

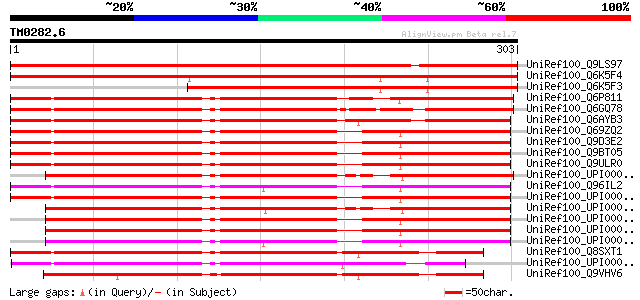
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0282.6
(303 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LS97 Dbj|BAA86474.1 [Arabidopsis thaliana] 491 e-137
UniRef100_Q6K5F4 2 coiled coil domains of eukaryotic origin (31.... 440 e-122
UniRef100_Q6K5F3 2 coiled coil domains of eukaryotic origin (31.... 271 2e-71
UniRef100_Q6P811 Hypothetical protein MGC76019 [Xenopus tropicalis] 268 2e-70
UniRef100_Q6GQ78 MGC80278 protein [Xenopus laevis] 266 4e-70
UniRef100_Q6AYB3 Hypothetical protein [Rattus norvegicus] 265 1e-69
UniRef100_Q69ZQ2 MKIAA1160 protein [Mus musculus] 264 2e-69
UniRef100_Q9D3E2 Mus musculus adult male thymus cDNA, RIKEN full... 263 6e-69
UniRef100_Q9BT05 Hypothetical protein KIAA1160 [Homo sapiens] 261 1e-68
UniRef100_Q9ULR0 KIAA1160 protein [Homo sapiens] 261 1e-68
UniRef100_UPI00003AA82B UPI00003AA82B UniRef100 entry 261 2e-68
UniRef100_Q96IL2 KIAA1160 protein [Homo sapiens] 251 1e-65
UniRef100_UPI0000457170 UPI0000457170 UniRef100 entry 250 3e-65
UniRef100_UPI00003AA82C UPI00003AA82C UniRef100 entry 249 8e-65
UniRef100_UPI000036B740 UPI000036B740 UniRef100 entry 247 3e-64
UniRef100_UPI000036B73E UPI000036B73E UniRef100 entry 247 3e-64
UniRef100_UPI000036B73F UPI000036B73F UniRef100 entry 237 3e-61
UniRef100_Q8SXT1 RE31520p [Drosophila melanogaster] 230 4e-59
UniRef100_UPI00002DA619 UPI00002DA619 UniRef100 entry 215 1e-54
UniRef100_Q9VHV6 CG9667-PA [Drosophila melanogaster] 213 7e-54
>UniRef100_Q9LS97 Dbj|BAA86474.1 [Arabidopsis thaliana]
Length = 300
Score = 491 bits (1264), Expect = e-137
Identities = 241/304 (79%), Positives = 270/304 (88%), Gaps = 5/304 (1%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKAQSMLNRFIT K EKKKPKERRP+LA+ECRDL+EADKWRQQI+REIG KVAE
Sbjct: 1 MARNEEKAQSMLNRFITQKESEKKKPKERRPYLASECRDLAEADKWRQQILREIGSKVAE 60
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQNEGLGEHRLRDLNDEINKL+RE+ HWERRIVELGG NY++HSAKMTDL+GNI+DVPNP
Sbjct: 61 IQNEGLGEHRLRDLNDEINKLLRERYHWERRIVELGGHNYSKHSAKMTDLEGNIIDVPNP 120
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
SGRGPGYRYFGAAKKLPGVRELF+KPPELRKR+TRYDIYKRIDA+YYGYRDDEDGILE+L
Sbjct: 121 SGRGPGYRYFGAAKKLPGVRELFEKPPELRKRKTRYDIYKRIDASYYGYRDDEDGILEKL 180
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKS-GEVAEVSAAVREILREEEEDVVEEERM 239
E +E MR+ + EEW RLD +R+EARKG V +AA RE+L EEEEDVVEEERM
Sbjct: 181 ERKSEGGMRKRSVEEWRRLDEVRKEARKGASEVVSVGAAAAAAREVLFEEEEDVVEEERM 240
Query: 240 RGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
+E+ EE EREFVVHVPLPDEKEIEKMVLEKKKM+LL+KY SEDL+++Q+EAK ML
Sbjct: 241 E----REKEEEKEREFVVHVPLPDEKEIEKMVLEKKKMDLLSKYASEDLVEQQTEAKSML 296
Query: 300 NIHR 303
NIHR
Sbjct: 297 NIHR 300
>UniRef100_Q6K5F4 2 coiled coil domains of eukaryotic origin (31.3 kD)-like protein
[Oryza sativa]
Length = 311
Score = 440 bits (1131), Expect = e-122
Identities = 221/311 (71%), Positives = 258/311 (82%), Gaps = 8/311 (2%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKAQSMLNRFITMK EEK+KP+ERRP+LA+ECRDL++A++WR +I+REIG KVAE
Sbjct: 1 MARNEEKAQSMLNRFITMKQEEKRKPRERRPYLASECRDLADAERWRSEILREIGAKVAE 60
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAK--MTDLDGNIVDVP 118
IQNEGLGEHRLRDLNDEINKL+RE+ HWERRIVELGG +++R S MTDLDGNIV +P
Sbjct: 61 IQNEGLGEHRLRDLNDEINKLLRERGHWERRIVELGGRDHSRSSNAPLMTDLDGNIVAIP 120
Query: 119 NPSGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILE 178
NPSGRGPGYRYFGAAKKLPGVRELFDKPPE+RKRRTRY+I+KRI+A YYGY DDEDG+LE
Sbjct: 121 NPSGRGPGYRYFGAAKKLPGVRELFDKPPEVRKRRTRYEIHKRINAGYYGYYDDEDGMLE 180
Query: 179 RLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSA----AVREILREEEEDVV 234
RLE AE MR E EW+R++R+R+EA KGV SGEVA A RE+L EE E+ V
Sbjct: 181 RLEAVAEKRMRNEVITEWHRVERVRREAMKGVVSGEVASAGGRGGEAAREVLFEEVEEEV 240
Query: 235 EEERMRGREMKERL--EENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQ 292
EEER E +ER EE +EF+ HVPLPDEKEIE+MVLE+KK ELL+KY S+ L EQ
Sbjct: 241 EEERRLEEEKREREKGEEAGKEFIAHVPLPDEKEIERMVLERKKKELLSKYTSDALQVEQ 300
Query: 293 SEAKDMLNIHR 303
EAK+MLN+ R
Sbjct: 301 EEAKEMLNVRR 311
>UniRef100_Q6K5F3 2 coiled coil domains of eukaryotic origin (31.3 kD)-like protein
[Oryza sativa]
Length = 203
Score = 271 bits (693), Expect = 2e-71
Identities = 138/203 (67%), Positives = 160/203 (77%), Gaps = 6/203 (2%)
Query: 107 MTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAY 166
MTDLDGNIV +PNPSGRGPGYRYFGAAKKLPGVRELFDKPPE+RKRRTRY+I+KRI+A Y
Sbjct: 1 MTDLDGNIVAIPNPSGRGPGYRYFGAAKKLPGVRELFDKPPEVRKRRTRYEIHKRINAGY 60
Query: 167 YGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSA----AV 222
YGY DDEDG+LERLE AE MR E EW+R++R+R+EA KGV SGEVA A
Sbjct: 61 YGYYDDEDGMLERLEAVAEKRMRNEVITEWHRVERVRREAMKGVVSGEVASAGGRGGEAA 120
Query: 223 REILREEEEDVVEEERMRGREMKERL--EENEREFVVHVPLPDEKEIEKMVLEKKKMELL 280
RE+L EE E+ VEEER E +ER EE +EF+ HVPLPDEKEIE+MVLE+KK ELL
Sbjct: 121 REVLFEEVEEEVEEERRLEEEKREREKGEEAGKEFIAHVPLPDEKEIERMVLERKKKELL 180
Query: 281 NKYVSEDLMDEQSEAKDMLNIHR 303
+KY S+ L EQ EAK+MLN+ R
Sbjct: 181 SKYTSDALQVEQEEAKEMLNVRR 203
>UniRef100_Q6P811 Hypothetical protein MGC76019 [Xenopus tropicalis]
Length = 282
Score = 268 bits (684), Expect = 2e-70
Identities = 154/305 (50%), Positives = 195/305 (63%), Gaps = 27/305 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECNELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKLIREK HWE RI ELGGP+Y R KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLIREKGHWEVRIKELGGPDYGRIGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K IDA YYGYRD++DG+L L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKSIDAEYYGYRDEDDGVLVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEE----DVVEE 236
E E EA E+W RQE + +G+ E EEEE V +
Sbjct: 174 EQEEEKKAIGEALEKW------RQEKEARLANGDRNE----------EEEEVNIYAVAAD 217
Query: 237 ERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAK 296
E + EE +++F+ HVP+P +KEIE+ ++ +KKMELL KY SE L+ + +AK
Sbjct: 218 ESGDDSDSGAEGEEGQQKFIAHVPVPTQKEIEEALIRRKKMELLQKYASETLLAQSEDAK 277
Query: 297 DMLNI 301
+L I
Sbjct: 278 RLLGI 282
>UniRef100_Q6GQ78 MGC80278 protein [Xenopus laevis]
Length = 284
Score = 266 bits (681), Expect = 4e-70
Identities = 150/302 (49%), Positives = 201/302 (65%), Gaps = 19/302 (6%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEDGKV-KERRPFLASECSELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE ++RD+NDEINKLIREK HWE RI ELGGP+Y R KM D +G V P
Sbjct: 60 IQNAGLGEFKIRDVNDEINKLIREKGHWEVRIKELGGPDYGRIGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K IDA YYGYRD++DG+L L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRTELMKSIDAEYYGYRDEDDGVLVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEE-EDVVEEERM 239
E E EA E+W RL++ + G K+ E EV+ + + +E ED ++E
Sbjct: 174 EQEQEKKAIAEALEKW-RLEK-EERLANGDKNEEEEEVN--IYAVAADESGEDSDDDEGA 229
Query: 240 RGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
G EE +++F+ HVP+P +KE+E+ ++ +KKMELL KY SE L+ + +AK +L
Sbjct: 230 EG-------EEGQQKFIAHVPVPSQKEMEEALVRRKKMELLQKYASETLLAQSEDAKRLL 282
Query: 300 NI 301
+
Sbjct: 283 GV 284
>UniRef100_Q6AYB3 Hypothetical protein [Rattus norvegicus]
Length = 284
Score = 265 bits (676), Expect = 1e-69
Identities = 146/301 (48%), Positives = 195/301 (64%), Gaps = 21/301 (6%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEAR--KGVKSGEVAEVSAAVREILREEEEDVVEEER 238
E E +R E E+W + +EAR +G K E E + + EE ++ +E+
Sbjct: 174 EQEYEKRLRAELVEKW----KAEREARLARGEKEEEEEEEEINIYAVTEEESDEEGSQEK 229
Query: 239 MRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDM 298
E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EAK +
Sbjct: 230 AG--------EDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEAKRL 281
Query: 299 L 299
L
Sbjct: 282 L 282
>UniRef100_Q69ZQ2 MKIAA1160 protein [Mus musculus]
Length = 300
Score = 264 bits (674), Expect = 2e-69
Identities = 147/304 (48%), Positives = 194/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 16 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 74
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 75 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 130
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 131 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 188
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 189 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 234
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 235 EESDEEGNQEKAGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 294
Query: 296 KDML 299
K +L
Sbjct: 295 KRLL 298
>UniRef100_Q9D3E2 Mus musculus adult male thymus cDNA, RIKEN full-length enriched
library, clone:5830446M03 product:hypothetical protein,
full insert sequence [Mus musculus]
Length = 285
Score = 263 bits (671), Expect = 6e-69
Identities = 146/304 (48%), Positives = 194/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGV+ELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVKELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGNQEKAGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
K +L
Sbjct: 280 KRLL 283
>UniRef100_Q9BT05 Hypothetical protein KIAA1160 [Homo sapiens]
Length = 285
Score = 261 bits (668), Expect = 1e-68
Identities = 145/304 (47%), Positives = 195/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
+ +L
Sbjct: 280 RRLL 283
>UniRef100_Q9ULR0 KIAA1160 protein [Homo sapiens]
Length = 351
Score = 261 bits (668), Expect = 1e-68
Identities = 145/304 (47%), Positives = 195/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 21 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 79
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 80 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 135
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 136 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 193
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 194 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 239
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 240 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 299
Query: 296 KDML 299
+ +L
Sbjct: 300 RRLL 303
>UniRef100_UPI00003AA82B UPI00003AA82B UniRef100 entry
Length = 271
Score = 261 bits (666), Expect = 2e-68
Identities = 143/282 (50%), Positives = 191/282 (67%), Gaps = 24/282 (8%)
Query: 22 EKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKL 81
E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+IQN GLGE R+RDLNDEINKL
Sbjct: 12 EEGKVKERRPFLASECNELPKAEKWRRQIIGEISKKVAQIQNAGLGEFRIRDLNDEINKL 71
Query: 82 IREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRE 141
+REK HWE RI ELGGP+Y R +M D +G V P R GY+YFGAAK LPGVRE
Sbjct: 72 LREKGHWEVRIRELGGPDYARIGPRMLDHEGKEV----PGNR--GYKYFGAAKDLPGVRE 125
Query: 142 LFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDR 201
LF+K P R+TR ++ K IDA YYGYRD++DGILE LE E + EA E+W +
Sbjct: 126 LFEKEPLPPPRKTRAELMKSIDAEYYGYRDEDDGILEPLEQEHEKKVIAEAVEKW----K 181
Query: 202 IRQEARKGVKSGEVAEVSAAVREILREEEEDV--VEEERMRGREMKERLEENEREFVVHV 259
+ +EAR + GE E EEEE++ V++E ++ E+ +++F+ HV
Sbjct: 182 MEREAR--LARGEEEE----------EEEENIYAVDDESDEEGGKEKEGEDGQQKFIAHV 229
Query: 260 PLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDMLNI 301
P+P ++EIE+ ++ +KKMELL KY SE LM + EAK +L +
Sbjct: 230 PVPSQQEIEEALVRRKKMELLQKYASETLMAQSEEAKRLLGL 271
>UniRef100_Q96IL2 KIAA1160 protein [Homo sapiens]
Length = 307
Score = 251 bits (642), Expect = 1e-65
Identities = 145/326 (44%), Positives = 197/326 (59%), Gaps = 48/326 (14%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELR----------------------KRRTRYDI 158
R GY+YFGAAK LPGVRELF+K ++R R+TR ++
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKERQVRWLMPVIPALWEAEAGGSQALPPPRKTRAEL 173
Query: 159 YKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEV 218
K ID YYGY D++DG++ LE E +R E E+W K+ A +
Sbjct: 174 MKAIDFEYYGYLDEDDGVIVPLEQEYEKKLRAELVEKW--------------KAEREARL 219
Query: 219 SAAVREILREEEED-----VVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLE 273
+ +E EEEE+ V EEE ++ ++++++F+ HVP+P ++EIE+ ++
Sbjct: 220 ARGEKEEEEEEEEEINIYAVTEEESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVR 279
Query: 274 KKKMELLNKYVSEDLMDEQSEAKDML 299
+KKMELL KY SE L + EA+ +L
Sbjct: 280 RKKMELLQKYASETLQAQSEEARRLL 305
>UniRef100_UPI0000457170 UPI0000457170 UniRef100 entry
Length = 351
Score = 250 bits (639), Expect = 3e-65
Identities = 140/304 (46%), Positives = 192/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
M + E ++ L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 21 MTCSAEGFKTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 79
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 80 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 135
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 136 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 193
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 194 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 239
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 240 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 299
Query: 296 KDML 299
+ +L
Sbjct: 300 RRLL 303
>UniRef100_UPI00003AA82C UPI00003AA82C UniRef100 entry
Length = 289
Score = 249 bits (635), Expect = 8e-65
Identities = 143/299 (47%), Positives = 190/299 (62%), Gaps = 43/299 (14%)
Query: 22 EKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKL 81
E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+IQN GLGE R+RDLNDEINKL
Sbjct: 12 EEGKVKERRPFLASECNELPKAEKWRRQIIGEISKKVAQIQNAGLGEFRIRDLNDEINKL 71
Query: 82 IREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRE 141
+REK HWE RI ELGGP+Y R +M D +G V P R GY+YFGAAK LPGVRE
Sbjct: 72 LREKGHWEVRIRELGGPDYARIGPRMLDHEGKEV----PGNR--GYKYFGAAKDLPGVRE 125
Query: 142 LFDKPPELRK-------------------RRTRYDIYKRIDAAYYGYRDDEDGILERLEP 182
LF+K K R+TR ++ K IDA YYGYRD++DGILE LE
Sbjct: 126 LFEKEQMFCKLSKVDLLERWSSSSALPPPRKTRAELMKSIDAEYYGYRDEDDGILEPLEQ 185
Query: 183 SAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDV--VEEERMR 240
E + EA E+W ++ +EAR + GE E EEEE++ V++E
Sbjct: 186 EHEKKVIAEAVEKW----KMEREAR--LARGEEEE----------EEEENIYAVDDESDE 229
Query: 241 GREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE LM + EAK +L
Sbjct: 230 EGGKEKEGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLMAQSEEAKRLL 288
>UniRef100_UPI000036B740 UPI000036B740 UniRef100 entry
Length = 302
Score = 247 bits (630), Expect = 3e-64
Identities = 134/283 (47%), Positives = 182/283 (63%), Gaps = 25/283 (8%)
Query: 22 EKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKL 81
E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+IQN GLGE R+RDLNDEINKL
Sbjct: 39 EEGKVKERRPFLASECTELPKAEKWRRQIIGEISKKVAQIQNAGLGEFRIRDLNDEINKL 98
Query: 82 IREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRE 141
+REK HWE RI ELGGP+Y + KM D +G V P R GY+YFGAAK LPGVRE
Sbjct: 99 LREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----PGNR--GYKYFGAAKDLPGVRE 152
Query: 142 LFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDR 201
LF+K P R+TR ++ K ID YYGY D++DG++ LE E R E E+W
Sbjct: 153 LFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPLEQEYEKKRRVELVEKW----- 207
Query: 202 IRQEARKGVKSGEVAEVSAAVREILREEEED-----VVEEERMRGREMKERLEENEREFV 256
K+ A ++ +E EEEE+ V EEE ++ ++++++F+
Sbjct: 208 ---------KAEREARLARGEKEEEEEEEEEINIYAVTEEESDEEGSQEKGGDDSQQKFI 258
Query: 257 VHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
HVP+P ++EIE+ ++ +KKMELL KY SE L + EA+ +L
Sbjct: 259 AHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEARRLL 301
>UniRef100_UPI000036B73E UPI000036B73E UniRef100 entry
Length = 277
Score = 247 bits (630), Expect = 3e-64
Identities = 134/283 (47%), Positives = 182/283 (63%), Gaps = 25/283 (8%)
Query: 22 EKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKL 81
E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+IQN GLGE R+RDLNDEINKL
Sbjct: 13 EEGKVKERRPFLASECTELPKAEKWRRQIIGEISKKVAQIQNAGLGEFRIRDLNDEINKL 72
Query: 82 IREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRE 141
+REK HWE RI ELGGP+Y + KM D +G V P R GY+YFGAAK LPGVRE
Sbjct: 73 LREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----PGNR--GYKYFGAAKDLPGVRE 126
Query: 142 LFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDR 201
LF+K P R+TR ++ K ID YYGY D++DG++ LE E R E E+W
Sbjct: 127 LFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPLEQEYEKKRRVELVEKW----- 181
Query: 202 IRQEARKGVKSGEVAEVSAAVREILREEEED-----VVEEERMRGREMKERLEENEREFV 256
K+ A ++ +E EEEE+ V EEE ++ ++++++F+
Sbjct: 182 ---------KAEREARLARGEKEEEEEEEEEINIYAVTEEESDEEGSQEKGGDDSQQKFI 232
Query: 257 VHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
HVP+P ++EIE+ ++ +KKMELL KY SE L + EA+ +L
Sbjct: 233 AHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEARRLL 275
>UniRef100_UPI000036B73F UPI000036B73F UniRef100 entry
Length = 298
Score = 237 bits (604), Expect = 3e-61
Identities = 134/305 (43%), Positives = 184/305 (59%), Gaps = 47/305 (15%)
Query: 22 EKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKL 81
E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+IQN GLGE R+RDLNDEINKL
Sbjct: 12 EEGKVKERRPFLASECTELPKAEKWRRQIIGEISKKVAQIQNAGLGEFRIRDLNDEINKL 71
Query: 82 IREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRE 141
+REK HWE RI ELGGP+Y + KM D +G V P R GY+YFGAAK LPGVRE
Sbjct: 72 LREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----PGNR--GYKYFGAAKDLPGVRE 125
Query: 142 LFDKPPELR----------------------KRRTRYDIYKRIDAAYYGYRDDEDGILER 179
LF+K ++R R+TR ++ K ID YYGY D++DG++
Sbjct: 126 LFEKERQVRWLMPVIPALWEAEAGGSQALPPPRKTRAELMKAIDFEYYGYLDEDDGVIVP 185
Query: 180 LEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VV 234
LE E R E E+W K+ A ++ +E EEEE+ V
Sbjct: 186 LEQEYEKKRRVELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVT 231
Query: 235 EEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSE 294
EEE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + E
Sbjct: 232 EEESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEE 291
Query: 295 AKDML 299
A+ +L
Sbjct: 292 ARRLL 296
>UniRef100_Q8SXT1 RE31520p [Drosophila melanogaster]
Length = 272
Score = 230 bits (586), Expect = 4e-59
Identities = 128/285 (44%), Positives = 179/285 (61%), Gaps = 20/285 (7%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L R+ K E + KERRP+LA+EC DL +K+R +I+R+I +KVA+
Sbjct: 1 MARNAEKAMTTLARWRAAKEVESGE-KERRPYLASECHDLPRCEKFRLEIIRDISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE +I LGGP+Y R+ KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKRHWENQISSLGGPHYRRYGPKMFDAEGREV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF++ P R++R ++ K IDA YYGYRDDEDG+L L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEQDPPPPPRKSRAELMKDIDAEYYGYRDDEDGVLIPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEAR--KGVKSGEVAEVSAAVREILREEEEDVVEEER 238
E E A ++A +EW +++ ++ R + E+ +S RE + + + E+
Sbjct: 174 EERIERAAIQKAVQEW--REKMARDGRIIDDEEEDEIYPLSGPAREAIEDAKARAAVEDP 231
Query: 239 MRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKY 283
K F HVP+P ++++++ +L ++K ELL KY
Sbjct: 232 HGLLASK---------FTAHVPVPTQQDVQEALLRQRKRELLEKY 267
>UniRef100_UPI00002DA619 UPI00002DA619 UniRef100 entry
Length = 366
Score = 215 bits (548), Expect = 1e-54
Identities = 124/280 (44%), Positives = 166/280 (59%), Gaps = 27/280 (9%)
Query: 2 ARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEI 61
ARN EKA + L RF + EE K KERRPFLA+EC DL +A+KWR+QI+ EI +KVA+I
Sbjct: 31 ARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECSDLPKAEKWRRQIISEISKKVAQI 89
Query: 62 QNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPS 121
QN GLGE +RDLNDEINKL+REK HWE RI ELG M D +G V P
Sbjct: 90 QNAGLGEFTIRDLNDEINKLLREKGHWEFRISELGRTQLPVFGPMMLDHEGKEV----PG 145
Query: 122 GRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLE 181
R GY+YFGAA+ LPGVRELF+K P R+TR ++ K +DA YYGYRD++DG+L LE
Sbjct: 146 NR--GYKYFGAARDLPGVRELFEKEPAPAPRKTRAELMKDVDAEYYGYRDEDDGVLLPLE 203
Query: 182 PSAEAAMRREAAEEWY---------RLDRIRQEARKGVKSGEVAEVSAAVREILREEEED 232
E EA ++W + + +Q+ ++ + E + + E+L +EE
Sbjct: 204 AQYEKQAVTEAVQKWRTEASHLSGDKPQQQQQQQQQQEEEEEEESIYSVHSEVLEDEESG 263
Query: 233 VVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVL 272
+E+ EE F+ HVP+P +KE+ + L
Sbjct: 264 FEQED-----------EEGGVGFIAHVPVPSQKEVGALTL 292
>UniRef100_Q9VHV6 CG9667-PA [Drosophila melanogaster]
Length = 274
Score = 213 bits (541), Expect = 7e-54
Identities = 118/269 (43%), Positives = 167/269 (61%), Gaps = 23/269 (8%)
Query: 21 EEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQN----EGLGEHRLRDLND 76
E + KERRP+LA+EC DL +K+R +I+R+I +KVA+IQN GLGE R+RDLND
Sbjct: 18 EVESGEKERRPYLASECHDLPRCEKFRLEIIRDISKKVAQIQNGTYFAGLGEFRIRDLND 77
Query: 77 EINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKL 136
EINKL+REK HWE +I LGGP+Y R+ KM D +G V P RG Y+YFGAAK L
Sbjct: 78 EINKLLREKRHWENQISSLGGPHYRRYGPKMFDAEGREV----PGNRG--YKYFGAAKDL 131
Query: 137 PGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEW 196
PGVRELF++ P R++R ++ K IDA YYGYRDDEDG+L LE E A ++A +EW
Sbjct: 132 PGVRELFEQDPPPPPRKSRAELMKDIDAEYYGYRDDEDGVLIPLEERIERAAIQKAVQEW 191
Query: 197 YRLDRIRQEAR--KGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENERE 254
+++ ++ R + E+ +S RE + + + E+ K
Sbjct: 192 --REKMARDGRIIDDEEEDEIYPLSGPAREAIEDAKARAAVEDPHGLLASK--------- 240
Query: 255 FVVHVPLPDEKEIEKMVLEKKKMELLNKY 283
F HVP+P ++++++ +L ++K ELL KY
Sbjct: 241 FTAHVPVPTQQDVQEALLRQRKRELLEKY 269
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,282,819
Number of Sequences: 2790947
Number of extensions: 23778204
Number of successful extensions: 130941
Number of sequences better than 10.0: 2153
Number of HSP's better than 10.0 without gapping: 280
Number of HSP's successfully gapped in prelim test: 2004
Number of HSP's that attempted gapping in prelim test: 112795
Number of HSP's gapped (non-prelim): 9283
length of query: 303
length of database: 848,049,833
effective HSP length: 127
effective length of query: 176
effective length of database: 493,599,564
effective search space: 86873523264
effective search space used: 86873523264
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0282.6


