

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
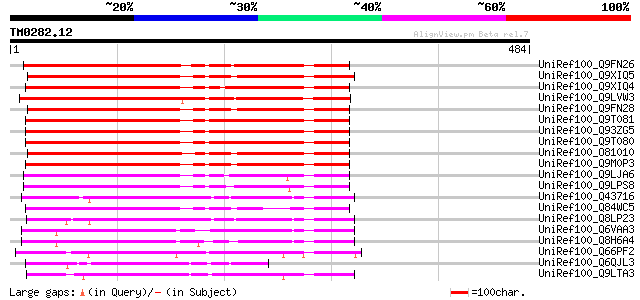
Query= TM0282.12
(484 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FN26 Flavonol 3-O-glucosyltransferase-like protein [... 291 4e-77
UniRef100_Q9XIQ5 Similar to Flavonol 3-O-Glucosyltransferase [Ar... 287 4e-76
UniRef100_Q9XIQ4 Similar to Flavonol 3-O-Glucosyltransferase [Ar... 286 1e-75
UniRef100_Q9LVW3 Flavonol 3-O-glucosyltransferase-like [Arabidop... 285 2e-75
UniRef100_Q9FN28 Putative flavonol 3-O-glucosyltransferase [Arab... 284 3e-75
UniRef100_Q9T081 UDP rhamnose--anthocyanidin-3-glucoside rhamnos... 280 7e-74
UniRef100_Q93ZG5 AT4g27560/T29A15_50 [Arabidopsis thaliana] 277 4e-73
UniRef100_Q9T080 Putative UDP rhamnose-anthocyanidin-3-glucoside... 277 4e-73
UniRef100_O81010 Putative flavonol 3-O-glucosyltransferase [Arab... 266 8e-70
UniRef100_Q9M0P3 Hypothetical protein AT4g09500 [Arabidopsis tha... 266 8e-70
UniRef100_Q9LJA6 UDP-glycose: flavonoid glucosyltransferase-like... 254 3e-66
UniRef100_Q9LPS8 Putative glucosyl transferase [Arabidopsis thal... 242 2e-62
UniRef100_Q43716 Flavonol 3-O-glucosyltransferase [Petunia hybrida] 219 1e-55
UniRef100_Q84WC5 Hypothetical protein At4g09500 [Arabidopsis tha... 218 2e-55
UniRef100_Q8LP23 Rhamnosyl transferase [Nierembergia sp. NB17] 208 2e-52
UniRef100_Q6VAA3 UDP-glycosyltransferase 79A2 [Stevia rebaudiana] 155 3e-36
UniRef100_Q8H6A4 UDP-glucosyltransferase [Stevia rebaudiana] 153 9e-36
UniRef100_Q66PF2 Putative UDP-rhamnose:rhamnosyltransferase [Fra... 145 2e-33
UniRef100_Q6QJL3 Glucosyl-transferase [Fagopyrum esculentum] 139 2e-31
UniRef100_Q9LTA3 Putative anthocyanidin-3-glucoside rhamnosyltra... 123 1e-26
>UniRef100_Q9FN26 Flavonol 3-O-glucosyltransferase-like protein [Arabidopsis
thaliana]
Length = 453
Score = 291 bits (744), Expect = 4e-77
Identities = 144/304 (47%), Positives = 199/304 (65%), Gaps = 20/304 (6%)
Query: 14 SKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHII 73
SK H MFPWF GHMT ++HLAN+LA++ HKITFLLP KAR L LN +P+ I F +
Sbjct: 3 SKFHAFMFPWFGFGHMTAFLHLANKLAEKDHKITFLLPKKARKQLESLNLFPDCIVFQTL 62
Query: 74 TVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEA 133
T+P+VDGLP G ET S+IP+SL L AMD+TR QV++ + KP + FD A+W+PE
Sbjct: 63 TIPSVDGLPDGAETTSDIPISLGSFLASAMDRTRIQVKEAVSVGKPDLIFFDFAHWIPEI 122
Query: 134 ARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRY 193
AR+ G+K++ + +SA +A+SFVP + + +D+ PPGYPS SK++LR +
Sbjct: 123 AREYGVKSVNFITISAACVAISFVPGR--------SQDDLGSTPPGYPS---SKVLLRGH 171
Query: 194 EAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLT 253
E F S PFG+ G + Y+R GL+ CD I+I+T E+EG FCD+I Q+ + VLLT
Sbjct: 172 ETNSLSFLSYPFGD-GTSFYERIMIGLKNCDVISIRTCQEMEGKFCDFIENQFQRKVLLT 230
Query: 254 GPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGL 313
GP+L E + LE++W +WL G V+YCA GSQ+IL K QFQE+ LG+
Sbjct: 231 GPMLPEPDNSKPLEDQWRQWLS--------KFDPGSVIYCALGSQIILEKDQFQELCLGM 282
Query: 314 EISG 317
E++G
Sbjct: 283 ELTG 286
>UniRef100_Q9XIQ5 Similar to Flavonol 3-O-Glucosyltransferase [Arabidopsis thaliana]
Length = 447
Score = 287 bits (735), Expect = 4e-76
Identities = 145/305 (47%), Positives = 198/305 (64%), Gaps = 26/305 (8%)
Query: 17 HIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIITVP 76
H MFPWFA GHMTPY+HLAN+LA+RGH+ITFL+P KA+ L HLN +P+ I FH +T+P
Sbjct: 6 HAFMFPWFAFGHMTPYLHLANKLAERGHRITFLIPKKAQKQLEHLNLFPDSIVFHSLTIP 65
Query: 77 NVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAARK 136
+VDGLP G ET S+IP+ L L A+D TR+QVE + + P +LFD+A WVPE A++
Sbjct: 66 HVDGLPAGAETFSDIPMPLWKFLPPAIDLTRDQVEAAVSALSPDLILFDIASWVPEVAKE 125
Query: 137 LGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYEAE 196
+K++ Y+++SA S+A FVP ++ PPPGYPS SKL+ R+++A
Sbjct: 126 YRVKSMLYNIISATSIAHDFVPG-----------GELGVPPPGYPS---SKLLYRKHDAH 171
Query: 197 QQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTGPV 256
+ S+ + R TGL CD I+I+T E+EG FC+Y+ QYHK V LTGP+
Sbjct: 172 ALLSFSVYYKR----FSHRLITGLMNCDFISIRTCKEIEGKFCEYLERQYHKKVFLTGPM 227
Query: 257 LSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLEIS 316
L E +G+ LE+RW+ WL+ G G VV+CA GSQV L K QFQE+ LG+E++
Sbjct: 228 LPEPNKGKPLEDRWSHWLN--------GFEQGSVVFCALGSQVTLEKDQFQELCLGIELT 279
Query: 317 GFSIF 321
G F
Sbjct: 280 GLPFF 284
>UniRef100_Q9XIQ4 Similar to Flavonol 3-O-Glucosyltransferase [Arabidopsis thaliana]
Length = 452
Score = 286 bits (731), Expect = 1e-75
Identities = 146/303 (48%), Positives = 197/303 (64%), Gaps = 26/303 (8%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIIT 74
K+H MFPWFA GHMTPY+HL N+LA++GH++TFLLP KA+ L H N +P+ I FH +
Sbjct: 4 KIHAFMFPWFAFGHMTPYLHLGNKLAEKGHRVTFLLPKKAQKQLEHQNLFPHGIVFHPLV 63
Query: 75 VPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAA 134
+P+VDGLP G ETAS+IP+SL L +AMD TR+Q+E + + +P +LFD+A+WVPE A
Sbjct: 64 IPHVDGLPAGAETASDIPISLVKFLSIAMDLTRDQIEAAIGALRPDLILFDLAHWVPEMA 123
Query: 135 RKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYE 194
+ L +K++ Y+V+SA S+A VP ++ PPGYPS SK + R ++
Sbjct: 124 KALKVKSMLYNVMSATSIAHDLVPG-----------GELGVAPPGYPS---SKALYREHD 169
Query: 195 AEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTG 254
A A + F Y R TTGL CD I+I+T E+EG FCDYI QY K VLLTG
Sbjct: 170 AH----ALLTFSGFYKRFYHRFTTGLMNCDFISIRTCEEIEGKFCDYIESQYKKKVLLTG 225
Query: 255 PVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLE 314
P+L E + + LE++W+ WL G G G VV+CA GSQ IL K QFQE+ LG+E
Sbjct: 226 PMLPEPDKSKPLEDQWSHWLS--------GFGQGSVVFCALGSQTILEKNQFQELCLGIE 277
Query: 315 ISG 317
++G
Sbjct: 278 LTG 280
>UniRef100_Q9LVW3 Flavonol 3-O-glucosyltransferase-like [Arabidopsis thaliana]
Length = 468
Score = 285 bits (730), Expect = 2e-75
Identities = 150/313 (47%), Positives = 207/313 (65%), Gaps = 16/313 (5%)
Query: 10 SSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLIT 69
S+ +S + I M+PW A GHMTP++HL+N+LA++GHKI FLLP KA L LN YPNLIT
Sbjct: 6 SNESSSMSIVMYPWLAFGHMTPFLHLSNKLAEKGHKIVFLLPKKALNQLEPLNLYPNLIT 65
Query: 70 FHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYW 129
FH I++P V GLP G ET S++P L HLL VAMD+TR +VE + ++ KP V +D A+W
Sbjct: 66 FHTISIPQVKGLPPGAETNSDVPFFLTHLLAVAMDQTRPEVETIFRTIKPDLVFYDSAHW 125
Query: 130 VPEAARKLGIKTICYHVVSAMSMAMSFVPA--KIIPKDRPLTVEDMSQPPPGYPSSSNSK 187
+PE A+ +G KT+C+++VSA S+A+S VP+ + + + ++ E++++ P GYPS SK
Sbjct: 126 IPEIAKPIGAKTVCFNIVSAASIALSLVPSAEREVIDGKEMSGEELAKTPLGYPS---SK 182
Query: 188 LVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYH 247
+VLR +EA+ F G + +D T +R CDAIAI+T E EG FCDYI+ QY
Sbjct: 183 VVLRPHEAKSLSFVWRKHEAIG-SFFDGKVTAMRNCDAIAIRTCRETEGKFCDYISRQYS 241
Query: 248 KNVLLTGPVL-SEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKA-Q 305
K V LTGPVL + L+ +WA+WL G VV+CAFGSQ ++ K Q
Sbjct: 242 KPVYLTGPVLPGSQPNQPSLDPQWAEWL--------AKFNHGSVVFCAFGSQPVVNKIDQ 293
Query: 306 FQEILLGLEISGF 318
FQE+ LGLE +GF
Sbjct: 294 FQELCLGLESTGF 306
>UniRef100_Q9FN28 Putative flavonol 3-O-glucosyltransferase [Arabidopsis thaliana]
Length = 447
Score = 284 bits (727), Expect = 3e-75
Identities = 145/301 (48%), Positives = 194/301 (64%), Gaps = 26/301 (8%)
Query: 17 HIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIITVP 76
H MFPWFA GHMTPY+HLAN+LA +GH++TFLLP KA+ L H N +P+ I FH +T+P
Sbjct: 6 HAFMFPWFAFGHMTPYLHLANKLAAKGHRVTFLLPKKAQKQLEHHNLFPDRIIFHSLTIP 65
Query: 77 NVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAARK 136
+VDGLP G ETAS+IP+SL L AMD TR+QVE +++ +P + FD AYWVPE A++
Sbjct: 66 HVDGLPAGAETASDIPISLGKFLTAAMDLTRDQVEAAVRALRPDLIFFDTAYWVPEMAKE 125
Query: 137 LGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYEAE 196
+K++ Y V+SA S+A VP ++ PPPGYPS SK++ R ++A
Sbjct: 126 HRVKSVIYFVISANSIAHELVPG-----------GELGVPPPGYPS---SKVLYRGHDAH 171
Query: 197 QQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTGPV 256
+ SI + L+ R TTGL+ CD I+I+T E+EG FCDYI QY + VLLTGP+
Sbjct: 172 ALLTFSIFYER----LHYRITTGLKNCDFISIRTCKEIEGKFCDYIERQYQRKVLLTGPM 227
Query: 257 LSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLEIS 316
L E LE+RW WL+ G V+YCA GSQ+ L K QFQE+ LG+E++
Sbjct: 228 LPEPDNSRPLEDRWNHWLN--------QFKPGSVIYCALGSQITLEKDQFQELCLGMELT 279
Query: 317 G 317
G
Sbjct: 280 G 280
>UniRef100_Q9T081 UDP rhamnose--anthocyanidin-3-glucoside rhamnosyltransferase-like
protein [Arabidopsis thaliana]
Length = 453
Score = 280 bits (716), Expect = 7e-74
Identities = 148/304 (48%), Positives = 194/304 (63%), Gaps = 23/304 (7%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIIT 74
K H+ M+PWFA GHMTP++ LAN+LA++GH +TFLLP K+ L H N +P+ I F +T
Sbjct: 5 KFHVLMYPWFATGHMTPFLFLANKLAEKGHTVTFLLPKKSLKQLEHFNLFPHNIVFRSVT 64
Query: 75 VPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAA 134
VP+VDGLP+GTETASEIP++ LL+ AMD TR+QVE V+++ +P + FD A+W+PE A
Sbjct: 65 VPHVDGLPVGTETASEIPVTSTDLLMSAMDLTRDQVEAVVRAVEPDLIFFDFAHWIPEVA 124
Query: 135 RKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYE 194
R G+KT+ Y VVSA ++A VP ++ PPPGYPS SK++LR+ +
Sbjct: 125 RDFGLKTVKYVVVSASTIASMLVPG-----------GELGVPPPGYPS---SKVLLRKQD 170
Query: 195 A-EQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLT 253
A + + G L +R TT L D IAI+T E+EG FCDYI K VLLT
Sbjct: 171 AYTMKKLEPTNTIDVGPNLLERVTTSLMNSDVIAIRTAREIEGNFCDYIEKHCRKKVLLT 230
Query: 254 GPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGL 313
GPV E + ELEERW KWL G VV+CA GSQVIL K QFQE+ LG+
Sbjct: 231 GPVFPEPDKTRELEERWVKWLS--------GYEPDSVVFCALGSQVILEKDQFQELCLGM 282
Query: 314 EISG 317
E++G
Sbjct: 283 ELTG 286
>UniRef100_Q93ZG5 AT4g27560/T29A15_50 [Arabidopsis thaliana]
Length = 461
Score = 277 bits (709), Expect = 4e-73
Identities = 148/304 (48%), Positives = 194/304 (63%), Gaps = 23/304 (7%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIIT 74
K H+ M+PWFA GHMTP++ LAN+LA++GH +TFL+P KA L +LN +P+ I F +T
Sbjct: 5 KFHVLMYPWFATGHMTPFLFLANKLAEKGHTVTFLIPKKALKQLENLNLFPHNIAFRSVT 64
Query: 75 VPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAA 134
VP+VDGLP+GTET SEIP++ LL+ AMD TR+QVE V+++ +P + FD A+W+PE A
Sbjct: 65 VPHVDGLPVGTETVSEIPVTSADLLMSAMDLTRDQVEGVVRAVEPDLIFFDFAHWIPEVA 124
Query: 135 RKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYE 194
R G+KT+ Y VVSA ++A VP ++ PPPGYPS SK++LR+ +
Sbjct: 125 RDFGLKTVKYVVVSASTIASMLVPG-----------GELGVPPPGYPS---SKVLLRKQD 170
Query: 195 A-EQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLT 253
A + S G L +R TT L D IAI+T E+EG FCDYI K VLLT
Sbjct: 171 AYTMKNLESTNTINVGPNLLERVTTSLMNSDVIAIRTAREIEGNFCDYIEKHCRKKVLLT 230
Query: 254 GPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGL 313
GPV E + ELEERW KWL G VV+CA GSQVIL K QFQE+ LG+
Sbjct: 231 GPVFPEPDKTRELEERWVKWLS--------GYEPDSVVFCALGSQVILEKDQFQELCLGM 282
Query: 314 EISG 317
E++G
Sbjct: 283 ELTG 286
>UniRef100_Q9T080 Putative UDP rhamnose-anthocyanidin-3-glucoside
rhamnosyltransferase [Arabidopsis thaliana]
Length = 455
Score = 277 bits (709), Expect = 4e-73
Identities = 148/304 (48%), Positives = 194/304 (63%), Gaps = 23/304 (7%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIIT 74
K H+ M+PWFA GHMTP++ LAN+LA++GH +TFL+P KA L +LN +P+ I F +T
Sbjct: 5 KFHVLMYPWFATGHMTPFLFLANKLAEKGHTVTFLIPKKALKQLENLNLFPHNIVFRSVT 64
Query: 75 VPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAA 134
VP+VDGLP+GTET SEIP++ LL+ AMD TR+QVE V+++ +P + FD A+W+PE A
Sbjct: 65 VPHVDGLPVGTETVSEIPVTSADLLMSAMDLTRDQVEGVVRAVEPDLIFFDFAHWIPEVA 124
Query: 135 RKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYE 194
R G+KT+ Y VVSA ++A VP ++ PPPGYPS SK++LR+ +
Sbjct: 125 RDFGLKTVKYVVVSASTIASMLVPG-----------GELGVPPPGYPS---SKVLLRKQD 170
Query: 195 A-EQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLT 253
A + S G L +R TT L D IAI+T E+EG FCDYI K VLLT
Sbjct: 171 AYTMKNLESTNTINVGPNLLERVTTSLMNSDVIAIRTAREIEGNFCDYIEKHCRKKVLLT 230
Query: 254 GPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGL 313
GPV E + ELEERW KWL G VV+CA GSQVIL K QFQE+ LG+
Sbjct: 231 GPVFPEPDKTRELEERWVKWLS--------GYEPDSVVFCALGSQVILEKDQFQELCLGM 282
Query: 314 EISG 317
E++G
Sbjct: 283 ELTG 286
>UniRef100_O81010 Putative flavonol 3-O-glucosyltransferase [Arabidopsis thaliana]
Length = 442
Score = 266 bits (681), Expect = 8e-70
Identities = 136/301 (45%), Positives = 196/301 (64%), Gaps = 26/301 (8%)
Query: 17 HIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIITVP 76
H MFPWFA GHM P++HLAN+LA++GH+ITFLLP KA+ L H N +P+ I FH +T+P
Sbjct: 6 HAFMFPWFAFGHMIPFLHLANKLAEKGHQITFLLPKKAQKQLEHHNLFPDSIVFHPLTIP 65
Query: 77 NVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAARK 136
+V+GLP G ET S+I +S+++LL A+D TR+QVE +++ +P + FD A+W+PE A++
Sbjct: 66 HVNGLPAGAETTSDISISMDNLLSEALDLTRDQVEAAVRALRPDLIFFDFAHWIPEIAKE 125
Query: 137 LGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYEAE 196
IK++ Y +VSA ++A +F P + + PPPGYPS SK++ R +A
Sbjct: 126 HMIKSVSYMIVSATTIAYTFAPGGV-----------LGVPPPGYPS---SKVLYRENDAH 171
Query: 197 QQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTGPV 256
SI + LY + TTG + CD IA++T E+EG FCDYI+ QYHK VLLTGP+
Sbjct: 172 ALATLSIFYKR----LYHQITTGFKSCDIIALRTCNEIEGKFCDYISSQYHKKVLLTGPM 227
Query: 257 LSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLEIS 316
L E+ + LEE+ + +L VV+CA GSQ++L K QFQE+ LG+E++
Sbjct: 228 LPEQDTSKPLEEQLSHFLS--------RFPPRSVVFCALGSQIVLEKDQFQELCLGMELT 279
Query: 317 G 317
G
Sbjct: 280 G 280
>UniRef100_Q9M0P3 Hypothetical protein AT4g09500 [Arabidopsis thaliana]
Length = 442
Score = 266 bits (681), Expect = 8e-70
Identities = 140/303 (46%), Positives = 193/303 (63%), Gaps = 26/303 (8%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIIT 74
K H MFPWFA GHM P++HLAN+LA++GH++TFLLP KA+ L H N +P+ I FH +T
Sbjct: 4 KFHAFMFPWFAFGHMIPFLHLANKLAEKGHRVTFLLPKKAQKQLEHHNLFPDSIVFHPLT 63
Query: 75 VPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAA 134
VP V+GLP G ET S+IP+SL++LL A+D TR+QVE +++ +P + FD A W+P+ A
Sbjct: 64 VPPVNGLPAGAETTSDIPISLDNLLSKALDLTRDQVEAAVRALRPDLIFFDFAQWIPDMA 123
Query: 135 RKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYE 194
++ IK++ Y +VSA ++A + VP + PPGYPS SK++ R +
Sbjct: 124 KEHMIKSVSYIIVSATTIAHTHVPG-----------GKLGVRPPGYPS---SKVMFREND 169
Query: 195 AEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTG 254
SI + LY + TTGL+ CD IA++T EVEG FCD+I+ QYHK VLLTG
Sbjct: 170 VHALATLSIFYKR----LYHQITTGLKSCDVIALRTCKEVEGMFCDFISRQYHKKVLLTG 225
Query: 255 PVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLE 314
P+ E + LEERW +L G VV+C+ GSQVIL K QFQE+ LG+E
Sbjct: 226 PMFPEPDTSKPLEERWNHFLS--------GFAPKSVVFCSPGSQVILEKDQFQELCLGME 277
Query: 315 ISG 317
++G
Sbjct: 278 LTG 280
>UniRef100_Q9LJA6 UDP-glycose: flavonoid glucosyltransferase-like protein
[Arabidopsis thaliana]
Length = 448
Score = 254 bits (650), Expect = 3e-66
Identities = 131/306 (42%), Positives = 184/306 (59%), Gaps = 28/306 (9%)
Query: 14 SKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHII 73
SK H ++PWF GHM PY+HLAN+LA++GH++TFL P KA+ L LN +PN I F +
Sbjct: 3 SKFHAFLYPWFGFGHMIPYLHLANKLAEKGHRVTFLAPKKAQKQLEPLNLFPNSIHFENV 62
Query: 74 TVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEA 133
T+P+VDGLP+G ET +++P S +L AMD R Q+E ++S KP + FD W+P+
Sbjct: 63 TLPHVDGLPVGAETTADLPNSSKRVLADAMDLLREQIEVKIRSLKPDLIFFDFVDWIPQM 122
Query: 134 ARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRY 193
A++LGIK++ Y ++SA +AM F P ++ PPPG+PS SK+ LR +
Sbjct: 123 AKELGIKSVSYQIISAAFIAMFFAPR-----------AELGSPPPGFPS---SKVALRGH 168
Query: 194 EAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLT 253
+A F L+DR TTGL+ CD IAI+T E+EG CD+I Q + VLLT
Sbjct: 169 DANIYSL----FANTRKFLFDRVTTGLKNCDVIAIRTCAEIEGNLCDFIERQCQRKVLLT 224
Query: 254 GPVL--SEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILL 311
GP+ + G+ LE+RW WL+ G VVYCAFG+ QFQE+ L
Sbjct: 225 GPMFLDPQGKSGKPLEDRWNNWLN--------GFEPSSVVYCAFGTHFFFEIDQFQELCL 276
Query: 312 GLEISG 317
G+E++G
Sbjct: 277 GMELTG 282
>UniRef100_Q9LPS8 Putative glucosyl transferase [Arabidopsis thaliana]
Length = 448
Score = 242 bits (618), Expect = 2e-62
Identities = 130/307 (42%), Positives = 180/307 (58%), Gaps = 31/307 (10%)
Query: 14 SKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHII 73
SK H M+PWF GHM PY+HLAN+LA++GH++TF LP KA L LN +P+ I F +
Sbjct: 3 SKFHAFMYPWFGFGHMIPYLHLANKLAEKGHRVTFFLPKKAHKQLQPLNLFPDSIVFEPL 62
Query: 74 TVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEA 133
T+P VDGLP G ETAS++P S + VAMD R+Q+E +++ KP + FD +WVPE
Sbjct: 63 TLPPVDGLPFGAETASDLPNSTKKPIFVAMDLLRDQIEAKVRALKPDLIFFDFVHWVPEM 122
Query: 134 ARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRY 193
A + GIK++ Y ++SA +AM P ++ PPP YP SK+ LR +
Sbjct: 123 AEEFGIKSVNYQIISAACVAMVLAPR-----------AELGFPPPDYPL---SKVALRGH 168
Query: 194 EAEQ-QMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLL 252
EA +FA+ L+ T GL+ CD ++I+T E+EG C +I + K +LL
Sbjct: 169 EANVCSLFAN------SHELFGLITKGLKNCDVVSIRTCVELEGKLCGFIEKECQKKLLL 222
Query: 253 TGPVLSE--EAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEIL 310
TGP+L E G+ LE+RW WL+ G G VV+CAFG+Q K QFQE
Sbjct: 223 TGPMLPEPQNKSGKFLEDRWNHWLN--------GFEPGSVVFCAFGTQFFFEKDQFQEFC 274
Query: 311 LGLEISG 317
LG+E+ G
Sbjct: 275 LGMELMG 281
>UniRef100_Q43716 Flavonol 3-O-glucosyltransferase [Petunia hybrida]
Length = 473
Score = 219 bits (559), Expect = 1e-55
Identities = 127/314 (40%), Positives = 188/314 (59%), Gaps = 19/314 (6%)
Query: 12 SNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPN-KARLALHHLNRYPNLITF 70
SN LH+ MFP+FA GH++P+V LAN+L+ G K++F + A LN P T
Sbjct: 8 SNDALHVVMFPFFAFGHISPFVQLANKLSSYGVKVSFFTASGNASRVKSMLNSAP---TT 64
Query: 71 HII--TVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAY 128
HI+ T+P+V+GLP G E+ +E+ + LL VA+D + Q++ ++ KPHFVLFD A
Sbjct: 65 HIVPLTLPHVEGLPPGAESTAELTPASAELLKVALDLMQPQIKTLLSHLKPHFVLFDFAQ 124
Query: 129 -WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSK 187
W+P+ A LGIKT+ Y VV A+S A PA+++ + ++EDM +PP G+P +S +
Sbjct: 125 EWLPKMANGLGIKTVYYSVVVALSTAFLTCPARVLEPKKYPSLEDMKKPPLGFPQTSVTS 184
Query: 188 LVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYH 247
+ R +EA ++ F G TLYDR +GLR C AI KT +++EG + Y+ Q++
Sbjct: 185 V--RTFEARDFLYVFKSF-HNGPTLYDRIQSGLRGCSAILAKTCSQMEGPYIKYVEAQFN 241
Query: 248 KNVLLTGPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQ 307
K V L GPV+ + G +LEE+WA WL+ G V+YC+FGS+ L Q +
Sbjct: 242 KPVFLIGPVVPDPPSG-KLEEKWATWLN--------KFEGGTVIYCSFGSETFLTDDQVK 292
Query: 308 EILLGLEISGFSIF 321
E+ LGLE +G F
Sbjct: 293 ELALGLEQTGLPFF 306
>UniRef100_Q84WC5 Hypothetical protein At4g09500 [Arabidopsis thaliana]
Length = 417
Score = 218 bits (556), Expect = 2e-55
Identities = 124/303 (40%), Positives = 174/303 (56%), Gaps = 51/303 (16%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNLITFHIIT 74
K H MFPWFA GHM P++HLAN+LA++GH++TFLLP KA+ L H N +P+ I FH +T
Sbjct: 4 KFHAFMFPWFAFGHMIPFLHLANKLAEKGHRVTFLLPKKAQKQLEHHNLFPDSIVFHPLT 63
Query: 75 VPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAYWVPEAA 134
VP V+GLP G ET S+IP+SL++LL A+D TR+QVE +++ +P + FD A W+P+ A
Sbjct: 64 VPPVNGLPAGAETTSDIPISLDNLLSKALDLTRDQVEAAVRALRPDLIFFDFAQWIPDMA 123
Query: 135 RKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVLRRYE 194
++ IK++ Y +VSA ++A + VP + PPGYPS SK++ R +
Sbjct: 124 KEHMIKSVSYIIVSATTIAHTHVPG-----------GKLGVRPPGYPS---SKVMFREND 169
Query: 195 AEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNVLLTG 254
SI + LY + TTGL+ CD IA++T EVE
Sbjct: 170 VHALATLSIFYKR----LYHQITTGLKSCDVIALRTCKEVED------------------ 207
Query: 255 PVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEILLGLE 314
+ LEERW +L G VV+C+ GSQVIL K QFQE+ LG+E
Sbjct: 208 -------TSKPLEERWNHFLS--------GFAPKSVVFCSPGSQVILEKDQFQELCLGME 252
Query: 315 ISG 317
++G
Sbjct: 253 LTG 255
>UniRef100_Q8LP23 Rhamnosyl transferase [Nierembergia sp. NB17]
Length = 465
Score = 208 bits (530), Expect = 2e-52
Identities = 120/311 (38%), Positives = 184/311 (58%), Gaps = 18/311 (5%)
Query: 16 LHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLP--NKARLALHHLNRYPNLITFHII 73
LH+ MFP+FA GH++P+ LAN+L+ G K++F N +RL LN P I+
Sbjct: 7 LHVVMFPFFAFGHISPFAQLANKLSSHGVKVSFFTASGNASRLR-SMLNSAPTTTHIDIV 65
Query: 74 --TVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVAY-WV 130
T+P+V+GLP G+E+ +E+ LL VA+D + Q++ ++ KPHFVLFD A W+
Sbjct: 66 PLTLPHVEGLPPGSESTAELTPVTAELLKVALDLMQPQIKTLLSHLKPHFVLFDFAQEWL 125
Query: 131 PEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKLVL 190
P+ A +LGIKT+ Y V A+S A PA++ + T+EDM +PP G+P +S + +
Sbjct: 126 PKMADELGIKTVFYSVFVALSTAFLTCPARVTEPKKYPTLEDMKKPPLGFPHTSITSV-- 183
Query: 191 RRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHKNV 250
+ +EA+ ++ F T+YDR +GL+ C AI KT +++EG + +Y+ Q+ K V
Sbjct: 184 KTFEAQDFLYIFKSFNNRP-TVYDRVLSGLKGCSAILAKTCSQMEGPYIEYVKSQFKKPV 242
Query: 251 LLTGPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQEIL 310
LL GPV+ + G +LEE+W WL+ G V+YC+FGS+ L Q +E+
Sbjct: 243 LLVGPVVPDPPSG-KLEEKWDAWLN--------KFEAGTVIYCSFGSETFLKDDQIKELA 293
Query: 311 LGLEISGFSIF 321
LGLE +G F
Sbjct: 294 LGLEQTGLPFF 304
>UniRef100_Q6VAA3 UDP-glycosyltransferase 79A2 [Stevia rebaudiana]
Length = 454
Score = 155 bits (391), Expect = 3e-36
Identities = 103/313 (32%), Positives = 163/313 (51%), Gaps = 28/313 (8%)
Query: 12 SNSKLHIAMFPWFAMGHMTPYVHLANELAKR--GHKITFLLPNKARLALHHLNRYPNLIT 69
++ +LH+ MFP+FA GH+TP+V L+N+++ G KITFL + + + +
Sbjct: 6 NDKELHLVMFPFFAFGHITPFVQLSNKISSLYPGVKITFLAASASVSRIETMLNPSTNTK 65
Query: 70 FHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFD-VAY 128
+T+P VDGLP G E ++ + LLVVA+D + Q++ ++ + KP FV+FD V +
Sbjct: 66 VIPLTLPRVDGLPEGVENTADASPATIGLLVVAIDLMQPQIKTLLANLKPDFVIFDFVHW 125
Query: 129 WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKL 188
W+PE A +LGIKTI + V A + S + ++P TVED+ K
Sbjct: 126 WLPEIASELGIKTIYFSVYMANIVMPS---TSKLTGNKPSTVEDI-------------KA 169
Query: 189 VLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYHK 248
+ + Y + F +I + +D + C+ + IK+ E+EG D + Q +
Sbjct: 170 LQQSYGIPVKTFEAISLMNVFKSFHDWMDKCINGCNLMLIKSCREMEGSRIDDVTKQSTR 229
Query: 249 NVLLTGPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQFQE 308
V L GPV+ E G EL+E WA WL+ V+YC+FGS+ L Q +E
Sbjct: 230 PVFLIGPVVPEPHSG-ELDETWANWLN--------RFPAKSVIYCSFGSETFLTDDQIRE 280
Query: 309 ILLGLEISGFSIF 321
+ LGLE++G F
Sbjct: 281 LALGLELTGLPFF 293
>UniRef100_Q8H6A4 UDP-glucosyltransferase [Stevia rebaudiana]
Length = 454
Score = 153 bits (387), Expect = 9e-36
Identities = 106/315 (33%), Positives = 164/315 (51%), Gaps = 32/315 (10%)
Query: 12 SNSKLHIAMFPWFAMGHMTPYVHLANELAKR--GHKITFLLPNKARLALHHLNRYPNLIT 69
++ +LH+ MFP+FA GH+TP+V L+N+++ G KITFL + + + +
Sbjct: 6 NDKELHLVMFPFFAFGHITPFVQLSNKISSLYPGVKITFLAASASVSRIETMLNPSTNTK 65
Query: 70 FHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFD-VAY 128
+T+P VDGLP G E ++ + LLVVA+D + Q++ ++ + KP FV+FD V +
Sbjct: 66 VIPLTLPRVDGLPEGVENTADASPATIGLLVVAIDLMQPQIKTLLANLKPDFVIFDFVHW 125
Query: 129 WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMS--QPPPGYPSSSNS 186
W+PE A +LGIKTI + V A + S + ++P TVED+ Q G P
Sbjct: 126 WLPEIASELGIKTIYFSVYMANIVMPS---TSKLTGNKPSTVEDIKALQQSDGIP----- 177
Query: 187 KLVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQY 246
++ +EA M F +D + C+ + IK+ E+EG D + Q
Sbjct: 178 ---VKTFEAISLMNVFKSF-------HDWMDKCINGCNLMLIKSCREMEGSRIDDVTKQS 227
Query: 247 HKNVLLTGPVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKAQF 306
+ V L GPV+ E G EL+E WA WL+ V+YC+FGS+ L Q
Sbjct: 228 TRPVFLIGPVVPEPHSG-ELDETWANWLN--------RFPAKSVIYCSFGSETFLTDDQI 278
Query: 307 QEILLGLEISGFSIF 321
+E+ LGLE++G F
Sbjct: 279 RELALGLELTGLPFF 293
>UniRef100_Q66PF2 Putative UDP-rhamnose:rhamnosyltransferase [Fragaria ananassa]
Length = 478
Score = 145 bits (367), Expect = 2e-33
Identities = 102/341 (29%), Positives = 170/341 (48%), Gaps = 31/341 (9%)
Query: 6 SKNTSSSNSKLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYP 65
S ++++ KLHIA+FPW A GH+ P++ +A +A++GHK++F+ + + L + P
Sbjct: 2 SSSSATKRKKLHIALFPWLAFGHIIPFLEVAKHIARKGHKVSFISTPR---NIQRLPKIP 58
Query: 66 NLITFHI----ITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHF 121
+T I I +P+V+ LP E ++P + L +A D + + ++++ P +
Sbjct: 59 ETLTPLINLVQIPLPHVENLPENAEATMDVPHDVIPYLKIAHDGLEQGISEFLQAQSPDW 118
Query: 122 VLFDVA-YWVPEAARKLGIKTICYHVVSAMSMAM--SFVPAKIIPKDRPLTVEDMSQPPP 178
++ D A +W+P A KLGI + + +A SM S P ++ +E + PP
Sbjct: 119 IIHDFAPHWLPPIATKLGISNAHFSIFNASSMCFFGSTSPNRVSRYAPRKKLEQFTSPPE 178
Query: 179 GYPSSSNSKLVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCF 238
P SK+ R +EA++ M ++ GVT R + ++ C I++ E+EG +
Sbjct: 179 WIPFP--SKIYHRPFEAKRLMDGTLTPNASGVTDRFRLESTIQGCQVYFIRSCREIEGEW 236
Query: 239 CDYIAIQYHKNVLL-TG---PVLSEEAEGEELEERWAK---WLDGLGLGLGLGLGLGPVV 291
D + + K ++L TG P L E + W+K WLD G VV
Sbjct: 237 LDLLEDLHEKPIVLPTGLLPPSLPRSDEDGGKDSNWSKIAVWLD--------KQEKGKVV 288
Query: 292 YCAFGSQVILGKAQFQEILLGLEISGFSIF----GGSEGSG 328
Y AFGS++ L + F E+ LGLE+SG F S GSG
Sbjct: 289 YAAFGSELNLSQEVFNELALGLELSGLPFFWVLRKPSHGSG 329
>UniRef100_Q6QJL3 Glucosyl-transferase [Fagopyrum esculentum]
Length = 240
Score = 139 bits (350), Expect = 2e-31
Identities = 86/233 (36%), Positives = 135/233 (57%), Gaps = 15/233 (6%)
Query: 15 KLHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPN----KARLALHHLNRYPNLITF 70
+LH+AMFPWFA GH+ P++HL+N+L+ G +I+FL + K R +L N +I
Sbjct: 17 RLHVAMFPWFAFGHINPFIHLSNKLSSHGVQISFLSASGNIPKIRNSLIQSNN-TQIIPL 75
Query: 71 HIITVPNVDGLPLGTETASEI-PLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVA-Y 128
HI P V+GLP G E S + S+ LL +A+D+ + Q++ ++ + KPH V FD A Y
Sbjct: 76 HI---PTVEGLPPGRENTSNMTDDSMPELLKLALDQMQPQIKTLLANLKPHLVFFDFAQY 132
Query: 129 WVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSKL 188
W+P+ A +L I+T+ + V S++ + + + A+ P TVE++ PP G+P S L
Sbjct: 133 WLPDLASELNIRTVHFCVYSSILRSFTLLAAEATKMTVP-TVEELKIPPKGHPKPS---L 188
Query: 189 VLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDY 241
L+ +EAE+ + F GG + R + DAIA K+ E+EG + DY
Sbjct: 189 SLKTFEAEKMLHPFRSF-HGGPSPIARGYLSTKNSDAIAFKSCNEIEGPYIDY 240
>UniRef100_Q9LTA3 Putative anthocyanidin-3-glucoside rhamnosyltransferase
[Arabidopsis thaliana]
Length = 460
Score = 123 bits (308), Expect = 1e-26
Identities = 88/317 (27%), Positives = 151/317 (46%), Gaps = 29/317 (9%)
Query: 16 LHIAMFPWFAMGHMTPYVHLANELAKRGHKITFLLPNKARLALHHLNRYPNL-------I 68
+H+AMFPW AMGH+ P++ L+ LA++GHKI+F+ + ++ R P L I
Sbjct: 9 MHVAMFPWLAMGHLLPFLRLSKLLAQKGHKISFISTPR------NIERLPKLQSNLASSI 62
Query: 69 TFHIITVPNVDGLPLGTETASEIPLSLNHLLVVAMDKTRNQVEQVMKSKKPHFVLFDVA- 127
TF +P + GLP +E++ ++P + L A D + +++ ++ P ++++D A
Sbjct: 63 TFVSFPLPPISGLPPSSESSMDVPYNKQQSLKAAFDLLQPPLKEFLRRSSPDWIIYDYAS 122
Query: 128 YWVPEAARKLGIKTICYHVVSAMSMAMSFVPAKIIPKDRPLTVEDMSQPPPGYPSSSNSK 187
+W+P A +LGI + + +A ++ + +I + R T ED + PP P SN
Sbjct: 123 HWLPSIAAELGISKAFFSLFNAATLCFMGPSSSLIEEIRS-TPEDFTVVPPWVPFKSN-- 179
Query: 188 LVLRRYEAEQQMFASIPFGEGGVTLYDRTTTGLRECDAIAIKTTTEVEGCFCDYIAIQYH 247
++ RY + GV+ R + E DA+ +++ E E + + Y
Sbjct: 180 -IVFRYHEVTRYVEKTEEDVTGVSDSVRFGYSIDESDAVFVRSCPEFEPEWFGLLKDLYR 238
Query: 248 KNVLLTG---PVLSEEAEGEELEERWAKWLDGLGLGLGLGLGLGPVVYCAFGSQVILGKA 304
K V G PV+ ++ + R KWLD L VVY + G++ L
Sbjct: 239 KPVFPIGFLPPVIEDDDAVDTTWVRIKKWLD--------KQRLNSVVYVSLGTEASLRHE 290
Query: 305 QFQEILLGLEISGFSIF 321
+ E+ LGLE S F
Sbjct: 291 EVTELALGLEKSETPFF 307
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.334 0.148 0.485
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 804,883,879
Number of Sequences: 2790947
Number of extensions: 34854135
Number of successful extensions: 124132
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 111
Number of HSP's successfully gapped in prelim test: 107
Number of HSP's that attempted gapping in prelim test: 123692
Number of HSP's gapped (non-prelim): 310
length of query: 484
length of database: 848,049,833
effective HSP length: 131
effective length of query: 353
effective length of database: 482,435,776
effective search space: 170299828928
effective search space used: 170299828928
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0282.12


