: 132 REKKECEQREKRLKKEKQKRDKEERKERERRERQLKKEREKQEKRESKQREKQEKQERQK 191
Query: 211 YNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSY 270
K +E+ E +Q + E+KEKE+ + EK + E KE ++ +++ + +
Sbjct: 192 REK-EERKERKQRDKREKKEKEEREKREKKEKEEREKREKKEKEEREKREKKEKEEREKR 250
Query: 271 EKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRER 324
EK ++E + + +K K + R K EKE R K KK + K++ +E+
Sbjct: 251 EKKEKE-----EREKREKKEKEEREKREKKEKEEREKREKKEKEERDKREKKEK 299
Score = 46.2 bits (108), Expect = 0.002
Identities = 55/243 (22%), Positives = 101/243 (40%), Gaps = 37/243 (15%)
Query: 86 DSLEIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRERIKRRTSEAL----RE 141
D E + RK ++ER R + K R + + E +ER +R E RE
Sbjct: 35 DQNERQKRERKEMEEREERERN--ERKKREEHERNEREKQENKERKEREKQERKEREERE 92
Query: 142 PKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGC 201
K RK+ + R K +E+ R+ +ER K +R + + +K C
Sbjct: 93 NKERKEREKQER----KEREEREKKERKEREEREKKERKKRE-----------EREKKEC 137
Query: 202 GQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSG 261
Q E ++K++ + + + +E+E+ + EK +E ++
Sbjct: 138 EQRE------KRLKKEKQKRDKEERKERERRERQLKKEREK----------QEKRESKQR 181
Query: 262 EKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDL 321
EK+ + ++ +EE Q + +K K + R K EKE R K KK ++K++
Sbjct: 182 EKQEKQERQKREKEERKERKQRDKREKKEKEEREKREKKEKEEREKREKKEKEEREKREK 241
Query: 322 RER 324
+E+
Sbjct: 242 KEK 244
Score = 41.6 bits (96), Expect = 0.043
Identities = 59/240 (24%), Positives = 103/240 (42%), Gaps = 27/240 (11%)
Query: 89 EIESLSRKAVKERMRRFKIGLANKGRVPWNKGRKHSAETRE-RIKRRTSEALREPKVRKK 147
E E +K KER R K K R + K E RE R+K+ + +E + ++
Sbjct: 106 EREEREKKERKEREEREK-----KERKKREEREKKECEQREKRLKKEKQKRDKEERKERE 160
Query: 148 MAEHPRFHSDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELD 207
E + +EK R +++ + KR +E+ ++ +K + E
Sbjct: 161 RRERQLKKEREKQEKRESKQREKQEKQERQKREKEERKERKQRDKREKKEKEEREKREKK 220
Query: 208 WDSYNKIKEQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDW 267
+ +E+ E ++ + E+KEKE+ + EK +E +K EKE
Sbjct: 221 EKEEREKREKKEKEEREKREKKEKEEREKREKKEK----------EEREKREKKEKEERE 270
Query: 268 DSYEKIQEEMFLLNQIRRTTEKAKAKER-ARTKAEKEARIKAIKKVMLNQKKKDLRERIK 326
+K +EE R EK + +ER R K EKE R K KK +++++ +E++K
Sbjct: 271 KREKKEKEE-------REKREKKEKEERDKREKKEKEEREKQEKK---EKEEREKQEKMK 320
Score = 41.6 bits (96), Expect = 0.043
Identities = 49/226 (21%), Positives = 96/226 (41%), Gaps = 35/226 (15%)
Query: 119 KGRKHSAETRERIKRRTSEALREPKVRKKMAEHPRFHSDKIKEKISYSLRRVWQERLKSK 178
K ++ + RER + E RE RKK EH R +K + K + + + K
Sbjct: 33 KDDQNERQKRERKEMEEREE-RERNERKKREEHERNEREKQENK---------ERKEREK 82
Query: 179 RFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIKEQLELQQLLQAEEKEKEKLMAIA 238
+ R++ +EE + NK +++ E Q+ + EE+EK++
Sbjct: 83 QERKE-------------------REERE----NKERKEREKQERKEREEREKKERKERE 119
Query: 239 RAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQEEMFLLNQIRRTTEKAKAKERART 298
EK + E +E K+ EK L + ++ +EE + R +K + K+ R
Sbjct: 120 EREKKERKKRE--EREKKECEQREKRLKKEKQKRDKEERKERERRERQLKKEREKQEKRE 177
Query: 299 KAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQLCKNANEDGE 344
++E + K ++ +++K+ ++R K K + K ++ E
Sbjct: 178 SKQREKQEKQERQKREKEERKERKQRDKREKKEKEEREKREKKEKE 223
>UniRef100_Q8H8F6 Hypothetical protein OSJNBa0041L14.16 [Oryza sativa]
Length = 551
Score = 57.0 bits (136), Expect = 1e-06
Identities = 48/211 (22%), Positives = 88/211 (40%), Gaps = 8/211 (3%)
Query: 156 SDKIKEKISYSLRRVWQERLKSKRFREQIFLSWEQCIANAAKKGGCGQEELDWDSYNKIK 215
S++ + KIS +RR W RL+ ++ F+ W IA+AA+KG G L W+SY +
Sbjct: 139 SEETRIKISMGVRRGWNLRLQKLMIQDGCFVEWRDMIADAARKGFAGGISLQWNSYKILT 198
Query: 216 EQLELQQLLQAEEKEKEKLMAIARAEKFIQSWSECLAKEAKKGGSGEKELDWDSYEKIQE 275
EQ+ + L E+ +K + M + E K A+ K LD + E++
Sbjct: 199 EQMRQEWL---EKVQKRRSMPRPTGNRRAPKSPEQRRKIAE--AIAAKWLDKEYRERVCS 253
Query: 276 EMFLLNQIRRTTEKAKAKERARTKAEKEARIKAIKKVMLNQKKKDLRERIKGRGNIKSQL 335
+ T+ K + R+ E ++ +KK + + L + +K +
Sbjct: 254 G---IASYHGTSSGTKVPRKPRSAREPGSKRDTVKKKPIQSRSAGLEDACGTTPTVKRKK 310
Query: 336 CKNANEDGEAALEITQQFKLDTKLTKINISK 366
+D A ++ K+ + + I K
Sbjct: 311 SATPYKDPMAGEKLEMITKIRAQRAALEIEK 341
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 650,869,762
Number of Sequences: 2790947
Number of extensions: 26307910
Number of successful extensions: 117895
Number of sequences better than 10.0: 2709
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 2750
Number of HSP's that attempted gapping in prelim test: 107420
Number of HSP's gapped (non-prelim): 7254
length of query: 420
length of database: 848,049,833
effective HSP length: 130
effective length of query: 290
effective length of database: 485,226,723
effective search space: 140715749670
effective search space used: 140715749670
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0340.11


