

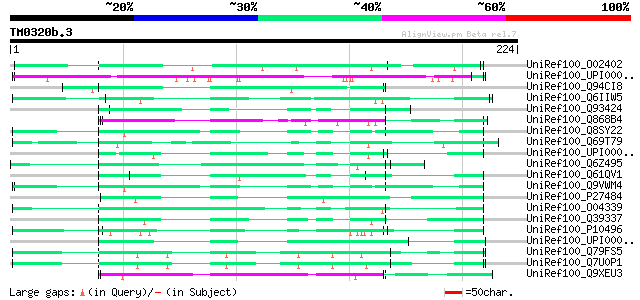
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0320b.3
(224 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O02402 Insoluble protein [Pinctada fucata] 74 3e-12
UniRef100_UPI00002216D1 UPI00002216D1 UniRef100 entry 74 4e-12
UniRef100_Q94CI8 Glycine-rich protein LeGRP1 [Lycopersicon escul... 70 4e-11
UniRef100_Q6IIW5 HDC16773 [Drosophila melanogaster] 65 1e-09
UniRef100_Q93424 Hypothetical protein E02A10.2 [Caenorhabditis e... 62 8e-09
UniRef100_Q868B4 Hypothetical protein ZK643.8 [Caenorhabditis el... 61 2e-08
UniRef100_Q8SY22 RE09269p [Drosophila melanogaster] 61 2e-08
UniRef100_Q69T79 Putative glycine-rich cell wall structural prot... 60 4e-08
UniRef100_UPI00002A4C48 UPI00002A4C48 UniRef100 entry 60 5e-08
UniRef100_Q6Z495 Putative glycine-rich cell wall structural prot... 59 1e-07
UniRef100_Q61QV1 Hypothetical protein CBG06865 [Caenorhabditis b... 59 1e-07
UniRef100_Q9VWM4 CG14191-PA [Drosophila melanogaster] 59 1e-07
UniRef100_P27484 Glycine-rich protein 2 [Nicotiana sylvestris] 59 1e-07
UniRef100_O04339 Putative glycine-rich protein [Arabidopsis thal... 58 2e-07
UniRef100_Q39337 Glycine-rich_protein_(Aa1-291) precursor [Brass... 58 2e-07
UniRef100_P10496 Glycine-rich cell wall structural protein 1.8 p... 58 2e-07
UniRef100_UPI0000332877 UPI0000332877 UniRef100 entry 57 3e-07
UniRef100_Q79FS5 PE-PGRS FAMILY PROTEIN [Mycobacterium tuberculo... 57 4e-07
UniRef100_Q7U0P1 PE-PGRS FAMILY PROTEIN [Mycobacterium bovis] 57 4e-07
UniRef100_Q9XEU3 Hypothetical protein [Oryza sativa] 56 6e-07
>UniRef100_O02402 Insoluble protein [Pinctada fucata]
Length = 738
Score = 73.9 bits (180), Expect = 3e-12
Identities = 67/208 (32%), Positives = 77/208 (36%), Gaps = 41/208 (19%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGGG GG L A + G G GGGG A+AA + G GGGG
Sbjct: 449 GGGGGGGAGALAAALAAAGAGGGL-------------GGGGGGGALAAALAAAGAGGGGF 495
Query: 63 GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATI 122
GG + GG GGG AA AA + + L
Sbjct: 496 GGLGGL---------------------GGLGGGSAAAAAAAAAAASGGGGRALRRALRRQ 534
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG--GGDYGGGGDKVVVAAIMVVVAT 180
GGGS A AA GG W G G G+GG GG +GGG AA A
Sbjct: 535 MRGGGSAAAAAAAAAAAAGGGWGGGMGGGFGVGLGGGFGGGFGGGSSAAAAAA-----AA 589
Query: 181 PVVTVGRWWRRRRRR*SDGGGGGGGANS 208
G RR R R G G G GA++
Sbjct: 590 AAAGFGGGGRRGRGRGRGGDGDGNGASA 617
Score = 53.5 bits (127), Expect = 4e-06
Identities = 56/189 (29%), Positives = 64/189 (33%), Gaps = 52/189 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G A AA GGG GG + A GGG +
Sbjct: 190 GAGGAGGAAAAAAAAAAAAGGGVGGAAAAAAAAAAA----------------AGGGAGRL 233
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G AA + A AA L G GGG GG GGG GG G+GG
Sbjct: 234 GGAAAAAAAAAAAAGGAGGLG--GLGGGL--------GGLGGGL-------GGLGGLGGL 276
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR-------------------*SDGG 200
G YGG AA + VG + R RR + GG
Sbjct: 277 GGYGGSAAAAAAAAAAAAGGGGLGGVGFYGGRGGRRGRGRGGRRRAAAAAAAAAAAAAGG 336
Query: 201 GGGGGANSG 209
GGGGG G
Sbjct: 337 GGGGGGGGG 345
Score = 52.8 bits (125), Expect = 7e-06
Identities = 52/158 (32%), Positives = 58/158 (35%), Gaps = 33/158 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWW----RRLWWWWR*GGGCGGG 95
G GGG V GGG GGG GGG + A + RR R G G G G
Sbjct: 558 GGMGGGFGVGL---GGGFGGGFGGGSSAAAAAAAAAAAGFGGGGRRGRGRGRGGDGDGNG 614
Query: 96 RAAVVGVAATSVVAV-----VAAMVVVVLATIGCG---------------------GGSK 129
+AV AA + A VAA A G G GG
Sbjct: 615 ASAVAAAAAAAAAAGGSAADVAAAAAAAAAMYGDGADGPDFDNGFGGGNGNGGGGSGGGG 674
Query: 130 AVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGD 167
+ GGG GGG GGG G GGG GGGG+
Sbjct: 675 SGGGGSGGGSGGGGGSGGSGGGGGSGGSGGGGSGGGGN 712
Score = 44.7 bits (104), Expect = 0.002
Identities = 51/193 (26%), Positives = 57/193 (29%), Gaps = 29/193 (15%)
Query: 1 VGGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGG 60
+GG GG GG A G G GG A AA G G G
Sbjct: 188 LGGAGGAGGAAAAAAAAAAAAGG----------------GVGGAAAAAAAAAAAAGGGAG 231
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLA 120
GG + A L GG GGG + G+ AA A
Sbjct: 232 RLGGAAAAAAAAAAAAGGAGGLGGLGGGLGGLGGGLGGLGGLGGLGGYGGSAAAAAAAAA 291
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*-------------GGGCGVGGGGDYGGGGD 167
GGG V G G G + GGG G GGGG GGG
Sbjct: 292 AAAGGGGLGGVGFYGGRGGRRGRGRGGRRRAAAAAAAAAAAAAGGGGGGGGGGGGGGGAG 351
Query: 168 KVVVAAIMVVVAT 180
AA A+
Sbjct: 352 AAAAAAAAAASAS 364
Score = 44.7 bits (104), Expect = 0.002
Identities = 48/139 (34%), Positives = 52/139 (36%), Gaps = 42/139 (30%)
Query: 40 GSGGGGKAVAATVEGGGNGG-GGGGGDRVVVVVSVVAIEWWRRLW---------WWWR*G 89
G GGGG A A GG +GG GGGGGD W + W W
Sbjct: 66 GVGGGGGAWGAGGAGGADGGRGGGGGD-------------WEYDYDDDSDDDDEWDWDDD 112
Query: 90 GGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ 149
GG G G G A A G G G+ A A G G GGG
Sbjct: 113 GGMGAGAGGGAGGGAGGGAG----------AGAGAGAGAGAGAG-LGLGLGGGL------ 155
Query: 150 *GGGC-GVGGGGDYGGGGD 167
GGG G+GG G GGG D
Sbjct: 156 -GGGLGGLGGLGGLGGGDD 173
Score = 43.5 bits (101), Expect = 0.004
Identities = 53/191 (27%), Positives = 66/191 (33%), Gaps = 47/191 (24%)
Query: 40 GSGGGGK--------AVAATVEGGGNGGGGGGGD------RVVVVVSVVAIEWWRRLWW- 84
G G GG+ A AA GGG GGGGGGG + + R++
Sbjct: 313 GRGRGGRRRAAAAAAAAAAAAAGGGGGGGGGGGGGGGAGAAAAAAAAAASASASRQMSGI 372
Query: 85 ---------WWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA** 135
R G A VA+T V+ LA + S + +A
Sbjct: 373 RDALGDIKDLLRSNGASAKASAKASAVASTKSQIDDLKDVLKDLAGLLKSSASASASASA 432
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR 195
GGG GGG GG G GGGG + AA+ A +
Sbjct: 433 SASAGGG--------GGGGNGGGNGGGGGGGAGALAAALAAAGAGGGL------------ 472
Query: 196 *SDGGGGGGGA 206
GGGGGGGA
Sbjct: 473 ---GGGGGGGA 480
Score = 34.7 bits (78), Expect = 1.9
Identities = 36/128 (28%), Positives = 40/128 (31%), Gaps = 73/128 (57%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGGG GGG+GGGG GG G G GGG
Sbjct: 664 GNGGGGSG------GGGSGGGGSGG------------------------GSGGGGGSG-- 691
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GG GGG GG GG G GGG
Sbjct: 692 -----------------------GSGG---------GGGSGG---------SGGGGSGGG 710
Query: 160 GDYGGGGD 167
G+ G G +
Sbjct: 711 GNNGWGNN 718
>UniRef100_UPI00002216D1 UPI00002216D1 UniRef100 entry
Length = 375
Score = 73.6 bits (179), Expect = 4e-12
Identities = 74/228 (32%), Positives = 93/228 (40%), Gaps = 42/228 (18%)
Query: 2 GGGGGNGGRLVLVA---VVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNG 58
G GG G LV V V V E WW W R W W G+GGGG + G G+G
Sbjct: 136 GQGGIGGSELVKVESEMVKVESEDQSWWRWNRRWSRWNRRIGAGGGG--IGDGQGGIGDG 193
Query: 59 GGGGGGDRVVVVVS-VVAIEWWRRLWWWW---------R*GGGCGGGRAAVVGVAATSVV 108
GG GG +V V S +V +E R WW W R G G GG G+ + +V
Sbjct: 194 QGGIGGSELVEVESEMVKVESEDRSWWRWNRRWSRWNRRIGAGGGGIGDGQGGIGGSELV 253
Query: 109 AVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWW---W--WQ*---GGGCGVGGGG 160
V + MV V ++ WW W W W GG G+G G
Sbjct: 254 KVESEMVKVESEMDKVESEDRS------------WWRWNRRWSRWNRRIGAGGGGIGDGQ 301
Query: 161 DYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRR----R*SDGGGGGG 204
GG + V + + MV V + + WWR RR + G GGGG
Sbjct: 302 GEIGGSELVEMESEMVKVESDDRS---WWRWNRRWSIWNRTIGAGGGG 346
Score = 73.2 bits (178), Expect = 5e-12
Identities = 79/245 (32%), Positives = 96/245 (38%), Gaps = 65/245 (26%)
Query: 2 GGGGGNGGRLVLVA---VVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNG 58
G GG G LV V V V E WW W R W W G+GGGG G+G
Sbjct: 86 GQGGIGGSELVEVESEMVKVESEDRSWWRWNRRWSRWNRRIGAGGGGI---------GDG 136
Query: 59 GGGGGGDRVVVVVS-VVAIE-----WWR--RLWWWWR-----*GGGCGGGRAAV----VG 101
GG GG +V V S +V +E WWR R W W GGG G G+ + G
Sbjct: 137 QGGIGGSELVKVESEMVKVESEDQSWWRWNRRWSRWNRRIGAGGGGIGDGQGGIGDGQGG 196
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWW---W--WQ---*GGG 153
+ + +V V + MV V + WW W W W GG
Sbjct: 197 IGGSELVEVESEMVKV-------------------ESEDRSWWRWNRRWSRWNRRIGAGG 237
Query: 154 CGVGGGGDYGGGGDKVVVAAIMVVVATPVVTV----GRWWRRRRR-----R*SDGGGGGG 204
G+G G GG + V V + MV V + + V WWR RR R GGGG
Sbjct: 238 GGIGDGQGGIGGSELVKVESEMVKVESEMDKVESEDRSWWRWNRRWSRWNRRIGAGGGGI 297
Query: 205 GANSG 209
G G
Sbjct: 298 GDGQG 302
Score = 54.3 bits (129), Expect = 2e-06
Identities = 63/223 (28%), Positives = 83/223 (36%), Gaps = 56/223 (25%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
G G G R V + + G W W R G+GGGG G+G G
Sbjct: 9 GAGALGSRAVGAGALGS---GCWSRWNRRI-------GAGGGGI---------GDGQGEI 49
Query: 63 GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV----GVAATSVVAVVAAMVVVV 118
GG +V + S +W W GGG G G+ + G+ + +V V + MV V
Sbjct: 50 GGSELVEMDSEDRSQW---NWIIGAGGGGIGDGQGGIGDGQGGIGGSELVEVESEMVKVE 106
Query: 119 LATIGCGGGSKAVAA**GGGDGGGWWWW---W--WQ*---GGGCGVGGGGDYGGGGDKVV 170
+ WW W W W GG G+G G GG + V
Sbjct: 107 -------------------SEDRSWWRWNRRWSRWNRRIGAGGGGIGDGQGGIGGSELVK 147
Query: 171 VAAIMVVVATPVVTVGRW---WRRRRRR*SDGGGGGGGANSGV 210
V + MV V + + RW W R RR GGGG G G+
Sbjct: 148 VESEMVKVESEDQSWWRWNRRWSRWNRRIGAGGGGIGDGQGGI 190
>UniRef100_Q94CI8 Glycine-rich protein LeGRP1 [Lycopersicon esculentum]
Length = 284
Score = 70.1 bits (170), Expect = 4e-11
Identities = 51/147 (34%), Positives = 53/147 (35%), Gaps = 54/147 (36%)
Query: 24 WWWWWQRWWLWW----W***GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWW 79
WWWWW WLWW W GGG A AA GG GGGG G
Sbjct: 8 WWWWWTWRWLWWWERFWGRNARAGGGGAYAAGGHDGGYAGGGGSGSGAG----------- 56
Query: 80 RRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGD 139
GG GGG A G GGGS A A GG D
Sbjct: 57 ---------GGHAGGGYAG------------------------GGGGGSGAGGAHAGGHD 83
Query: 140 GGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
GG + GGG G G GG + GGG
Sbjct: 84 GG------YASGGGEGGGAGGGHAGGG 104
Score = 50.1 bits (118), Expect = 4e-05
Identities = 45/130 (34%), Positives = 47/130 (35%), Gaps = 24/130 (18%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GG G A GGG G GGGGG V GGG GG A
Sbjct: 139 GYGGAGSADGGYAAGGGGGSGGGGGGAVA--------------------GGGAHGGGAYA 178
Query: 100 VGVAATSVVAVVAAMVVVVLATIG---CGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV 156
G + A G GGG A A GG GGG+ GGG G
Sbjct: 179 GGSGGGTGGGYGAGGAPGGGYGGGGGHGGGGGSAYAGGEGGASGGGYGSGGGA-GGGAGG 237
Query: 157 GGGGDYGGGG 166
GG YGGGG
Sbjct: 238 AHGGAYGGGG 247
Score = 46.6 bits (109), Expect = 5e-04
Identities = 39/126 (30%), Positives = 43/126 (33%), Gaps = 48/126 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG + A EGG +GGG G G GGG GG
Sbjct: 204 GHGGGGGSAYAGGEGGASGGGYGSGGGA---------------------GGGAGGAHGGA 242
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G S G GGG GG GG+ GGG G G G
Sbjct: 243 YGGGGGS----------------GAGGGGAYA----GGEHAGGY-------GGGAGGGEG 275
Query: 160 GDYGGG 165
G +GGG
Sbjct: 276 GGHGGG 281
Score = 43.5 bits (101), Expect = 0.004
Identities = 40/134 (29%), Positives = 44/134 (31%), Gaps = 51/134 (38%)
Query: 79 WRRLWWW---WR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA** 135
WR LWWW W GGG A G GGGS
Sbjct: 14 WRWLWWWERFWGRNARAGGGGAYAAGGHDGGYAG---------------GGGS------- 51
Query: 136 GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR 195
G G GGG + GGG G G GG + GG D G +
Sbjct: 52 GSGAGGGHAGGGYAGGGGGGSGAGGAHAGGHD------------------GGY------- 86
Query: 196 *SDGGGGGGGANSG 209
+ GGG GGGA G
Sbjct: 87 -ASGGGEGGGAGGG 99
Score = 34.3 bits (77), Expect = 2.5
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 9/38 (23%)
Query: 138 GDGGGWWW-------WWWQ--*GGGCGVGGGGDYGGGG 166
G+ WWW WWW+ G GGGG Y GG
Sbjct: 3 GEVRRWWWWWTWRWLWWWERFWGRNARAGGGGAYAAGG 40
Score = 32.3 bits (72), Expect = 9.4
Identities = 23/59 (38%), Positives = 24/59 (39%), Gaps = 24/59 (40%)
Query: 40 GSG-GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRA 97
GSG GGG A A GG GGG GGG+ GGG GGG A
Sbjct: 248 GSGAGGGGAYAGGEHAGGYGGGAGGGE-----------------------GGGHGGGYA 283
>UniRef100_Q6IIW5 HDC16773 [Drosophila melanogaster]
Length = 241
Score = 65.1 bits (157), Expect = 1e-09
Identities = 65/218 (29%), Positives = 79/218 (35%), Gaps = 70/218 (32%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGG----N 57
GGGGG+GG + + V E G G GGGG ++ GGG +
Sbjct: 82 GGGGGHGGGGDVQIIKVITESGSS--------------GGGGGGGGWSSGGGGGGGGWSS 127
Query: 58 GGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVV 117
GGGGGGG W GGG GG + G + + V
Sbjct: 128 GGGGGGGG--------------------WSSGGGGGGWSSGGGGGSGSDVK--------- 158
Query: 118 VLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYG--GGGDKVVVAAIM 175
++ I GGG GG GGG W GGG GG G +G GGGD V+ I
Sbjct: 159 LIKIISLGGGG-------GGHSGGGGGWSSGGGGGGWSSGGSGGHGSSGGGDTKVIKIIK 211
Query: 176 VVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSWQ 213
+ GGGGG G G WQ
Sbjct: 212 L--------------SSGGHGGAGGGGGHGGGGGGGWQ 235
Score = 53.1 bits (126), Expect = 5e-06
Identities = 51/174 (29%), Positives = 62/174 (35%), Gaps = 61/174 (35%)
Query: 43 GGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGV 102
GGG + ++ GGG GGGGG GG GGG ++ V
Sbjct: 63 GGGSSWSSGGSGGGWSSGGGGG------------------------GGHGGGGDVQIIKV 98
Query: 103 AATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG-- 160
S + G GGG ++ GGG GGGW GGG GGGG
Sbjct: 99 ITES-------------GSSGGGGGGGGWSS-GGGGGGGGWSSGGGGGGGGWSSGGGGGG 144
Query: 161 --DYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
GGGG V I ++ S GGGGGG + G W
Sbjct: 145 WSSGGGGGSGSDVKLIKII-------------------SLGGGGGGHSGGGGGW 179
>UniRef100_Q93424 Hypothetical protein E02A10.2 [Caenorhabditis elegans]
Length = 385
Score = 62.4 bits (150), Expect = 8e-09
Identities = 49/138 (35%), Positives = 50/138 (35%), Gaps = 57/138 (41%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GG GGGG GGGCGGG
Sbjct: 121 GCGGGGGGCGGGCGGGGGGGCGGGG------------------------GGGCGGGGGGC 156
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GCGGG GGG GGG GGGCG GGG
Sbjct: 157 GGGGG------------------GCGGG--------GGGCGGGG-------GGGCGGGGG 183
Query: 160 GDYGGGGDKVVVAAIMVV 177
G GGG + AI VV
Sbjct: 184 GGCGGGCGRKKREAINVV 201
Score = 59.3 bits (142), Expect = 7e-08
Identities = 46/127 (36%), Positives = 46/127 (36%), Gaps = 47/127 (37%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGG G A GGGCGGG
Sbjct: 25 GGGGGGGCGGGCGGGGGCGGGGGCGGGCAPPPPPPAC------------GGGCGGGGG-- 70
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GCGGG GGG GGG GGGCG GGG
Sbjct: 71 -----------------------GCGGGGGGCGG--GGGCGGG--------GGGCGGGGG 97
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 98 GCGGGGG 104
Score = 55.1 bits (131), Expect = 1e-06
Identities = 45/127 (35%), Positives = 45/127 (35%), Gaps = 54/127 (42%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG G GGGGG GGGCGGG A
Sbjct: 73 GGGGGGCGGGGGCGGGGGGCGGGGGG--------------------CGGGGGCGGGCAPP 112
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
A CGGG GG GGG GGGCG GGG
Sbjct: 113 PPPPA-------------------CGGGC--------GGGGGGC-------GGGCGGGGG 138
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 139 GGCGGGG 145
Score = 49.3 bits (116), Expect = 7e-05
Identities = 42/122 (34%), Positives = 43/122 (34%), Gaps = 35/122 (28%)
Query: 45 GKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAA 104
G +VA GGGGGGG GGGCGGG G A
Sbjct: 11 GLSVATAFLFPSGGGGGGGGG----------------CGGGCGGGGGCGGGGGCGGGCAP 54
Query: 105 TSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGG 164
GCGGG GGG GG GGGCG GGGG GG
Sbjct: 55 PPPPPACGG---------GCGGGGGGCGG--GGGGCGG--------GGGCGGGGGGCGGG 95
Query: 165 GG 166
GG
Sbjct: 96 GG 97
>UniRef100_Q868B4 Hypothetical protein ZK643.8 [Caenorhabditis elegans]
Length = 774
Score = 61.2 bits (147), Expect = 2e-08
Identities = 49/126 (38%), Positives = 55/126 (42%), Gaps = 25/126 (19%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
SGGGG AVA + GG GGG GG V S GGGCGGG ++
Sbjct: 176 SGGGGYAVAPSGGGGCGGGGSSGGGGYAVAPSG---------------GGGCGGGGSSGG 220
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGG 160
G A++ AT G GGGS A GG GG + GGG GGGG
Sbjct: 221 GGYASAPSGGGG------YATSG-GGGSGGYAT---GGSSGGGYSSGGSSGGGYSTGGGG 270
Query: 161 DYGGGG 166
Y GGG
Sbjct: 271 GYAGGG 276
Score = 58.2 bits (139), Expect = 2e-07
Identities = 55/171 (32%), Positives = 58/171 (33%), Gaps = 75/171 (43%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG G G GGGG GG GGGCGGG
Sbjct: 102 GCGGGGGGCGGGGGGCGGGGGGCGGG-----------------------GGGCGGGGGGG 138
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG-G 158
G ++ GCGGGS GGG GGG GGG G G G
Sbjct: 139 CGGGSSG----------------GCGGGSSGGC---GGGGGGGC-------GGGGGGGCG 172
Query: 159 GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG GGGG V S GGG GGG +SG
Sbjct: 173 GGSSGGGGYAVAP-------------------------SGGGGCGGGGSSG 198
Score = 56.6 bits (135), Expect = 5e-07
Identities = 46/125 (36%), Positives = 46/125 (36%), Gaps = 56/125 (44%)
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GGGG A GGG GGGGGG GGGCGGG G
Sbjct: 68 GGGGCAPPPAPCGGGCGGGGGGCGGG---------------------GGGCGGGGGCGGG 106
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD 161
GCGGG GGG GGG GGGCG GGGG
Sbjct: 107 GG-------------------GCGGG--------GGGCGGG--------GGGCGGGGGGC 131
Query: 162 YGGGG 166
GGGG
Sbjct: 132 GGGGG 136
Score = 56.2 bits (134), Expect = 6e-07
Identities = 53/172 (30%), Positives = 53/172 (30%), Gaps = 94/172 (54%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGG A GG GGGGG G GGGCGGG
Sbjct: 67 GGGGGCAPPPAPCGGGCGGGGGGCGGG----------------------GGGCGGGG--- 101
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GCGGG GGG GGG GGGCG GGG
Sbjct: 102 -----------------------GCGGG--------GGGCGGG--------GGGCGGGGG 122
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVS 211
G GGGG GGGGGGG G S
Sbjct: 123 GCGGGGG------------------------------GCGGGGGGGCGGGSS 144
Score = 54.7 bits (130), Expect = 2e-06
Identities = 59/181 (32%), Positives = 65/181 (35%), Gaps = 50/181 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GG GGGG GGGCGGG +
Sbjct: 111 GGGGGGCGGGGGGCGGGGGGCGGGG------------------------GGGCGGGSSGG 146
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSK-----AVAA**GGGDGGGW----WWWWWQ* 150
G ++ GCGGGS AVA GGG GGG +
Sbjct: 147 CGGGSSGGCGGGGGGGCGGGGGGGCGGGSSGGGGYAVAPSGGGGCGGGGSSGGGGYAVAP 206
Query: 151 GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
GG G GGGG GGGG A+ G + S GGG GG A G
Sbjct: 207 SGGGGCGGGGSSGGGG-----------YASAPSGGGGYAT------SGGGGSGGYATGGS 249
Query: 211 S 211
S
Sbjct: 250 S 250
Score = 52.8 bits (125), Expect = 7e-06
Identities = 44/127 (34%), Positives = 45/127 (34%), Gaps = 37/127 (29%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GG GG GGGG GGGCGGG
Sbjct: 26 GGGGGGGGCG----GGCGGGCGGGGGCAPPPAPC---------------GGGCGGGGGGC 66
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + GCGGG GGG GG GGGCG GGG
Sbjct: 67 GGGGGCAPPPAPCGG--------GCGGGGGGCGG--GGGGCGG--------GGGCGGGGG 108
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 109 GCGGGGG 115
Score = 50.8 bits (120), Expect = 3e-05
Identities = 45/128 (35%), Positives = 45/128 (35%), Gaps = 51/128 (39%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG G GGGGG GGGCGGG
Sbjct: 91 GGGGGGCGGGGGCGGGGGGCGGGGGG-------------------CGGGGGGCGGGGGGC 131
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GCGGGS GG GGG GGCG GGG
Sbjct: 132 GGGGGG-----------------GCGGGSS-------GGCGGGS-------SGGCGGGGG 160
Query: 160 G-DYGGGG 166
G GGGG
Sbjct: 161 GGCGGGGG 168
Score = 40.4 bits (93), Expect = 0.034
Identities = 33/87 (37%), Positives = 33/87 (37%), Gaps = 26/87 (29%)
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
GCGGG G G GGG GGGCG GGGG GGGG A P
Sbjct: 33 GCGGGCGG-----GCGGGGGCAPPPAPCGGGCGGGGGGCGGGGG-----------CAPPP 76
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGGGG G
Sbjct: 77 APCG----------GGCGGGGGGCGGG 93
Score = 37.0 bits (84), Expect = 0.38
Identities = 38/133 (28%), Positives = 44/133 (32%), Gaps = 40/133 (30%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GS GGG + GGG G GGGG GGG GG A
Sbjct: 258 GSSGGGYST-----GGGGGYAGGGGG-----------------------GGGSSGGYAGS 289
Query: 100 VGVAATSVVAVV-----AAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC 154
G S A A + GGG ++ GGG G + GG
Sbjct: 290 SGGGGYSAPAAAPPPPPPPPPPPAPAPVSSGGGYSEQSS--GGGGGSS-----YSGGGEA 342
Query: 155 GVGGGGDYGGGGD 167
GG Y GGG+
Sbjct: 343 SSSSGGGYSGGGE 355
Score = 33.9 bits (76), Expect = 3.2
Identities = 26/72 (36%), Positives = 26/72 (36%), Gaps = 28/72 (38%)
Query: 138 GDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*S 197
G GGG GGGCG G GG GGGG A P G
Sbjct: 26 GGGGG--------GGGCGGGCGGGCGGGGG----------CAPPPAPCG----------G 57
Query: 198 DGGGGGGGANSG 209
GGGGGG G
Sbjct: 58 GCGGGGGGCGGG 69
>UniRef100_Q8SY22 RE09269p [Drosophila melanogaster]
Length = 218
Score = 60.8 bits (146), Expect = 2e-08
Identities = 54/170 (31%), Positives = 66/170 (38%), Gaps = 46/170 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGG GG + ++ V+ + GGG GGG
Sbjct: 74 GHGGGGGYGGGGYGGGGYGGGGHGGGSTIKIIKVITDSGAGGGGY----GGGYGGGHGGG 129
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGGS + GG GGGW G G GGG
Sbjct: 130 YGG--------------------GYGGGSSGGYS---GGHGGGW-------SSGGGYGGG 159
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G YGGGG+ ++ I ++ G GGG GGG +SG
Sbjct: 160 G-YGGGGNVKIIKVISDAGSSGGYGGGY-----------GGGHGGGYSSG 197
Score = 46.2 bits (108), Expect = 6e-04
Identities = 48/166 (28%), Positives = 58/166 (34%), Gaps = 52/166 (31%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVA-ATVEGGGNGGG 60
GGGG GG + + V+T G+GGGG GGG GGG
Sbjct: 92 GGGGHGGGSTIKIIKVITDS------------------GAGGGGYGGGYGGGHGGGYGGG 133
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLA 120
GGG + + W GGG GGG G ++ V++
Sbjct: 134 YGGGS---------SGGYSGGHGGGWSSGGGYGGGGYGGGG----------NVKIIKVIS 174
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GG GGG GGG GGG GG Y GG
Sbjct: 175 DAGSSGGY-------GGGYGGGH-------GGGYSSGGASSYSAGG 206
Score = 35.4 bits (80), Expect = 1.1
Identities = 50/209 (23%), Positives = 60/209 (27%), Gaps = 110/209 (52%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGGG GG + GW SGGGG + GG GGGG
Sbjct: 45 GGGGAGGS--------ALSSGW---------------SSGGGGGGYSGGYSGGHGGGGGY 81
Query: 63 GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATI 122
G GGG GGG G S + ++ V+
Sbjct: 82 G-------------------------GGGYGGGGYGGGGHGGGSTIKIIK-----VITDS 111
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GG GG GGG+ GG +GGG
Sbjct: 112 GAGG----------GGYGGGY---------------GGGHGGGY---------------- 130
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSGVS 211
GGG GGG++ G S
Sbjct: 131 ----------------GGGYGGGSSGGYS 143
Score = 33.9 bits (76), Expect = 3.2
Identities = 20/44 (45%), Positives = 22/44 (49%), Gaps = 5/44 (11%)
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GGS + GG GGG+ G G GGGG YGGGG
Sbjct: 47 GGAGGSALSSGWSSGGGGGGY-----SGGYSGGHGGGGGYGGGG 85
>UniRef100_Q69T79 Putative glycine-rich cell wall structural protein [Oryza sativa]
Length = 239
Score = 60.1 bits (144), Expect = 4e-08
Identities = 57/188 (30%), Positives = 66/188 (34%), Gaps = 80/188 (42%)
Query: 40 GSGGGGK-------AVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGC 92
G GGGG +VA +GGG GGGGGG S W W W G
Sbjct: 54 GGGGGGNGTSYNATSVAGRKDGGGGGGGGGG--------SSGGSSW--SYGWGWGWGTDS 103
Query: 93 GGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG 152
GGG ++ GGG GGG GGG GG
Sbjct: 104 GGGGSS--------------------------GGGG-------GGGGGGG--------GG 122
Query: 153 GCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWR----RRRRR*SDGGGGGGGANS 208
G G GGGG GGGG + WW RRR + GGGGG
Sbjct: 123 GGGGGGGGGGGGGGRRC------------------WWGCGNGRRRHKGGKEGGGGGEGRK 164
Query: 209 GVSWQQ*R 216
G+ ++ R
Sbjct: 165 GIRTERER 172
Score = 54.7 bits (130), Expect = 2e-06
Identities = 62/213 (29%), Positives = 67/213 (31%), Gaps = 107/213 (50%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG GG + + GW W W SGGGG + GGG GGGG
Sbjct: 75 GGGGGGGGG--GSSGGSSWSYGWGWGWGT---------DSGGGGSS------GGGGGGGG 117
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG GGG GGG
Sbjct: 118 GGG------------------------GGGGGGG-------------------------- 127
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG-----GGGDYGGGGDKVVVAAIMV 176
GGG GGG GG WW GCG G GG + GGGG+
Sbjct: 128 ---GGG--------GGGGGGRRCWW------GCGNGRRRHKGGKEGGGGGE--------- 161
Query: 177 VVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GR R R GGGG G G
Sbjct: 162 ---------GRKGIRTEREREGVGGGGAGRRHG 185
Score = 43.1 bits (100), Expect = 0.005
Identities = 46/168 (27%), Positives = 50/168 (29%), Gaps = 75/168 (44%)
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GG G+ ++E GGGGG G S A R + GGG GGG
Sbjct: 39 GGDGRVQVQSLERPVGGGGGGNG------TSYNATSVAGR-----KDGGGGGGG------ 81
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD 161
GGGS G GW W W GG G GGG
Sbjct: 82 -----------------------GGGSSG-----GSSWSYGWGWGWGTDSGGGGSSGGGG 113
Query: 162 YGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGG GGGGGGG G
Sbjct: 114 GGGGG------------------------------GGGGGGGGGGGGG 131
>UniRef100_UPI00002A4C48 UPI00002A4C48 UniRef100 entry
Length = 206
Score = 59.7 bits (143), Expect = 5e-08
Identities = 52/170 (30%), Positives = 61/170 (35%), Gaps = 69/170 (40%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG+ + GG GGG GGG GGGC GG
Sbjct: 5 GDGGGGEGGGSAGGGGKGGGGDGGGGE----------------------GGGCDGGGGGR 42
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + + V G GGG + GGGDGGG GG GVGGG
Sbjct: 43 AGSPSGTAGGVTGGG--------GDGGGGRG-----GGGDGGGG-------EGGGGVGGG 82
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G+ GGGGD + +GGGG GG + G
Sbjct: 83 GE-GGGGD--------------------------GKGGEGGGGDGGGSKG 105
Score = 57.0 bits (136), Expect = 4e-07
Identities = 44/126 (34%), Positives = 50/126 (38%), Gaps = 42/126 (33%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG+ + GGG GGGG GG GGGC GG
Sbjct: 123 GDGGGGEG-GGSAGGGGKGGGGDGGGGE---------------------GGGCDGGGGGR 160
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + + V G GGG + GGGDGGG GG GVGGG
Sbjct: 161 AGSPSGTAGGVTGGG--------GDGGGGRG-----GGGDGGGG-------EGGGGVGGG 200
Query: 160 GDYGGG 165
G+ GGG
Sbjct: 201 GEGGGG 206
Score = 53.5 bits (127), Expect = 4e-06
Identities = 48/130 (36%), Positives = 50/130 (37%), Gaps = 45/130 (34%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG+ GGG+GGGG GG V GGG GGG
Sbjct: 58 GDGGGGRG------GGGDGGGGEGGGGVG--------------------GGGEGGGGDGK 91
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG-- 157
G G GGGSK GGGDGGG GGG G G
Sbjct: 92 GGEGGG-----------------GDGGGSKGGGDDGGGGDGGGGDGSGGDGGGGEGGGSA 134
Query: 158 GGGDYGGGGD 167
GGG GGGGD
Sbjct: 135 GGGGKGGGGD 144
Score = 46.2 bits (108), Expect = 6e-04
Identities = 53/173 (30%), Positives = 55/173 (31%), Gaps = 73/173 (42%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGG---GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGR 96
G G GG + +GGG GGGG GGD GGG GGG
Sbjct: 96 GGGDGGGSKGGGDDGGGGDGGGGDGSGGDG----------------------GGGEGGGS 133
Query: 97 AAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV 156
A GGG K GGGDGGG GGGC
Sbjct: 134 A---------------------------GGGGKG-----GGGDGGGGE------GGGCDG 155
Query: 157 GGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GGGG G A V G R DGGGG GG G
Sbjct: 156 GGGGRAGSPSG----------TAGGVTGGGGDGGGGRGGGGDGGGGEGGGGVG 198
>UniRef100_Q6Z495 Putative glycine-rich cell wall structural protein [Oryza sativa]
Length = 296
Score = 58.5 bits (140), Expect = 1e-07
Identities = 48/129 (37%), Positives = 49/129 (37%), Gaps = 33/129 (25%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG EGGG GGG GGG GGG GGG
Sbjct: 88 GGGGGGGLGGGGCEGGGFGGGVGGGSGA---------------------GGGLGGGGGGG 126
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + V A G GGGS GGG GGG GGG G GGG
Sbjct: 127 FGGGSGGGVGGGGGQGGGFGAGGGVGGGSGT-----GGGLGGGG-------GGGFGGGGG 174
Query: 160 GDYGGGGDK 168
G GGGG K
Sbjct: 175 GGIGGGGGK 183
Score = 53.5 bits (127), Expect = 4e-06
Identities = 58/185 (31%), Positives = 66/185 (35%), Gaps = 63/185 (34%)
Query: 1 VGGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGG 60
VGGGGG GG G G GG + GGG GGG
Sbjct: 135 VGGGGGQGG------------------------------GFGAGGGVGGGSGTGGGLGGG 164
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLA 120
GGGG + GGG GGG G A V A
Sbjct: 165 GGGG-------------------FGGGGGGGIGGGGGKGGGFGAGGGVGGAAGGG----G 201
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG--GCGVGGGGDYGGGGDKVVVAAIMVVV 178
+G GGG GGG GGG+ GG G G GGGG GGGG +V +V +
Sbjct: 202 GMGSGGGGGFGG---GGGKGGGFGA-----GGVMGGGAGGGGGLGGGGGGGMVEVSVVEL 253
Query: 179 ATPVV 183
A +V
Sbjct: 254 AEVLV 258
Score = 52.4 bits (124), Expect = 9e-06
Identities = 46/127 (36%), Positives = 48/127 (37%), Gaps = 28/127 (22%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GG GGGG GGG GGG A
Sbjct: 75 GGGGGGLGGGGGFGGGGGGGLGGGGCE------------------GGGFGGGVGGGSGAG 116
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ V G GGG A GGG GGG GGG G GGG
Sbjct: 117 GGLGGGGGGGFGGGSGGGVGGGGGQGGGFGA-----GGGVGGGSGT-----GGGLGGGGG 166
Query: 160 GDYGGGG 166
G +GGGG
Sbjct: 167 GGFGGGG 173
Score = 44.7 bits (104), Expect = 0.002
Identities = 54/170 (31%), Positives = 57/170 (32%), Gaps = 45/170 (26%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G G GGG G GGGGG R + GGG GGG
Sbjct: 48 GPGFGHDCRFGRCHGGGGGFGGGGGFRGGGGGGLGG-------------GGGFGGGGGGG 94
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
+G G GGGS A GGG GGG GGG G G G
Sbjct: 95 LGGGGCEGGGFGG----------GVGGGSGA-----GGGLGGGG-------GGGFGGGSG 132
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG + V T G GGGGGGG G
Sbjct: 133 GGVGGGGGQGGGFGAGGGVGGGSGTGG----------GLGGGGGGGFGGG 172
Score = 33.1 bits (74), Expect = 5.5
Identities = 36/122 (29%), Positives = 41/122 (33%), Gaps = 39/122 (31%)
Query: 1 VGGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGG---- 56
+GGGGG GG V G GSGGGG +GGG
Sbjct: 177 IGGGGGKGGGFGAGGGVGGAAGGGGGM------------GSGGGGGFGGGGGKGGGFGAG 224
Query: 57 --NGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAM 114
GGG GGG + GGG GGG V V V+ AA
Sbjct: 225 GVMGGGAGGGGGL---------------------GGGGGGGMVEVSVVELAEVLVKAAAA 263
Query: 115 VV 116
V+
Sbjct: 264 VL 265
>UniRef100_Q61QV1 Hypothetical protein CBG06865 [Caenorhabditis briggsae]
Length = 646
Score = 58.5 bits (140), Expect = 1e-07
Identities = 54/170 (31%), Positives = 55/170 (31%), Gaps = 89/170 (52%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GG GGG GGG GGGCGGG A
Sbjct: 82 GGGGGGCGGGC----GGGGGGCGGG------------------------GGGCGGGGGAC 113
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GCGGG GGG GGG GGGCG GGG
Sbjct: 114 GGGGG------------------GCGGG--------GGGCGGG--------GGGCGGGGG 139
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GGGG GGGG GG +SG
Sbjct: 140 GGCGGGGGGC---------------------------GGGGGGCGGGSSG 162
Score = 55.1 bits (131), Expect = 1e-06
Identities = 42/127 (33%), Positives = 42/127 (33%), Gaps = 58/127 (45%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GG GGGG GGCGGG
Sbjct: 100 GGGGGGCGGGGGACGGGGGGCGGGG-------------------------GGCGGGGG-- 132
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
GCGGG GGG GGG GGGCG G
Sbjct: 133 -----------------------GCGGGGGGGCGGGGGGCGGG--------GGGCGGGSS 161
Query: 160 GDYGGGG 166
G GGGG
Sbjct: 162 GGCGGGG 168
Score = 48.5 bits (114), Expect = 1e-04
Identities = 41/119 (34%), Positives = 42/119 (34%), Gaps = 39/119 (32%)
Query: 54 GGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV------GVAATSV 107
GGG GGG GGG GGGCGGG G V
Sbjct: 26 GGGGGGGCGGGCGG---------------------GGGCGGGCGPPPPPPCGGGCGGGGV 64
Query: 108 VAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
+ CGGG GGG GGG GGGCG GGGG GGGG
Sbjct: 65 CGGGGVCAPALPPPPPCGGGGGGC----GGGCGGG--------GGGCGGGGGGCGGGGG 111
Score = 45.8 bits (107), Expect = 8e-04
Identities = 39/118 (33%), Positives = 39/118 (33%), Gaps = 57/118 (48%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GG GGGG GGGCGGG
Sbjct: 114 GGGGGGCGGGGGGCGGGGGGCGGGG------------------------GGGCGGGGGGC 149
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG 157
G GCGGGS GG GGG GGGCG G
Sbjct: 150 GGGGG------------------GCGGGS-------SGGCGGG--------GGGCGGG 174
>UniRef100_Q9VWM4 CG14191-PA [Drosophila melanogaster]
Length = 193
Score = 58.5 bits (140), Expect = 1e-07
Identities = 53/170 (31%), Positives = 65/170 (38%), Gaps = 46/170 (27%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGG GG + ++ V+ + GGG GGG
Sbjct: 49 GHGGGGGYGGGGYGGGGYGGGGHGGGSTIKIIKVITDSGAGGGGY----GGGYGGGHGGG 104
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGGS + GG GGGW G G GGG
Sbjct: 105 YGG--------------------GYGGGSSGGYS---GGHGGGW-------SSGGGYGGG 134
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G YG GG+ ++ I ++ G GGG GGG +SG
Sbjct: 135 G-YGSGGNVKIIKVISDAGSSGGYGGGY-----------GGGHGGGYSSG 172
Score = 54.7 bits (130), Expect = 2e-06
Identities = 59/207 (28%), Positives = 66/207 (31%), Gaps = 87/207 (42%)
Query: 3 GGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGGG 62
GGGG GG + GW SGGGG + GG GGGG
Sbjct: 20 GGGGAGGS--------ALSSGW---------------SSGGGGGGYSGGYSGGHGGGGGY 56
Query: 63 GGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATI 122
G GGG GGG G S + ++ V+
Sbjct: 57 G-------------------------GGGYGGGGYGGGGHGGGSTIKIIK-----VITDS 86
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GGG+ GGG G G G Y GG
Sbjct: 87 GAGGGGYGGGY--GGGHGGGY-------GGGYGGGSSGGYSGGH---------------- 121
Query: 183 VTVGRWWRRRRRR*SDGGGGGGGANSG 209
G W S GG GGGG SG
Sbjct: 122 ---GGGWS------SGGGYGGGGYGSG 139
Score = 46.6 bits (109), Expect = 5e-04
Identities = 48/166 (28%), Positives = 58/166 (34%), Gaps = 52/166 (31%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVA-ATVEGGGNGGG 60
GGGG GG + + V+T G+GGGG GGG GGG
Sbjct: 67 GGGGHGGGSTIKIIKVITDS------------------GAGGGGYGGGYGGGHGGGYGGG 108
Query: 61 GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLA 120
GGG + + W GGG GGG G ++ V++
Sbjct: 109 YGGGS---------SGGYSGGHGGGWSSGGGYGGGGYGSGG----------NVKIIKVIS 149
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGG 166
G GG GGG GGG GGG GG Y GG
Sbjct: 150 DAGSSGGY-------GGGYGGGH-------GGGYSSGGASSYSAGG 181
>UniRef100_P27484 Glycine-rich protein 2 [Nicotiana sylvestris]
Length = 214
Score = 58.5 bits (140), Expect = 1e-07
Identities = 57/177 (32%), Positives = 61/177 (34%), Gaps = 59/177 (33%)
Query: 41 SGGGGKAVAATVEG-------GGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCG 93
SGG G+ A V G GG GGGGGGG R GGG G
Sbjct: 61 SGGDGRTKAVDVTGPDGAAVQGGRGGGGGGGGRG---------------------GGGYG 99
Query: 94 GGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GG-GDGGGWWWWWWQ*GG 152
GG G G GGS+ GG G GGG+ GG
Sbjct: 100 GGSGGYGG---------------------GGRGGSRGYGGGDGGYGGGGGYGGGSRYGGG 138
Query: 153 GCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G G GGGG YGGGG + G R GGGGGGG G
Sbjct: 139 GGGYGGGGGYGGGGSG---------GGSGCFKCGESGHFARDCSQSGGGGGGGRFGG 186
Score = 38.1 bits (87), Expect = 0.17
Identities = 44/143 (30%), Positives = 49/143 (33%), Gaps = 59/143 (41%)
Query: 40 GSGGGGKAVAATVEGG----GNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGG 95
G GGGG+ + GG G GGG GGG R + GGG GGG
Sbjct: 104 GYGGGGRGGSRGYGGGDGGYGGGGGYGGGSR------------------YGGGGGGYGGG 145
Query: 96 RAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAV-----------AA**GGGDGGGWW 144
G G GGGS + GGG GGG +
Sbjct: 146 GGYGGG---------------------GSGGGSGCFKCGESGHFARDCSQSGGGGGGGRF 184
Query: 145 WWWWQ*GGGCGVGGGGDYGGGGD 167
GGG G GGGG Y G D
Sbjct: 185 G-----GGGGGGGGGGCYKCGED 202
>UniRef100_O04339 Putative glycine-rich protein [Arabidopsis thaliana]
Length = 171
Score = 58.2 bits (139), Expect = 2e-07
Identities = 56/167 (33%), Positives = 64/167 (37%), Gaps = 52/167 (31%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGG GGR G GGGG A GG +GGGG
Sbjct: 17 GGGGSGGGR-----------------------------GGGGGGGAKGGCGGGGKSGGGG 47
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG +V S + R + GG GGG+ G + A +
Sbjct: 48 GGGGYMVAPGSNRSSYISRDNFESDPKGGSGGGGKGGGGGGGISGGGAGGKS-------- 99
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYG--GGG 166
GCGGG GGG GGG + GGGCG GGGG G GGG
Sbjct: 100 -GCGGGKS------GGGGGGG------KNGGGCGGGGGGKGGKSGGG 133
Score = 55.5 bits (132), Expect = 1e-06
Identities = 50/170 (29%), Positives = 57/170 (33%), Gaps = 60/170 (35%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGGK GGG GGGGGGG + GGCGGG +
Sbjct: 8 GSGGGGKGGGGGGSGGGRGGGGGGGAK-----------------------GGCGGGGKSG 44
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G + + ++ K + GGG GGG GG G G G
Sbjct: 45 GGGGGGGYMVAPGSNRSSYISRDNFESDPKGGSG--GGGKGGGGG------GGISGGGAG 96
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G G GG K GGGGGGG N G
Sbjct: 97 GKSGCGGGK-----------------------------SGGGGGGGKNGG 117
Score = 39.3 bits (90), Expect = 0.077
Identities = 32/103 (31%), Positives = 35/103 (33%), Gaps = 41/103 (39%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGGK GGG GGG GG GCGGG++
Sbjct: 76 GSGGGGKGGGG---GGGISGGGAGGK------------------------SGCGGGKSGG 108
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGG 142
G + GCGGG GGG GGG
Sbjct: 109 GGGGGKNGG--------------GCGGGGGGKGGKSGGGSGGG 137
Score = 39.3 bits (90), Expect = 0.077
Identities = 33/94 (35%), Positives = 37/94 (39%), Gaps = 20/94 (21%)
Query: 123 GCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPV 182
G GGG GGG GGG GGCG GG GGGG +VA P
Sbjct: 13 GKGGGGGGSGGGRGGGGGGGA-------KGGCGGGGKSGGGGGGGGYMVA--------PG 57
Query: 183 VTVGRWWRRRR-----RR*SDGGGGGGGANSGVS 211
+ R + S GGG GGG G+S
Sbjct: 58 SNRSSYISRDNFESDPKGGSGGGGKGGGGGGGIS 91
>UniRef100_Q39337 Glycine-rich_protein_(Aa1-291) precursor [Brassica napus]
Length = 291
Score = 57.8 bits (138), Expect = 2e-07
Identities = 54/172 (31%), Positives = 57/172 (32%), Gaps = 66/172 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGGNGGGGGGG GGG GGG A
Sbjct: 155 GYGGGGAGGHGGGGGGGNGGGGGGGGAH---------------------GGGYGGGEGAG 193
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG- 158
G GGG+ GGG GGG GGG G GG
Sbjct: 194 AGGGYG-------------------GGGAGGHGGGGGGGKGGGG-------GGGSGAGGA 227
Query: 159 -GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG YG GG G + GGGGGGG +G
Sbjct: 228 HGGGYGAGGG-----------------AGEGYGGGGGEGGHGGGGGGGGGAG 262
Score = 57.4 bits (137), Expect = 3e-07
Identities = 52/170 (30%), Positives = 53/170 (30%), Gaps = 79/170 (46%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
GSGGGG A GGG GGG G G GGG GGG A
Sbjct: 127 GSGGGGGGGAGGAHGGGYGGGEGAGA-----------------------GGGYGGGGAGG 163
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GG GGG+ GGG G G G
Sbjct: 164 HG---------------------GGGGGGNGGGGGGGGAHGGGY-------GGGEGAGAG 195
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G YGGGG GGGGGGG G
Sbjct: 196 GGYGGGGAG----------------------------GHGGGGGGGKGGG 217
Score = 45.8 bits (107), Expect = 8e-04
Identities = 45/129 (34%), Positives = 46/129 (34%), Gaps = 49/129 (37%)
Query: 40 GSGGGGKAVAATVEGGGNG--GGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRA 97
G GGG + EGGG G GGG GG GGG GGG
Sbjct: 95 GPGGGSGYGGGSGEGGGAGYGGGGAGGHGG---------------------GGGSGGGGG 133
Query: 98 AVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG 157
G A G GGG A A GGG GGG GG G G
Sbjct: 134 GGAGGAHGG----------------GYGGGEGAGA---GGGYGGGG-------AGGHGGG 167
Query: 158 GGGDYGGGG 166
GGG GGGG
Sbjct: 168 GGGGNGGGG 176
Score = 44.7 bits (104), Expect = 0.002
Identities = 40/127 (31%), Positives = 42/127 (32%), Gaps = 55/127 (43%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G G A GGG GGG G G + GGG GGG
Sbjct: 47 GYGIGVDAGVGVGGGGGEGGGAGYGGAEGI-------------------GGGGGGGHGG- 86
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GGG GGG GGG + GGG G GGG
Sbjct: 87 -----------------------GAGGGG-------GGGPGGGSGY-----GGGSGEGGG 111
Query: 160 GDYGGGG 166
YGGGG
Sbjct: 112 AGYGGGG 118
Score = 42.7 bits (99), Expect = 0.007
Identities = 41/127 (32%), Positives = 42/127 (32%), Gaps = 33/127 (25%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG G G A
Sbjct: 197 GYGGGGAGGHGGGGGGGKGGGGGGGSGAGGAH-----------------GGGYGAGGGAG 239
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG + G G G G G
Sbjct: 240 EGYGGGGGEG-------------GHGGGGGGGGGAGGGGGGGGGYA---AAGSGHGGGAG 283
Query: 160 GDYGGGG 166
GGGG
Sbjct: 284 RGEGGGG 290
Score = 37.4 bits (85), Expect = 0.29
Identities = 50/171 (29%), Positives = 51/171 (29%), Gaps = 95/171 (55%)
Query: 40 GSGGGGKAVAATVEGG--GNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRA 97
G GGGG GG G GGGGGGG GGG GGG
Sbjct: 57 GVGGGGGEGGGAGYGGAEGIGGGGGGGH-----------------------GGGAGGGGG 93
Query: 98 AVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVG 157
G GGGS GGG G G GG G G
Sbjct: 94 G------------------------GPGGGSGY-----GGGSGEG---------GGAGYG 115
Query: 158 GGG-D-YGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGA 206
GGG +GGGG GGGGGGGA
Sbjct: 116 GGGAGGHGGGG------------------------------GSGGGGGGGA 136
>UniRef100_P10496 Glycine-rich cell wall structural protein 1.8 precursor [Phaseolus
vulgaris]
Length = 465
Score = 57.8 bits (138), Expect = 2e-07
Identities = 52/170 (30%), Positives = 59/170 (34%), Gaps = 56/170 (32%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GGG A + GGG GG GGGG VV GGG GGG+
Sbjct: 39 GTGGGYGGAAGSYGGGGGGGSGGGGG-YAGEHGVVGY------------GGGSGGGQGGG 85
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
VG G G G A GGG+ GG+ GGG G G G
Sbjct: 86 VGYGGDQGAGYGG----------GGGSGGGGGVAYGGGGERGGY-------GGGQGGGAG 128
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G YG GG+ + GGGGG GA G
Sbjct: 129 GGYGAGGEHGI--------------------------GYGGGGGSGAGGG 152
Score = 53.1 bits (126), Expect = 5e-06
Identities = 52/177 (29%), Positives = 58/177 (32%), Gaps = 89/177 (50%)
Query: 40 GSGGGGKAVAATVEGGG-------NGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGC 92
G+GGGG A +GGG GGGGGGGD GGG
Sbjct: 148 GAGGGGGYNAGGAQGGGYGTGGGAGGGGGGGGDH----------------------GGGY 185
Query: 93 GGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG 152
GGG+ A G G GGG + GGG GGG G
Sbjct: 186 GGGQGAGGGAGG------------------GYGGGGEH-----GGGGGGGQGG-----GA 217
Query: 153 GCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G G G GG++GGG GGG GGGA G
Sbjct: 218 GGGYGAGGEHGGGA--------------------------------GGGQGGGAGGG 242
Score = 51.6 bits (122), Expect = 1e-05
Identities = 45/131 (34%), Positives = 48/131 (36%), Gaps = 51/131 (38%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GG +GGGGGGG GGG GGG AAV
Sbjct: 298 GGGQGGGAGGGYGAGGEHGGGGGGGQ-----------------------GGGAGGGYAAV 334
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ---*GGGCGV 156
G GGG GGGDGGG+ GGG G
Sbjct: 335 GEHGG------------------GYGGGQ-------GGGDGGGYGTGGEHGGGYGGGQGG 369
Query: 157 GGGGDYGGGGD 167
G GG YG GG+
Sbjct: 370 GAGGGYGTGGE 380
Score = 48.5 bits (114), Expect = 1e-04
Identities = 56/179 (31%), Positives = 63/179 (34%), Gaps = 63/179 (35%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGG--------GGKAVAATVE 53
GGGGG+GG V G GSGG GG A
Sbjct: 54 GGGGGSGGGGGYAGEHGVVGYGG---------------GSGGGQGGGVGYGGDQGAGYGG 98
Query: 54 GGGNGGG-----GGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVV 108
GGG+GGG GGGG+R GGG GGG G
Sbjct: 99 GGGSGGGGGVAYGGGGERGGY-------------------GGGQGGGAGGGYGAGGEH-- 137
Query: 109 AVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGC--GVGGGGDYGGG 165
IG GGG + A GG + GG + GGG G GGGGD+GGG
Sbjct: 138 ------------GIGYGGGGGSGAGGGGGYNAGGAQGGGYGTGGGAGGGGGGGGDHGGG 184
Score = 48.5 bits (114), Expect = 1e-04
Identities = 51/170 (30%), Positives = 56/170 (32%), Gaps = 55/170 (32%)
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GGGG A E GG GGG GGG + + G G GGG G
Sbjct: 104 GGGGVAYGGGGERGGYGGGQGGG-----AGGGYGAGGEHGIGYGGGGGSGAGGGGGYNAG 158
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG--VGGG 159
A G G G A GGGD GG + GGG G GGG
Sbjct: 159 GAQGG----------------GYGTGGGAGGGGGGGGDHGGGYGGGQGAGGGAGGGYGGG 202
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G++GGGG GGG GGGA G
Sbjct: 203 GEHGGGG--------------------------------GGGQGGGAGGG 220
Score = 48.1 bits (113), Expect = 2e-04
Identities = 50/172 (29%), Positives = 55/172 (31%), Gaps = 87/172 (50%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG+ GGG+G GGGGG + GG GGG
Sbjct: 130 GYGAGGEHGIGYGGGGGSGAGGGGG---------------------YNAGGAQGGGYGTG 168
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG- 158
G G GGG GGGD GG + GGG G GG
Sbjct: 169 GGA--------------------GGGGG--------GGGDHGGGY------GGGQGAGGG 194
Query: 159 -GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG YGGGG+ GGGGGGG G
Sbjct: 195 AGGGYGGGGE------------------------------HGGGGGGGQGGG 216
Score = 46.2 bits (108), Expect = 6e-04
Identities = 51/173 (29%), Positives = 54/173 (30%), Gaps = 77/173 (44%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GG +GGGGGGG GGG GGG A
Sbjct: 188 GQGAGGGAGGGYGGGGEHGGGGGGGQ-----------------------GGGAGGGYGAG 224
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GG---GCGV 156
G GGG GGG GGG+ GG G G
Sbjct: 225 GEHGG------------------GAGGGQ-------GGGAGGGYGAGGEHGGGAGGGQGG 259
Query: 157 GGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GG YG GG+ A GGG GGGA G
Sbjct: 260 GAGGGYGAGGEHGGGA--------------------------GGGQGGGAGGG 286
Score = 43.9 bits (102), Expect = 0.003
Identities = 48/170 (28%), Positives = 51/170 (29%), Gaps = 75/170 (44%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GGG GGG GGG GGG GGG+
Sbjct: 236 GGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAG-------------GEHGGGAGGGQGGG 282
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GG A GGG GGG G G G G
Sbjct: 283 AG------------------GGYGAGGEHGGGA---GGGQGGG---------AGGGYGAG 312
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G++GGGG GGG GGGA G
Sbjct: 313 GEHGGGG--------------------------------GGGQGGGAGGG 330
Score = 43.9 bits (102), Expect = 0.003
Identities = 50/170 (29%), Positives = 52/170 (30%), Gaps = 75/170 (44%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GGG GGG GGG GGG GGG+
Sbjct: 214 GGGAGGGYGAGGEHGGGAGGGQGGGAGGGYGAG-------------GEHGGGAGGGQGG- 259
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G GGG A GG GGG GGG G G G
Sbjct: 260 -----------------------GAGGGYGA-----GGEHGGGA-------GGGQGGGAG 284
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G YG GG+ A GGG GGGA G
Sbjct: 285 GGYGAGGEHGGGA--------------------------GGGQGGGAGGG 308
Score = 43.1 bits (100), Expect = 0.005
Identities = 45/134 (33%), Positives = 48/134 (35%), Gaps = 25/134 (18%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG A GGG GGG GGGD GGG GGG+
Sbjct: 324 GGGAGGGYAAVGEHGGGYGGGQGGGDGGGYGTG-------------GEHGGGYGGGQGGG 370
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG- 158
G + G G G AA GGG+GGG GGG G GG
Sbjct: 371 AGGGYGTGGEHGGGYGGGQGGGGGYGAGGDHGAAGYGGGEGGGGGS-----GGGYGDGGA 425
Query: 159 ------GGDYGGGG 166
GG GGGG
Sbjct: 426 HGGGYGGGAGGGGG 439
Score = 35.4 bits (80), Expect = 1.1
Identities = 51/166 (30%), Positives = 54/166 (31%), Gaps = 26/166 (15%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GGGGG GG V G + Q G G GG GGG GGG
Sbjct: 318 GGGGGQGGGAGGGYAAVGEHGGGYGGGQ----------GGGDGGGYGTGGEHGGGYGGGQ 367
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLAT 121
GGG GGG GGG+ G A A
Sbjct: 368 GGGAGGGYGTG-------------GEHGGGYGGGQGGGGGYGAGGDHGA-AGYGGGEGGG 413
Query: 122 IGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGV--GGGGDYGGG 165
G GGG A GG GG + GG G GGGG GGG
Sbjct: 414 GGSGGGYGDGGAHGGGYGGGAGGGGGYGAGGAHGGGYGGGGGIGGG 459
Score = 33.1 bits (74), Expect = 5.5
Identities = 15/25 (60%), Positives = 16/25 (64%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGG 64
G+GGGG A GGG GGGGG G
Sbjct: 433 GAGGGGGYGAGGAHGGGYGGGGGIG 457
>UniRef100_UPI0000332877 UPI0000332877 UniRef100 entry
Length = 127
Score = 57.4 bits (137), Expect = 3e-07
Identities = 53/171 (30%), Positives = 53/171 (30%), Gaps = 83/171 (48%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG GGG
Sbjct: 10 GGGGGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGGGGGG 45
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 46 GG---------------------GGGGGGGGGGGGGGGGGGGG--------GGGGGGGGG 76
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
G GGGG GGGGGGG GV
Sbjct: 77 GGGGGGG------------------------------GGGGGGGGGGGCGV 97
Score = 54.7 bits (130), Expect = 2e-06
Identities = 46/137 (33%), Positives = 47/137 (33%), Gaps = 45/137 (32%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GGGG GGG GGGGGGG GGG GGG
Sbjct: 11 GGGGGGGGGGGGGGGGGGGGGGGGG------------------------GGGGGGGGGGG 46
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G G GGG GGG GGG GGG G GGG
Sbjct: 47 GG---------------------GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 85
Query: 160 GDYGGGGDKVVVAAIMV 176
G GGGG V +V
Sbjct: 86 GGGGGGGGGCGVCVCVV 102
>UniRef100_Q79FS5 PE-PGRS FAMILY PROTEIN [Mycobacterium tuberculosis]
Length = 853
Score = 57.0 bits (136), Expect = 4e-07
Identities = 54/168 (32%), Positives = 64/168 (37%), Gaps = 43/168 (25%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GG GG+GG +L G GSGG G A +T GGNGG G
Sbjct: 289 GGAGGSGGHALLWGAGGAGGNG----------------GSGGTGGAGGSTAGAGGNGGAG 332
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGG-GRAAVVGVAATSVVAVVAAMVVVVLA 120
GGG ++ GG GG G AA G+AA + V+ +
Sbjct: 333 GGGGTGGLLFG----------------NGGAGGHGAAAGNGLAAGNGVS----------S 366
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDK 168
+ G G G A GG G G W GG G GG G GG G K
Sbjct: 367 SGGGGAGGTGGAGGDGGAGGAGGNARLWGVGGAGGAGGDGGAGGAGGK 414
Score = 52.0 bits (123), Expect = 1e-05
Identities = 58/194 (29%), Positives = 66/194 (33%), Gaps = 46/194 (23%)
Query: 40 GSGGGGKAVAATVEGG------GNGGGGGGGDRVV---VVVSVVAIEWWRRLWWWWR*GG 90
G+GGGG A AA GG GNGG GG G V V LW GG
Sbjct: 170 GAGGGGGAGAAGGNGGAGGWLYGNGGAGGAGGTSVIPGVAGGNGGAGGSAGLWGTGGAGG 229
Query: 91 GCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGG-------GSKAVAA**GGGDGG-- 141
G GR+ V VA ++ A + G GG G A + GDGG
Sbjct: 230 DGGNGRSGPVNVAGSAGGNGGAGGAAGLFGDAGAGGNGGKGGAGGAAFSINFTAGDGGAG 289
Query: 142 -----GWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR* 196
G W GG G GG G GG G A
Sbjct: 290 GAGGSGGHALLWGAGGAGGNGGSGGTGGAGGSTAGAG----------------------- 326
Query: 197 SDGGGGGGGANSGV 210
+GG GGGG G+
Sbjct: 327 GNGGAGGGGGTGGL 340
Score = 46.2 bits (108), Expect = 6e-04
Identities = 52/181 (28%), Positives = 59/181 (31%), Gaps = 53/181 (29%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G A AA GG GGGGG G G G GG
Sbjct: 514 GAGGNGGAAAAGGAGGQAGGGGGNG------------------------GNGGNGGNGGN 549
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGG-GSKAVAA**GGGDGGGWWWWWWQ*GGGC---- 154
G AT AT G GG G+ V++ GGG GG GG
Sbjct: 550 GGNGATGGWLYGNGGAGGQGATAGAGGAGANGVSSTNGGGTGGNGGIGGTGGSGGAGGNA 609
Query: 155 ------GVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANS 208
G GG G GG GD+ + SDGG GG G +
Sbjct: 610 GLLGVGGAGGHGASGGAGDRGGAGGTGFIS------------------SDGGAGGDGGDG 651
Query: 209 G 209
G
Sbjct: 652 G 652
Score = 46.2 bits (108), Expect = 6e-04
Identities = 54/170 (31%), Positives = 59/170 (33%), Gaps = 34/170 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G A V GG+GG GG G V A R G G GG
Sbjct: 438 GAGGEGGAAGLLVGTGGHGGDGGAGGAAVKGGDGGAAAGTGIAGAGGRGGAGGSGGSGGD 497
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A A + G GG A AA GG GG Q GGG G GG
Sbjct: 498 GGGGA-------AGPAGWLFGDGGAGGNGGAAAA---GGAGG-------QAGGGGGNGGN 540
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GG G A G W +GG GG GA +G
Sbjct: 541 GGNGGNGGNGGNGA-----------TGGWLY------GNGGAGGQGATAG 573
Score = 43.9 bits (102), Expect = 0.003
Identities = 48/174 (27%), Positives = 60/174 (33%), Gaps = 35/174 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GG A + GG GGGGG G W + G G GG + +
Sbjct: 154 GVAGGNGGAAGLIGNGGAGGGGGAG--------AAGGNGGAGGWLYGNGGAGGAGGTSVI 205
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKA----VAA**GGGDGGGWWWWWWQ*GGGCG 155
GVA + A +A + G GG ++ VA GG G G + G G
Sbjct: 206 PGVAGGNGGAGGSAGLWGTGGAGGDGGNGRSGPVNVAGSAGGNGGAGGAAGLF---GDAG 262
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG G GG G AA + DGG GG G + G
Sbjct: 263 AGGNGGKGGAGG----AAFSINFTA----------------GDGGAGGAGGSGG 296
Score = 42.7 bits (99), Expect = 0.007
Identities = 54/173 (31%), Positives = 59/173 (33%), Gaps = 32/173 (18%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
SGGGG T GG+GG GG G RLW GG G G A
Sbjct: 367 SGGGG--AGGTGGAGGDGGAGGAGGNA-------------RLWGVGGAGGAGGDGGAGGA 411
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG-G 159
G S ++ A G GG S GG+GG G G GG G
Sbjct: 412 GGKGGSGLSGNA--------NGGAGGDSGRGGTGGAGGEGGAA-------GLLVGTGGHG 456
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
GD G GG V T + G S GG GGGGA W
Sbjct: 457 GDGGAGGAAVKGGDGGAAAGTGIAGAGGRGGAGGSGGS-GGDGGGGAAGPAGW 508
Score = 40.8 bits (94), Expect = 0.026
Identities = 42/169 (24%), Positives = 50/169 (28%), Gaps = 55/169 (32%)
Query: 42 GGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVG 101
GG G + +GG G GG GG+ GG GG + G
Sbjct: 630 GGAGGTGFISSDGGAGGDGGDGGN------------------------GGAGGTGGLLFG 665
Query: 102 VAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGD 161
G GG+ + G G+GGG G G G G GGD
Sbjct: 666 AGGN--------------GGPGGSGGAADIGGNGGAGNGGGTDGNGGNGGSGGGAGSGGD 711
Query: 162 YGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
GG G G W GG GG GA G+
Sbjct: 712 GGGAGGN-----------------GAWLFGNGGAGGGGGKGGNGAGGGL 743
Score = 40.4 bits (93), Expect = 0.034
Identities = 48/174 (27%), Positives = 56/174 (31%), Gaps = 42/174 (24%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G + + G NGG GG R + L GG G G AAV
Sbjct: 407 GAGGAGGKGGSGLSGNANGGAGGDSGRGGTGGAGGEGGAAGLLVGTGGHGGDGGAGGAAV 466
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGW----WWWWWQ*GGGCG 155
G + A G GG A + GGDGGG W + GG G
Sbjct: 467 KGGDGGAAAGTGIA---------GAGGRGGAGGSGGSGGDGGGGAAGPAGWLFGDGGAGG 517
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG GG G + GGGGG G N G
Sbjct: 518 NGGAAAAGGAGGQA-----------------------------GGGGGNGGNGG 542
Score = 38.9 bits (89), Expect = 0.10
Identities = 55/218 (25%), Positives = 68/218 (30%), Gaps = 38/218 (17%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GG GGNGG G W + G+GG G ++ GGG GG G
Sbjct: 535 GGNGGNGGNGGNGGNGGNGATGGWLYGNGGAGGQGATAGAGGAGANGVSSTNGGGTGGNG 594
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGV-------AATSVVAVVAAM 114
G G G G GG A ++GV A+ A
Sbjct: 595 GIGGTG---------------------GSGGAGGNAGLLGVGGAGGHGASGGAGDRGGAG 633
Query: 115 VVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAI 174
+++ G GG G G GG + GG G GG D GG G
Sbjct: 634 GTGFISSDGGAGGDGGDGGNGGAGGTGGLLFGAGGNGGPGGSGGAADIGGNGG------- 686
Query: 175 MVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
T G + GG GGGA +W
Sbjct: 687 ---AGNGGGTDGNGGNGGSGGGAGSGGDGGGAGGNGAW 721
Score = 38.5 bits (88), Expect = 0.13
Identities = 47/175 (26%), Positives = 53/175 (29%), Gaps = 37/175 (21%)
Query: 42 GGGGKAVAATV------EGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGG 95
GG G A A T GG G GG GGD W + GG G G
Sbjct: 468 GGDGGAAAGTGIAGAGGRGGAGGSGGSGGDGGGGAAGPAG--------WLFGDGGAGGNG 519
Query: 96 RAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCG 155
AA G A G GG+ G G G W + GG G
Sbjct: 520 GAAAAGGAGGQAGGG------------GGNGGNGGNGGNGGNGGNGATGGWLYGNGGAGG 567
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
G GG G V + T G GG GG G N+G+
Sbjct: 568 QGATAGAGGAGANGVSS-----------TNGGGTGGNGGIGGTGGSGGAGGNAGL 611
Score = 38.1 bits (87), Expect = 0.17
Identities = 47/169 (27%), Positives = 58/169 (33%), Gaps = 48/169 (28%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG ++ T GG GG GG G ++ W GG G G +
Sbjct: 271 GAGGAAFSINFTAGDGGAGGAGGSGGHALL----------------WGAGGAGGNGGSGG 314
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A S T G GG A GGG G G + G G GG
Sbjct: 315 TGGAGGS--------------TAGAGGNGGA-----GGGGGTGGLLF-----GNGGAGGH 350
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANS 208
G G G +AA V ++ G DGG GG G N+
Sbjct: 351 GAAAGNG----LAAGNGVSSSGGGGAGGTGGAG----GDGGAGGAGGNA 391
Score = 38.1 bits (87), Expect = 0.17
Identities = 42/170 (24%), Positives = 48/170 (27%), Gaps = 59/170 (34%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG GG GGGG GG+ GGG GGG +
Sbjct: 712 GGGAGGNGAWLFGNGGAGGGGGKGGNGA---------------------GGGLGGGSFGL 750
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ + G GG G G GG G G G G
Sbjct: 751 PGLNGSG-------------GDGGDGGNGAPGGVLYGNGGAGGQGSSGGIGGPGATGGAG 797
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G G GGD ++ DGG GG G G
Sbjct: 798 GKGGDGGDAQLI-------------------------GDGGNGGNGGAGG 822
Score = 37.7 bits (86), Expect = 0.22
Identities = 36/128 (28%), Positives = 40/128 (31%), Gaps = 54/128 (42%)
Query: 40 GSGG-GGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAA 98
G+GG GG AA + G G G GGG D G G GG
Sbjct: 668 GNGGPGGSGGAADIGGNGGAGNGGGTD-----------------------GNGGNGGSGG 704
Query: 99 VVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG 158
G G G GGG GG W + G G G G
Sbjct: 705 ------------------------GAGSGGD------GGGAGGNGAWLFGNGGAGGGGGK 734
Query: 159 GGDYGGGG 166
GG+ GGG
Sbjct: 735 GGNGAGGG 742
Score = 37.4 bits (85), Expect = 0.29
Identities = 38/127 (29%), Positives = 46/127 (35%), Gaps = 43/127 (33%)
Query: 89 GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWW 148
G G GG + GVA + A ++ G GGG A AA GG+GG W +
Sbjct: 142 GNGGNGGTSTTAGVAGGNGGAAG------LIGNGGAGGGGGAGAA---GGNGGAGGWLYG 192
Query: 149 Q*GGGCGVGG--------GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGG 200
GG G GG GG+ G GG + G W GG
Sbjct: 193 N-GGAGGAGGTSVIPGVAGGNGGAGG-----------------SAGLW--------GTGG 226
Query: 201 GGGGGAN 207
GG G N
Sbjct: 227 AGGDGGN 233
>UniRef100_Q7U0P1 PE-PGRS FAMILY PROTEIN [Mycobacterium bovis]
Length = 850
Score = 57.0 bits (136), Expect = 4e-07
Identities = 54/168 (32%), Positives = 64/168 (37%), Gaps = 43/168 (25%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GG GG+GG +L G GSGG G A +T GGNGG G
Sbjct: 289 GGAGGSGGHALLWGAGGAGGNG----------------GSGGTGGAGGSTAGAGGNGGAG 332
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGG-GRAAVVGVAATSVVAVVAAMVVVVLA 120
GGG ++ GG GG G AA G+AA + V+ +
Sbjct: 333 GGGGTGGLLFG----------------NGGAGGHGAAAGNGLAAGNGVS----------S 366
Query: 121 TIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDK 168
+ G G G A GG G G W GG G GG G GG G K
Sbjct: 367 SGGGGAGGTGGAGGDGGAGGAGGNARLWGVGGAGGAGGDGGAGGAGGK 414
Score = 52.0 bits (123), Expect = 1e-05
Identities = 58/194 (29%), Positives = 66/194 (33%), Gaps = 46/194 (23%)
Query: 40 GSGGGGKAVAATVEGG------GNGGGGGGGDRVV---VVVSVVAIEWWRRLWWWWR*GG 90
G+GGGG A AA GG GNGG GG G V V LW GG
Sbjct: 170 GAGGGGGAGAAGGNGGAGGWLYGNGGAGGAGGTSVIPGVAGGNGGAGGSAGLWGTGGAGG 229
Query: 91 GCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGG-------GSKAVAA**GGGDGG-- 141
G GR+ V VA ++ A + G GG G A + GDGG
Sbjct: 230 DGGNGRSGPVNVAGSAGGNGGAGGAAGLFGDAGAGGNGGKGGAGGAAFSINFTAGDGGAG 289
Query: 142 -----GWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR* 196
G W GG G GG G GG G A
Sbjct: 290 GAGGSGGHALLWGAGGAGGNGGSGGTGGAGGSTAGAG----------------------- 326
Query: 197 SDGGGGGGGANSGV 210
+GG GGGG G+
Sbjct: 327 GNGGAGGGGGTGGL 340
Score = 46.6 bits (109), Expect = 5e-04
Identities = 53/178 (29%), Positives = 62/178 (34%), Gaps = 50/178 (28%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G A AA GG GGGGG G G G GG
Sbjct: 514 GAGGNGGAAAAGGAGGQAGGGGGNG------------------------GNGGNGGNGGN 549
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGG-GSKAVAA**GGGDGG-GWWWWWWQ*GGGCGV- 156
G AT AT G GG G+ V++ GGG+GG G GG G+
Sbjct: 550 GGNGATGGWLYGNGGAGGQGATAGAGGAGANGVSSTNGGGNGGIGGTGGSGGAGGNAGLL 609
Query: 157 -----GGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG G GG GD+ + SDGG GG G + G
Sbjct: 610 GVGGAGGHGASGGAGDRGGAGGTGFIS------------------SDGGAGGDGGDGG 649
Score = 46.2 bits (108), Expect = 6e-04
Identities = 54/170 (31%), Positives = 59/170 (33%), Gaps = 34/170 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G A V GG+GG GG G V A R G G GG
Sbjct: 438 GAGGEGGAAGLLVGTGGHGGDGGAGGAAVKGGDGGAAAGTGIAGAGGRGGAGGSGGSGGD 497
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A A + G GG A AA GG GG Q GGG G GG
Sbjct: 498 GGGGA-------AGPAGWLFGDGGAGGNGGAAAA---GGAGG-------QAGGGGGNGGN 540
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G GG G A G W +GG GG GA +G
Sbjct: 541 GGNGGNGGNGGNGA-----------TGGWLY------GNGGAGGQGATAG 573
Score = 43.9 bits (102), Expect = 0.003
Identities = 48/174 (27%), Positives = 60/174 (33%), Gaps = 35/174 (20%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GG A + GG GGGGG G W + G G GG + +
Sbjct: 154 GVAGGNGGAAGLIGNGGAGGGGGAG--------AAGGNGGAGGWLYGNGGAGGAGGTSVI 205
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKA----VAA**GGGDGGGWWWWWWQ*GGGCG 155
GVA + A +A + G GG ++ VA GG G G + G G
Sbjct: 206 PGVAGGNGGAGGSAGLWGTGGAGGDGGNGRSGPVNVAGSAGGNGGAGGAAGLF---GDAG 262
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG G GG G AA + DGG GG G + G
Sbjct: 263 AGGNGGKGGAGG----AAFSINFTA----------------GDGGAGGAGGSGG 296
Score = 42.7 bits (99), Expect = 0.007
Identities = 54/173 (31%), Positives = 59/173 (33%), Gaps = 32/173 (18%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
SGGGG T GG+GG GG G RLW GG G G A
Sbjct: 367 SGGGG--AGGTGGAGGDGGAGGAGGNA-------------RLWGVGGAGGAGGDGGAGGA 411
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGG-G 159
G S ++ A G GG S GG+GG G G GG G
Sbjct: 412 GGKGGSGLSGNA--------NGGAGGDSGRGGTGGAGGEGGAA-------GLLVGTGGHG 456
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
GD G GG V T + G S GG GGGGA W
Sbjct: 457 GDGGAGGAAVKGGDGGAAAGTGIAGAGGRGGAGGSGGS-GGDGGGGAAGPAGW 508
Score = 42.0 bits (97), Expect = 0.012
Identities = 55/218 (25%), Positives = 67/218 (30%), Gaps = 50/218 (22%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGGGG 61
GG GGNGG G W + G+GG G ++ GGGNGG G
Sbjct: 544 GGNGGNGGNGAT---------GGWLYGNGGAGGQGATAGAGGAGANGVSSTNGGGNGGIG 594
Query: 62 GGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAVVA-------AM 114
G G G G GG A ++GV A
Sbjct: 595 GTG------------------------GSGGAGGNAGLLGVGGAGGHGASGGAGDRGGAG 630
Query: 115 VVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGGGDKVVVAAI 174
+++ G GG G G GG + GG G GG D GG G
Sbjct: 631 GTGFISSDGGAGGDGGDGGNGGAGGTGGLLFGAGGNGGPGGSGGAADIGGNGG------- 683
Query: 175 MVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSW 212
T G + GG GGGA +W
Sbjct: 684 ---AGNGGGTDGNGGNGGSGGGAGSGGDGGGAGGNGAW 718
Score = 42.0 bits (97), Expect = 0.012
Identities = 58/236 (24%), Positives = 73/236 (30%), Gaps = 57/236 (24%)
Query: 2 GGGGGNGGRLVLVAVVVTVEIGWWWWWQRWWLWWW***GSGGGGKAVAATVEGGGNGG-- 59
GG GGNGG G W + G+GG G ++ GGGNGG
Sbjct: 535 GGNGGNGGNGGNGGNGGNGATGGWLYGNGGAGGQGATAGAGGAGANGVSSTNGGGNGGIG 594
Query: 60 -----GGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAATSVVAV---- 110
GG GG+ ++ V G G GG G T ++
Sbjct: 595 GTGGSGGAGGNAGLLGVGGAG-------------GHGASGGAGDRGGAGGTGFISSDGGA 641
Query: 111 -----------VAAMVVVVLATIGCG-----GGSKAVAA**GGGDGGGWWWWWWQ*GGGC 154
++ G G GG+ + G G+GGG G G
Sbjct: 642 GGDGGDGGNGGAGGTGGLLFGAGGNGGPGGSGGAADIGGNGGAGNGGGTDGNGGNGGSGG 701
Query: 155 GVGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
G G GGD GG G G W GG GG GA G+
Sbjct: 702 GAGSGGDGGGAGGN-----------------GAWLFGNGGAGGGGGKGGNGAGGGL 740
Score = 40.4 bits (93), Expect = 0.034
Identities = 48/174 (27%), Positives = 56/174 (31%), Gaps = 42/174 (24%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG G + + G NGG GG R + L GG G G AAV
Sbjct: 407 GAGGAGGKGGSGLSGNANGGAGGDSGRGGTGGAGGEGGAAGLLVGTGGHGGDGGAGGAAV 466
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGW----WWWWWQ*GGGCG 155
G + A G GG A + GGDGGG W + GG G
Sbjct: 467 KGGDGGAAAGTGIA---------GAGGRGGAGGSGGSGGDGGGGAAGPAGWLFGDGGAGG 517
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
GG GG G + GGGGG G N G
Sbjct: 518 NGGAAAAGGAGGQA-----------------------------GGGGGNGGNGG 542
Score = 38.1 bits (87), Expect = 0.17
Identities = 42/170 (24%), Positives = 48/170 (27%), Gaps = 59/170 (34%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G G GG GG GGGG GG+ GGG GGG +
Sbjct: 709 GGGAGGNGAWLFGNGGAGGGGGKGGNGA---------------------GGGLGGGSFGL 747
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G+ + G GG G G GG G G G G
Sbjct: 748 PGLNGSG-------------GDGGDGGNGAPGGVLYGNGGAGGQGSSGGIGGPGATGGAG 794
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSG 209
G G GGD ++ DGG GG G G
Sbjct: 795 GKGGDGGDAQLI-------------------------GDGGNGGNGGAGG 819
Score = 38.1 bits (87), Expect = 0.17
Identities = 47/169 (27%), Positives = 58/169 (33%), Gaps = 48/169 (28%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G+GG ++ T GG GG GG G ++ W GG G G +
Sbjct: 271 GAGGAAFSINFTAGDGGAGGAGGSGGHALL----------------WGAGGAGGNGGSGG 314
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGG 159
G A S T G GG A GGG G G + G G GG
Sbjct: 315 TGGAGGS--------------TAGAGGNGGA-----GGGGGTGGLLF-----GNGGAGGH 350
Query: 160 GDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANS 208
G G G +AA V ++ G DGG GG G N+
Sbjct: 351 GAAAGNG----LAAGNGVSSSGGGGAGGTGGAG----GDGGAGGAGGNA 391
Score = 37.4 bits (85), Expect = 0.29
Identities = 38/127 (29%), Positives = 46/127 (35%), Gaps = 43/127 (33%)
Query: 89 GGGCGGGRAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWW 148
G G GG + GVA + A ++ G GGG A AA GG+GG W +
Sbjct: 142 GNGGNGGTSTTAGVAGGNGGAAG------LIGNGGAGGGGGAGAA---GGNGGAGGWLYG 192
Query: 149 Q*GGGCGVGG--------GGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGG 200
GG G GG GG+ G GG + G W GG
Sbjct: 193 N-GGAGGAGGTSVIPGVAGGNGGAGG-----------------SAGLW--------GTGG 226
Query: 201 GGGGGAN 207
GG G N
Sbjct: 227 AGGDGGN 233
>UniRef100_Q9XEU3 Hypothetical protein [Oryza sativa]
Length = 934
Score = 56.2 bits (134), Expect = 6e-07
Identities = 45/129 (34%), Positives = 53/129 (40%), Gaps = 14/129 (10%)
Query: 41 SGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVV 100
SGGGG + G G GGG GGG VV + + + GGCG G
Sbjct: 717 SGGGGGQAESAGCGSGCGGGCGGGVAKVVEATKAGVGGHGK-------SGGCGSGCGGGC 769
Query: 101 GVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*G----GGCGV 156
G +AV ++ V GCG G GGG GGG + G GGCG
Sbjct: 770 GGGGCGAMAVESSKDDVHAKCAGCGSGCGGGC---GGGCGGGMVMEASKAGHVKSGGCGS 826
Query: 157 GGGGDYGGG 165
G GG GGG
Sbjct: 827 GCGGGCGGG 835
Score = 49.3 bits (116), Expect = 7e-05
Identities = 58/178 (32%), Positives = 71/178 (39%), Gaps = 31/178 (17%)
Query: 40 GSGGGGKAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAV 99
G GG GK+ GG GGGG G + V S + GGGCGGG
Sbjct: 751 GVGGHGKSGGCGSGCGGGCGGGGCG--AMAVESSKDDVHAKCAGCGSGCGGGCGGGCGGG 808
Query: 100 VGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*G----GGCG 155
+ + A+ A V GCGGG GGG GGG + G GGCG
Sbjct: 809 MVMEASK-----AGHVKSGGCGSGCGGGC-------GGGCGGGVAMESSKAGHVKSGGCG 856
Query: 156 VGGGGDYGGGGDKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGVSWQ 213
G GG GGG MV+ ++ V V + GGG GGG GV+ +
Sbjct: 857 SGCGGGCGGG-----CGGGMVMESSKAVHV--------KSGGCGGGCGGGCGGGVAME 901
Score = 48.1 bits (113), Expect = 2e-04
Identities = 46/135 (34%), Positives = 52/135 (38%), Gaps = 51/135 (37%)
Query: 40 GSGGGGKAV----AATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGG 95
G GGG A+ A V+ GG G G GGG GGGCGGG
Sbjct: 834 GGCGGGVAMESSKAGHVKSGGCGSGCGGGC-----------------------GGGCGGG 870
Query: 96 RAAVVGVAATSVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*G---- 151
V+ + V + + GCGGG GGG GGG G
Sbjct: 871 M-------------VMESSKAVHVKSGGCGGGC-------GGGCGGGVAMESSTVGHAKS 910
Query: 152 GGCGVGGGGDYGGGG 166
GGCG G GG GGGG
Sbjct: 911 GGCGSGCGGGCGGGG 925
Score = 35.4 bits (80), Expect = 1.1
Identities = 45/166 (27%), Positives = 50/166 (30%), Gaps = 69/166 (41%)
Query: 46 KAVAATVEGGGNGGGGGGGDRVVVVVSVVAIEWWRRLWWWWR*GGGCGGGRAAVVGVAAT 105
KAVAA+ GGG G G GGGCGGG A VV
Sbjct: 712 KAVAASGGGGGQAESAGCGSGC---------------------GGGCGGGVAKVVEATKA 750
Query: 106 SVVAVVAAMVVVVLATIGCGGGSKAVAA**GGGDGGGWWWWWWQ*GGGCGVGGGGDYGGG 165
G GG K+ GGCG G GG GGG
Sbjct: 751 -----------------GVGGHGKS---------------------GGCGSGCGGGCGGG 772
Query: 166 G-DKVVVAAIMVVVATPVVTVGRWWRRRRRR*SDGGGGGGGANSGV 210
G + V + V G GGG GGG G+
Sbjct: 773 GCGAMAVESSKDDVHAKCAGCGS---------GCGGGCGGGCGGGM 809
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.338 0.154 0.636
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 433,621,149
Number of Sequences: 2790947
Number of extensions: 23538664
Number of successful extensions: 533637
Number of sequences better than 10.0: 2198
Number of HSP's better than 10.0 without gapping: 814
Number of HSP's successfully gapped in prelim test: 1469
Number of HSP's that attempted gapping in prelim test: 343739
Number of HSP's gapped (non-prelim): 54556
length of query: 224
length of database: 848,049,833
effective HSP length: 123
effective length of query: 101
effective length of database: 504,763,352
effective search space: 50981098552
effective search space used: 50981098552
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0320b.3


