

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0311.7
(381 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
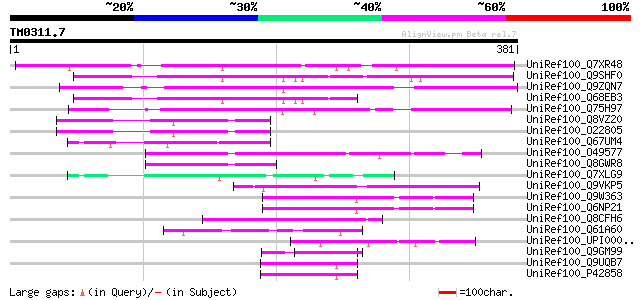
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XR48 OSJNBa0043A12.37 protein [Oryza sativa] 315 1e-84
UniRef100_Q9SHF0 T19E23.10 [Arabidopsis thaliana] 229 1e-58
UniRef100_Q9ZQN7 Hypothetical protein At2g35640 [Arabidopsis tha... 226 6e-58
UniRef100_Q68EB3 At1g31310 [Arabidopsis thaliana] 175 2e-42
UniRef100_Q75H97 Hypothetical protein OSJNBa0056E06.15 [Oryza sa... 169 9e-41
UniRef100_Q8VZ20 Hypothetical protein At2g33550 [Arabidopsis tha... 68 4e-10
UniRef100_O22805 Hypothetical protein At2g33550 [Arabidopsis tha... 68 4e-10
UniRef100_Q67UM4 Gt-2-related-like [Oryza sativa] 65 3e-09
UniRef100_O49577 Hypothetical protein AT4g31270 [Arabidopsis tha... 65 4e-09
UniRef100_Q8GWR8 Hypothetical protein At4g31270/F8F16_90 [Arabid... 63 1e-08
UniRef100_Q7XLG9 OSJNBa0039C07.7 protein [Oryza sativa] 63 2e-08
UniRef100_Q9VKP5 CG6700-PA [Drosophila melanogaster] 60 8e-08
UniRef100_Q9W363 CG12124-PA [Drosophila melanogaster] 58 5e-07
UniRef100_Q6NP21 LD32107p [Drosophila melanogaster] 58 5e-07
UniRef100_Q8CFH6 Salt inducible kinase 2 [Mus musculus] 57 1e-06
UniRef100_Q61A60 Hypothetical protein CBG13916 [Caenorhabditis b... 55 2e-06
UniRef100_UPI0000437276 UPI0000437276 UniRef100 entry 55 3e-06
UniRef100_Q9GM99 Huntingtin [Sus scrofa] 55 3e-06
UniRef100_Q9UQB7 Huntingtin [Homo sapiens] 55 3e-06
UniRef100_P42858 Huntingtin [Homo sapiens] 55 3e-06
>UniRef100_Q7XR48 OSJNBa0043A12.37 protein [Oryza sativa]
Length = 390
Score = 315 bits (808), Expect = 1e-84
Identities = 181/401 (45%), Positives = 233/401 (57%), Gaps = 54/401 (13%)
Query: 5 STTPIPPPPINIPDPHHHHH-HNHHLPLLHGTTTAPPLPS--SSSAPRDYRKGNWTIQET 61
ST P+ + H H H H P T APP PS S SAPRDYRKGNWT+ ET
Sbjct: 13 STAGAAVSPLALLRAHGHGHGHLTATPPSGATGPAPPPPSPASGSAPRDYRKGNWTLHET 72
Query: 62 LILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAASSSRTSGELRWKW 121
LILITA +LDD+RR +P TPR S E RWKW
Sbjct: 73 LILITANRLDDDRRAGVGGAAAGGGGAGSP----PTPR--------------SAEQRWKW 114
Query: 122 VENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESK--------SESKDHNYNFPSYWT 173
VENYCW +GCLRSQNQCNDKWDNLLRDYKKVRDYES+ + ++ PSYWT
Sbjct: 115 VENYCWKNGCLRSQNQCNDKWDNLLRDYKKVRDYESRVAAAAATGGAAAANSAPLPSYWT 174
Query: 174 LNKQQRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPP 233
+ + +RKD NLP+N+ E+Y A+S+VL R+ ++R T P PL + PPP
Sbjct: 175 MERHERKDCNLPTNLAPEVYDALSEVLSRRAARRGGATIAPTPPPP--PLALPLPPPPPP 232
Query: 234 QHPQPLQAQPL---------PPPPPPPPQ----PPATSITPAGSEYNHGDQCVKEKTESS 280
P+PL AQ PPPP PPP PPA P S V + E S
Sbjct: 233 SPPKPLVAQQQHHHHGHHHHPPPPQPPPSSLQLPPAVVAPPPAS--------VSAEEEMS 284
Query: 281 GSEQSDQEE--NNGSESSKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRHREMMELEQ 338
GS +S +EE + G +KRR++ +GSS++RSA+V+AR L +CEEK+E+RHRE+++LE+
Sbjct: 285 GSSESGEEEEGSGGEPEAKRRRLSRLGSSVVRSATVVARTLVACEEKRERRHRELLQLEE 344
Query: 339 RRIQMEENRNEVHRQGLATLVTAVSNLSGAIQSFINSQRHG 379
RR+++EE R EV RQG A L+ AV++LS AI + ++ R G
Sbjct: 345 RRLRLEEERTEVRRQGFAGLIAAVNSLSSAIHALVSDHRSG 385
Score = 39.3 bits (90), Expect = 0.18
Identities = 18/43 (41%), Positives = 20/43 (45%), Gaps = 8/43 (18%)
Query: 4 PSTTPIPPPPINIPDPHHHHHHNHHLPLLHGTTTAPPLPSSSS 46
P P PP P+ HHHH H+HH P PP P SS
Sbjct: 228 PPPPPSPPKPLVAQQQHHHHGHHHHPP--------PPQPPPSS 262
>UniRef100_Q9SHF0 T19E23.10 [Arabidopsis thaliana]
Length = 397
Score = 229 bits (583), Expect = 1e-58
Identities = 141/405 (34%), Positives = 205/405 (49%), Gaps = 96/405 (23%)
Query: 49 RDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAA 108
R+YRKGNWT+ ET++LI AK++DDERR++ S PPP
Sbjct: 12 REYRKGNWTLNETMVLIEAKRMDDERRMRRSIGLPPPEQQQDIR---------------- 55
Query: 109 SSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESK---------- 158
S ELRWKW+E+YCW GC+RSQNQCNDKWDNL+RDYKKVR+YE +
Sbjct: 56 --SNKPAELRWKWIEDYCWRKGCMRSQNQCNDKWDNLMRDYKKVREYERRRVESSITAGE 113
Query: 159 -SESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKN--SQRFITTTT-- 213
S S SYW + K +RK+ +LPSNM+ + YQA+ +V++ K S +T T
Sbjct: 114 SSSSSAPAGETASYWKMEKSERKERSLPSNMLPQTYQALFEVVESKTLPSSTAVTAVTAA 173
Query: 214 -------------------------------PQQQQ--QQQPLVTLVTSTPPPQHPQPLQ 240
P+ Q QQQP++ + PPP QPL
Sbjct: 174 VAAAAAAISSGNGSGGGQIQKVIQQGLGFVVPKVHQIIQQQPVLLPLQPPPPPPPSQPL- 232
Query: 241 AQPLPPPPPPPPQPPATSITPAGSEYNHGDQCVKEKTESSGSEQSDQEENNGSESSKRRK 300
+PL PPPPPP A I P + C+ + S SD E + + +KRR+
Sbjct: 233 PRPLLLPPPPPPSFHAQPILPTVVIW--FGLCLITWLKKDSSTDSDTSEYSDTSPAKRRR 290
Query: 301 ----------------VRNIGSS-----------IMRSASVLARALRSCEEKKEKRHREM 333
V +G S + RS SV+A A+R EE++++RH+E+
Sbjct: 291 TMPTTTTAGPSGGGVDVEEVGRSKRDEETTVAAALSRSVSVIANAIRESEERQDRRHKEV 350
Query: 334 MELEQRRIQMEENRNEVHRQGLATLVTAVSNLSGAIQSFINSQRH 378
M +++RR+++EE+ E++R+G+ LV A++ L+ +I + +S RH
Sbjct: 351 MNVQERRLKIEESNVEMNREGMNGLVEAINKLASSIFALASSSRH 395
>UniRef100_Q9ZQN7 Hypothetical protein At2g35640 [Arabidopsis thaliana]
Length = 340
Score = 226 bits (577), Expect = 6e-58
Identities = 127/369 (34%), Positives = 192/369 (51%), Gaps = 64/369 (17%)
Query: 38 APPLPSSSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTT 97
A P R+ RKGNWT+ ETL+LI AKK+DD+RR++ S P
Sbjct: 4 ADPSSGEQIVMRECRKGNWTVSETLVLIEAKKMDDQRRVRRSEKQPEG------------ 51
Query: 98 PRPSPAACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYE- 156
R PA ELRWKW+E YCW GC R+QNQCNDKWDNL+RDYKK+R+YE
Sbjct: 52 -RNKPA------------ELRWKWIEEYCWRRGCYRNQNQCNDKWDNLMRDYKKIREYER 98
Query: 157 SKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKN------------ 204
S+ ES + SYW ++K +RK+ NLPSNM+ +IY +S+++ RK
Sbjct: 99 SRVESSFNTVTSSSYWKMDKTERKEKNLPSNMLPQIYDVLSELVDRKTLPSSSSAAAAVG 158
Query: 205 ----------SQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQ-AQPLPPPPPPPPQ 253
Q+ + P Q + T + + PP PQ L + P PP PPP
Sbjct: 159 NGNGGQILRVCQQSLGFVAPMMAQPMHQIPTTIVLSLPPPPPQSLSLSLPSPPQPPPSSS 218
Query: 254 PPATSITP-AGSEYNHGDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRNIGSSIMRSA 312
A I P G+ + +T + G + +++ +G ++ R
Sbjct: 219 FHAEPIPPTVGTSSTKRRRTTPGETTAGGEREVEEDA--------------VGVALSRCT 264
Query: 313 SVLARALRSCEEKKEKRHREMMELEQRRIQMEENRNEVHRQGLATLVTAVSNLSGAIQSF 372
SV+ + +R EE +E+RH+E++ L++RR+++EE++ E++RQG+ LV A++ L+ +I +
Sbjct: 265 SVITQVIRENEEGQERRHKEVVRLQERRLKIEESKTEINRQGMNGLVDAINQLASSILAL 324
Query: 373 INSQRHGQR 381
+S H R
Sbjct: 325 ASSSCHNNR 333
>UniRef100_Q68EB3 At1g31310 [Arabidopsis thaliana]
Length = 256
Score = 175 bits (443), Expect = 2e-42
Identities = 102/261 (39%), Positives = 132/261 (50%), Gaps = 67/261 (25%)
Query: 49 RDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPAACSAA 108
R+YRKGNWT+ ET++LI AK++DDERR++ S PPP
Sbjct: 12 REYRKGNWTLNETMVLIEAKRMDDERRMRRSIGLPPPEQQQDIR---------------- 55
Query: 109 SSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESK---------- 158
S ELRWKW+E+YCW GC+RSQNQCNDKWDNL+RDYKKVR+YE +
Sbjct: 56 --SNKPAELRWKWIEDYCWRKGCMRSQNQCNDKWDNLMRDYKKVREYERRRVESSITAGE 113
Query: 159 -SESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKN--SQRFITTTT-- 213
S S SYW + K +RK+ +LPSNM+ + YQA+ +V++ K S +T T
Sbjct: 114 SSSSSAPAGETASYWKMEKSERKERSLPSNMLPQTYQALFEVVESKTLPSSTAVTAVTAA 173
Query: 214 -------------------------------PQQQQ--QQQPLVTLVTSTPPPQHPQPLQ 240
P+ Q QQQP++ + PPP QPL
Sbjct: 174 VAAAAAAISSGNGSGGGQIQKVIQQGLGFVVPKVHQIIQQQPVLLPLQPPPPPPPSQPL- 232
Query: 241 AQPLPPPPPPPPQPPATSITP 261
+PL PPPPPP A I P
Sbjct: 233 PRPLLLPPPPPPSFHAQPILP 253
>UniRef100_Q75H97 Hypothetical protein OSJNBa0056E06.15 [Oryza sativa]
Length = 345
Score = 169 bits (429), Expect = 9e-41
Identities = 115/360 (31%), Positives = 172/360 (46%), Gaps = 77/360 (21%)
Query: 45 SSAPRDYRKGNWTIQETLILITAKK-LDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPA 103
++A R+YR+GNWT+ ET++L+ AKK + D RR PA
Sbjct: 30 AAAAREYRRGNWTLPETMLLVEAKKRVSDGRR--------------------------PA 63
Query: 104 ACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYE-SKSESK 162
A G RW+WVE+YCW GC RSQNQCND+WDNL+RDYKKVR +E + +
Sbjct: 64 A--------DQGLARWRWVEDYCWRRGCRRSQNQCNDRWDNLMRDYKKVRAHELAAAGGG 115
Query: 163 DHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRK------------------N 204
SYW + + +RK+ LP+N++ EIY A+ +V++R+ +
Sbjct: 116 GGGGPAESYWVMGRTERKEKGLPANLLREIYDAMGEVVERRPMSSGGGGGGAVFLAGASS 175
Query: 205 SQRFITTTTPQQQQQQQPLVTLV------TSTPPPQHPQPLQAQP-LPPPPPPPPQPPAT 257
S P Q PL L+ T+ P+ + +P L PP PPAT
Sbjct: 176 SGSGGLADVPAMAMQASPLAQLLPRPLEATANCSSGSPERKRRRPSLDNEPPGGSTPPAT 235
Query: 258 SITPAGSEYNHGDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRNIGSSIMRSASVLAR 317
+ E++ D E E SD + +G +I R A++L+
Sbjct: 236 TGRQGHQEHDDDDD---EYAHHGADESSDDD-------------GGLGGAIGRCAAILSV 279
Query: 318 ALRSCEEKKEKRHREMMELEQRRIQMEENRNEVHRQGLATLVTAVSNLSGAIQSFINSQR 377
AL + E +E+RHRE++ E+RR + + R E Q +A L TAVS L+G++ + +R
Sbjct: 280 ALENREASEERRHREVVAAEERRGRARQARREAGEQCMAGLATAVSQLAGSMLALAAKRR 339
>UniRef100_Q8VZ20 Hypothetical protein At2g33550 [Arabidopsis thaliana]
Length = 314
Score = 68.2 bits (165), Expect = 4e-10
Identities = 43/163 (26%), Positives = 68/163 (41%), Gaps = 34/163 (20%)
Query: 36 TTAPPLPSSSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPS 95
++AP + R WT QE L+LI K++ + R +
Sbjct: 19 SSAPSNDGGDDGVKTARLPRWTRQEILVLIQGKRVAENRVRRG----------------- 61
Query: 96 TTPRPSPAACSAASSSRTSGELRWKW--VENYCWSHGCLRSQNQCNDKWDNLLRDYKKVR 153
AA + SG++ KW V +YC HG R QC +W NL DYKK++
Sbjct: 62 ----------RAAGMALGSGQMEPKWASVSSYCKRHGVNRGPVQCRKRWSNLAGDYKKIK 111
Query: 154 DYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAI 196
++ES+ + + SYW + R++ LP E+Y +
Sbjct: 112 EWESQIKEETE-----SYWVMRNDVRREKKLPGFFDKEVYDIV 149
>UniRef100_O22805 Hypothetical protein At2g33550 [Arabidopsis thaliana]
Length = 311
Score = 68.2 bits (165), Expect = 4e-10
Identities = 43/163 (26%), Positives = 68/163 (41%), Gaps = 34/163 (20%)
Query: 36 TTAPPLPSSSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPS 95
++AP + R WT QE L+LI K++ + R +
Sbjct: 19 SSAPSNDGGDDGVKTARLPRWTRQEILVLIQGKRVAENRVRRG----------------- 61
Query: 96 TTPRPSPAACSAASSSRTSGELRWKW--VENYCWSHGCLRSQNQCNDKWDNLLRDYKKVR 153
AA + SG++ KW V +YC HG R QC +W NL DYKK++
Sbjct: 62 ----------RAAGMALGSGQMEPKWASVSSYCKRHGVNRGPVQCRKRWSNLAGDYKKIK 111
Query: 154 DYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAI 196
++ES+ + + SYW + R++ LP E+Y +
Sbjct: 112 EWESQIKEETE-----SYWVMRNDVRREKKLPGFFDKEVYDIV 149
>UniRef100_Q67UM4 Gt-2-related-like [Oryza sativa]
Length = 374
Score = 65.1 bits (157), Expect = 3e-09
Identities = 47/166 (28%), Positives = 74/166 (44%), Gaps = 37/166 (22%)
Query: 44 SSSAPRDYRKGNWTIQETLILITAKKLDDER------RLKASSPPPPPTTTTTPHDPSTT 97
S APR R WT QE L+LI K++ + R R +A++
Sbjct: 20 SGRAPRLPR---WTRQEILVLIEGKRMVEGRGGGRGGRGRAAA----------------- 59
Query: 98 PRPSPAACSAASSSRTSGEL-------RWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYK 150
+ AA +AA++ + GE +W V YC HG R QC +W NL DYK
Sbjct: 60 ---AAAAAAAAAAGGSGGEAAVAALEPKWAAVAEYCRRHGVERGAVQCRKRWSNLAGDYK 116
Query: 151 KVRDYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAI 196
K++++E ++ + PS+W + R++ LP E+Y +
Sbjct: 117 KIKEWE-RAAAAAAPPREPSFWAMRNDARRERRLPGFFDREVYDIL 161
>UniRef100_O49577 Hypothetical protein AT4g31270 [Arabidopsis thaliana]
Length = 291
Score = 64.7 bits (156), Expect = 4e-09
Identities = 56/254 (22%), Positives = 108/254 (42%), Gaps = 21/254 (8%)
Query: 103 AACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESKSESK 162
AA A S+ S +W + C + R+ NQC KWD+L+ DY +++ +ES+
Sbjct: 32 AAVEADCSNALSSFQKWTMITENCNALDVSRNLNQCRRKWDSLMSDYNQIKKWESQYRGT 91
Query: 163 DHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQP 222
SYW+L+ +RK NLP ++ E+++AI+ V+ ++ + + + + Q
Sbjct: 92 GR-----SYWSLSSDKRKLLNLPGDIDIELFEAINAVVMIQDEKAGTESDSDPEAQDVVD 146
Query: 223 LVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNHGDQCVKEK--TESS 280
L + S Q ++ P Q T P ++ H ++ + EK E
Sbjct: 147 LSAELGSKRSRQRTMVMKETKKEEPRTSRVQ-VNTREKPITTKATHQNKTMGEKKPVEDM 205
Query: 281 GSEQSDQEENNGSESSKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRHREMMELEQRR 340
+++ + E N E + + + + I +++ R L E K+
Sbjct: 206 STDEEEDETMNIEEDVEVMEAK-LSYKIDLIHAIVGRNLAKDNETKD------------G 252
Query: 341 IQMEENRNEVHRQG 354
+ M++ V +QG
Sbjct: 253 VSMDDKLKSVRQQG 266
>UniRef100_Q8GWR8 Hypothetical protein At4g31270/F8F16_90 [Arabidopsis thaliana]
Length = 294
Score = 63.2 bits (152), Expect = 1e-08
Identities = 32/98 (32%), Positives = 54/98 (54%), Gaps = 5/98 (5%)
Query: 103 AACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYESKSESK 162
AA A S+ S +W + C + R+ NQC KWD+L+ DY +++ +ES+
Sbjct: 32 AAVEADCSNALSSFQKWTMITENCNALDVSRNLNQCRRKWDSLMSDYNQIKKWESQYRGT 91
Query: 163 DHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVL 200
SYW+L+ +RK NLP ++ E+++AI+ V+
Sbjct: 92 GR-----SYWSLSSDKRKLLNLPGDIDIELFEAINAVV 124
>UniRef100_Q7XLG9 OSJNBa0039C07.7 protein [Oryza sativa]
Length = 329
Score = 62.8 bits (151), Expect = 2e-08
Identities = 62/280 (22%), Positives = 95/280 (33%), Gaps = 77/280 (27%)
Query: 44 SSSAPRDYRKGNWTIQETLILITAKKLDDERRLKASSPPPPPTTTTTPHDPSTTPRPSPA 103
S APR R WT QE L+LI K++ + R
Sbjct: 8 SGRAPRLPR---WTRQEILVLIEGKRVVEGR--------------------------GRG 38
Query: 104 ACSAASSSRTSGELRWKWVENYCWSHGCLRSQNQCNDKWDNLLRDYKKVRDYE------S 157
+ +W V YC HG R QC +W NL DYKK+R++E S
Sbjct: 39 RGRGGGGGAAAEPTKWAAVAEYCRRHGLERGPVQCRKRWSNLAGDYKKIREWERSLSSPS 98
Query: 158 KSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQ 217
S + S+W + R++ LP E+Y D+L+ + ++
Sbjct: 99 SSSAAAGMGKEVSFWAMRNDARRERRLPGFFDREVY----DILEGRGGGNAAAAAAAGKE 154
Query: 218 QQQQPLVTLVT----------------------------STPPPQHPQPLQAQPLPPPPP 249
+++ + +TPP P A P PPP P
Sbjct: 155 GEEEKAAVFDSGRAAAGGGGGGGDDGLFSSSEEEEDDDEATPPATTP---AAAPAPPPAP 211
Query: 250 PPPQPPATSITPAGSEYNHGDQCVKEKTESSGSEQSDQEE 289
P P TS D ++ +E +G+ ++ Q E
Sbjct: 212 APAVPVLTS-------EKKSDPPRQDASEQAGTSRAKQPE 244
>UniRef100_Q9VKP5 CG6700-PA [Drosophila melanogaster]
Length = 874
Score = 60.5 bits (145), Expect = 8e-08
Identities = 42/190 (22%), Positives = 83/190 (43%), Gaps = 13/190 (6%)
Query: 169 PSYWTLNKQQRKDHNLPSNMV-----FEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPL 223
P + +QQ++ N P+ + + Y + Q++ Q + QQQQQQQ
Sbjct: 13 PQLQEMQQQQQQSAN-PAQLAQQWSNYAWYANQNAAYQQQYQQYYQYYVRYQQQQQQQVT 71
Query: 224 VTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNHGDQCVKEKTESSGSE 283
T+ TS PP L A P P PPPP PP P G +++ +
Sbjct: 72 STVSTSNAPPGFSNALAAPPPLPSGPPPPPPPQIGQPP-------GQNKQQQQQQQQQQL 124
Query: 284 QSDQEENNGSESSKRRKVRNIGSSIMRSASVLARALRSCEEKKEKRHREMMELEQRRIQM 343
Q Q++ + +++ I ++ + LA A +++++++H++ +Q+++
Sbjct: 125 QQQQQQQQQQQQQQQKNFGGIRFNLNLNQKRLAGAPNPLQQQQQQQHQQQQHQQQQQVMQ 184
Query: 344 EENRNEVHRQ 353
N N +++
Sbjct: 185 LYNNNNSNKK 194
>UniRef100_Q9W363 CG12124-PA [Drosophila melanogaster]
Length = 1837
Score = 57.8 bits (138), Expect = 5e-07
Identities = 45/161 (27%), Positives = 71/161 (43%), Gaps = 7/161 (4%)
Query: 191 EIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPP 250
E+ I + L Q T T+ +QQ + P + PPP P A P PPPPPP
Sbjct: 1302 EVRYPIRNWLTPSKEQAQETDTSGEQQTKPTPPKPVAAPVPPPPLPLTPPAAPPPPPPPP 1361
Query: 251 PPQPPATSI---TPAGSEYNHGDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRNIGSS 307
P +S+ T + S H Q + EK + ++ +Q + + RK N +
Sbjct: 1362 PEADDPSSVALPTASTSRQEHQRQQLNEKRQRVVAKFKEQPQ---PIKGQGRKTANKKLT 1418
Query: 308 IMRSASVLARALRSCEEKKEKRHREMMELEQRRIQMEENRN 348
MR + LA S + K ++H E ++ + + Q+E N N
Sbjct: 1419 AMRENAKLANP-SSQKSPKNRKHIEPLKTKAAKKQIEPNPN 1458
>UniRef100_Q6NP21 LD32107p [Drosophila melanogaster]
Length = 1777
Score = 57.8 bits (138), Expect = 5e-07
Identities = 45/161 (27%), Positives = 71/161 (43%), Gaps = 7/161 (4%)
Query: 191 EIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPP 250
E+ I + L Q T T+ +QQ + P + PPP P A P PPPPPP
Sbjct: 1242 EVRYPIRNWLTPSKEQAQETDTSGEQQTKPTPPKPVAAPVPPPPLPLTPPAAPPPPPPPP 1301
Query: 251 PPQPPATSI---TPAGSEYNHGDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRNIGSS 307
P +S+ T + S H Q + EK + ++ +Q + + RK N +
Sbjct: 1302 PEADDPSSVALPTASTSRQEHQRQQLNEKRQRVVAKFKEQPQ---PIKGQGRKTANKKLT 1358
Query: 308 IMRSASVLARALRSCEEKKEKRHREMMELEQRRIQMEENRN 348
MR + LA S + K ++H E ++ + + Q+E N N
Sbjct: 1359 AMRENAKLANP-SSQKSPKNRKHIEPLKTKAAKKQIEPNPN 1398
>UniRef100_Q8CFH6 Salt inducible kinase 2 [Mus musculus]
Length = 931
Score = 56.6 bits (135), Expect = 1e-06
Identities = 37/136 (27%), Positives = 59/136 (43%), Gaps = 2/136 (1%)
Query: 146 LRDYKKVRDYESKSESKDHNYNFPSYWTLNKQQRKDHNLPSNMVFEIYQAISDVLQRKNS 205
L+ +++ Y ++ + + +Y PS Q F + QA+S VL+ +
Sbjct: 724 LQKQSQLQAYFNQMQIAESSYPGPSQQLALPHQETPLTSQQPPSFSLTQALSPVLEPSSE 783
Query: 206 Q-RFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPPPPPQPPATSITPAGS 264
Q +F + + + Q QPL + PP P LQ PPPPPPPP P P
Sbjct: 784 QMQFSSFLSQYPEMQLQPLPSTPGPRAPPPLPSQLQQHQQPPPPPPPPPPQQPGAAPTSL 843
Query: 265 EYNHGDQCVKEKTESS 280
++++ C T SS
Sbjct: 844 QFSY-QTCELPSTTSS 858
>UniRef100_Q61A60 Hypothetical protein CBG13916 [Caenorhabditis briggsae]
Length = 1173
Score = 55.5 bits (132), Expect = 2e-06
Identities = 48/156 (30%), Positives = 65/156 (40%), Gaps = 24/156 (15%)
Query: 116 ELRWKWVENYCWSH--GC-LRSQNQCNDKWDNLLRDYKKVRDYESKSESKDHNYNFPSYW 172
E+ W+ E WS C S N + W L +Y K SKS FP
Sbjct: 208 EVCWQSFEGLTWSGLGDCQCTSSNSSDCHWIRLHTNYNKCIYEISKSG------RFPVLQ 261
Query: 173 TLNKQQRKDHNLPSNMVFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPP 232
TL ++ R+D M+ E YQ + +N Q+ + TTT T T+TP
Sbjct: 262 TLAQRTREDRENEQRMMSEQYQR-----RYENRQQNVPTTT-------STTTTTTTTTPR 309
Query: 233 PQHPQPLQAQPLPPP---PPPPPQPPATSITPAGSE 265
P +PPP PPPP PPAT++ P S+
Sbjct: 310 PTTTTTTTTTTIPPPTRSPPPPRAPPATTVPPWWSQ 345
>UniRef100_UPI0000437276 UPI0000437276 UniRef100 entry
Length = 571
Score = 55.1 bits (131), Expect = 3e-06
Identities = 48/163 (29%), Positives = 68/163 (41%), Gaps = 27/163 (16%)
Query: 212 TTPQQQQQQQPLVTLVTSTPP--PQHPQPLQAQPLPPPPPPPPQPPATSITPAGSEYNH- 268
TTP P L STPP P+HP P PL PP PPP+P S A E
Sbjct: 295 TTPAAPAHSNPPAVLPHSTPPTVPKHPTPAAPVPLKQPPMPPPKPVHHSSNTALQELEPR 354
Query: 269 ------GDQCVKEKTESSGSEQSDQEENNGSESSKRRKVRN---------------IGSS 307
D V E +GSE S++EE+ S+S + K R+ + S
Sbjct: 355 SRRGLVEDPQVAVIPEDAGSESSEEEEDE-SDSDQSIKYRDDNEEDDDEEDVPKSGLASR 413
Query: 308 IMRSASVLARALRSCEEKKEKRHREMMELEQRRIQMEENRNEV 350
+ R ++ + R +++KEK E + Q E RN++
Sbjct: 414 VKRKDTLALKLER--QQEKEKSQEEDSSTWNNKEQWEAVRNKI 454
>UniRef100_Q9GM99 Huntingtin [Sus scrofa]
Length = 3139
Score = 55.1 bits (131), Expect = 3e-06
Identities = 29/52 (55%), Positives = 30/52 (56%), Gaps = 1/52 (1%)
Query: 215 QQQQQQQPLVTLVTSTPPPQHPQPL-QAQPLPPPPPPPPQPPATSITPAGSE 265
QQQQQQQ PPPQ PQP Q QP P PPPPPP PP PA +E
Sbjct: 23 QQQQQQQQQQQQQLPPPPPQPPQPPPQTQPPPQPPPPPPPPPPPPPGPAVAE 74
Score = 52.0 bits (123), Expect = 3e-05
Identities = 29/72 (40%), Positives = 35/72 (48%), Gaps = 6/72 (8%)
Query: 190 FEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPLQAQPLPPPPP 249
FE ++ Q++ Q+ QQQQQQ P PPPQ P Q P PPPPP
Sbjct: 11 FESLKSFQQQQQQQQQQQ------QQQQQQQLPPPPPQPPQPPPQTQPPPQPPPPPPPPP 64
Query: 250 PPPQPPATSITP 261
PPP PA + P
Sbjct: 65 PPPPGPAVAEEP 76
>UniRef100_Q9UQB7 Huntingtin [Homo sapiens]
Length = 3144
Score = 55.1 bits (131), Expect = 3e-06
Identities = 32/80 (40%), Positives = 42/80 (52%), Gaps = 7/80 (8%)
Query: 189 VFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPL-QAQPL--- 244
+ + ++++ Q++ Q+ QQQQQQQ PPPQ PQP QAQPL
Sbjct: 7 LMKAFESLKSFQQQQQQQQQQQQQQQQQQQQQQQPPPPPPPPPPPQLPQPPPQAQPLLPQ 66
Query: 245 ---PPPPPPPPQPPATSITP 261
PPPPPPPP PA + P
Sbjct: 67 PQPPPPPPPPPPGPAVAEEP 86
>UniRef100_P42858 Huntingtin [Homo sapiens]
Length = 3144
Score = 55.1 bits (131), Expect = 3e-06
Identities = 32/80 (40%), Positives = 42/80 (52%), Gaps = 7/80 (8%)
Query: 189 VFEIYQAISDVLQRKNSQRFITTTTPQQQQQQQPLVTLVTSTPPPQHPQPL-QAQPL--- 244
+ + ++++ Q++ Q+ QQQQQQQ PPPQ PQP QAQPL
Sbjct: 7 LMKAFESLKSFQQQQQQQQQQQQQQQQQQQQQQQPPPPPPPPPPPQLPQPPPQAQPLLPQ 66
Query: 245 ---PPPPPPPPQPPATSITP 261
PPPPPPPP PA + P
Sbjct: 67 PQPPPPPPPPPPGPAVAEEP 86
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.126 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 745,934,763
Number of Sequences: 2790947
Number of extensions: 38091830
Number of successful extensions: 765215
Number of sequences better than 10.0: 7866
Number of HSP's better than 10.0 without gapping: 3499
Number of HSP's successfully gapped in prelim test: 4811
Number of HSP's that attempted gapping in prelim test: 541469
Number of HSP's gapped (non-prelim): 99805
length of query: 381
length of database: 848,049,833
effective HSP length: 129
effective length of query: 252
effective length of database: 488,017,670
effective search space: 122980452840
effective search space used: 122980452840
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0311.7


