

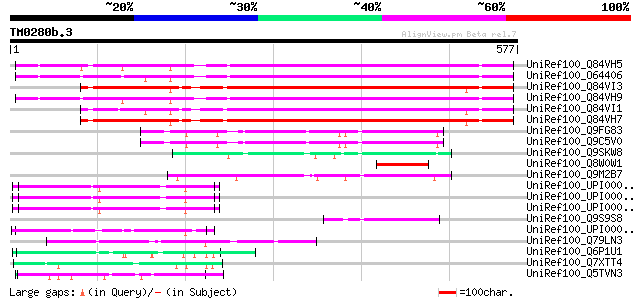
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0280b.3
(577 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84VH5 Envelope-like protein [Glycine max] 383 e-105
UniRef100_O64406 Envelope-like [Glycine max] 374 e-102
UniRef100_Q84VI3 Envelope-like protein [Glycine max] 367 e-100
UniRef100_Q84VH9 Envelope-like protein [Glycine max] 366 1e-99
UniRef100_Q84VI1 Envelope-like protein [Glycine max] 363 6e-99
UniRef100_Q84VH7 Envelope-like protein [Glycine max] 363 7e-99
UniRef100_Q9FG83 Gb|AAF18631.1 [Arabidopsis thaliana] 82 6e-14
UniRef100_Q9C5V0 Putative envelope protein [Arabidopsis thaliana] 82 6e-14
UniRef100_Q9SKW8 F5J5.2 [Arabidopsis thaliana] 81 1e-13
UniRef100_Q8W0W1 Putative envelope protein [Vicia faba] 78 8e-13
UniRef100_Q9M2B7 Hypothetical protein F23N14_70 [Arabidopsis tha... 70 1e-10
UniRef100_UPI00004307AD UPI00004307AD UniRef100 entry 62 6e-08
UniRef100_UPI00004307AC UPI00004307AC UniRef100 entry 62 6e-08
UniRef100_UPI00004307AB UPI00004307AB UniRef100 entry 62 6e-08
UniRef100_Q9S9S8 F28J9.1 [Arabidopsis thaliana] 60 2e-07
UniRef100_UPI00004307A7 UPI00004307A7 UniRef100 entry 59 4e-07
UniRef100_Q79LN3 Biofilm-associated surface protein [Staphylococ... 58 7e-07
UniRef100_Q6P1U1 LOC395027 protein [Xenopus tropicalis] 58 9e-07
UniRef100_Q7XTT4 OSJNBa0058K23.21 protein [Oryza sativa] 58 9e-07
UniRef100_Q5TVN3 ENSANGP00000027660 [Anopheles gambiae str. PEST] 58 9e-07
>UniRef100_Q84VH5 Envelope-like protein [Glycine max]
Length = 685
Score = 383 bits (984), Expect = e-105
Identities = 231/596 (38%), Positives = 336/596 (55%), Gaps = 55/596 (9%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
S K TS+ S+ S + P+ + + + ++ E+ P
Sbjct: 72 SPKETSAPGSPSVPSSPSSTKAPSNQ--ERPEFNIQPIQMIPGQAPVPEKLVPKRQQGVK 129
Query: 67 INEEETLMGSDRDV--------------------VPDAEASAPQDTNVSQENPIPENPDV 106
I+E +L S R+V VPDA+ P + P PD
Sbjct: 130 ISENPSLATSPREVDTEMDKKIRTIVSNILKDVSVPDADKDVP-----TSSTPNVSVPDA 184
Query: 107 E-DVIAEETVSTETPSSSKKGD---KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPL 162
E DV T + E SS K D + +++ET + + + D ++ + D+ P+
Sbjct: 185 EKDVPTSSTPNAEVLSSPSKEDSTEEEDQATEETPAPRAPEPAPGDLIDLEEVESDEEPI 244
Query: 163 FETLPESVGARLKRKRRTT----ASEAPSVPQKKTKP-SPTTPKSKKKDLKGKGKATAEK 217
L + RL+ ++ T + ++ QKK+ P +PTT + K + K
Sbjct: 245 ANKLAPGIAERLQSRKGKTPIKRSGRIKTMAQKKSTPITPTTSRRSKVAIPSK------- 297
Query: 218 SMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRRIPKNVPAAPLDNVSFHSEENA 277
K+ + S+SD DVE DVPDI A K +G+++P NVP APLDN+SFHS +N
Sbjct: 298 ------KRKEISSSDSDDDVELDVPDIKRA---KTSGKKVPGNVPDAPLDNISFHSIDNV 348
Query: 278 LKWKYVYQRRLATEREVGSDVLECKEVMALIEKAGLIKTVLQVGRCFERLVKEFVVNLSV 337
+WK+VYQRRLA ERE+G D L+CKE+M LI+ AGL+KTV ++G C+E LV+EF+VN+
Sbjct: 349 ERWKFVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPS 408
Query: 338 EVGLPESDEYRKVYVREKCVKFSPDVINKALGRSATAVTDEEPSLDVVAKELTAGQVQKW 397
++ +SDEY+KV+VR KCV+FSP VINK LGR V D S +AKE+TA QVQ W
Sbjct: 409 DITNRKSDEYQKVFVRGKCVRFSPAVINKYLGRPTDGVVDITVSEHQIAKEITAKQVQHW 468
Query: 398 PKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTSAISATLARLIYKIGTMVPVDFGNFVFE 457
PKK LS LSVKYAIL+RIGA NWVPT HTS ++ L + +Y +GT +FGN++F+
Sbjct: 469 PKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFD 528
Query: 458 QTLKHAETCAVKLPIYFPSLITEIILQQHPKIVRDDEEAMPKGSPITLDHRFFYGPHVPD 517
QT+KH+E+ AVKLPI FP+++ I+L QHP I+ + M + SP++L ++ F G HVPD
Sbjct: 529 QTVKHSESFAVKLPIAFPTVLCGIMLSQHPNILNYTDSVMKRESPLSLHYKLFEGTHVPD 588
Query: 518 IDVPSNKTFASVPVSKSGRRAIIDELQAVSKALQETITASTARKVKVDELLLKLQE 573
I S K AS VSK A+I EL+ K L+ TI A+T +K++++ L+ +L +
Sbjct: 589 IVSTSGKAAASGAVSKD---ALIAELKDTCKVLEATIKATTEKKMELERLIKRLSD 641
>UniRef100_O64406 Envelope-like [Glycine max]
Length = 648
Score = 374 bits (960), Expect = e-102
Identities = 228/578 (39%), Positives = 329/578 (56%), Gaps = 36/578 (6%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
S K TSS S+ S + P+ + + + ++ E+ P
Sbjct: 59 SPKDTSSPGSPSVPSSPSSTKAPSNQ--EQPEFHIQPIQMIPGQAPVPEKLVPKRQQGVK 116
Query: 67 INEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVE-DVIAEETVSTETPSSSKK 125
I+E ++ S R+V D E + VS PD + DV T + E SSS K
Sbjct: 117 ISENPSIATSPREV--DTEMDKKIRSIVSSILKNASVPDADKDVPTSSTPNAEVLSSSSK 174
Query: 126 GDKGSVSSKETHSDEPIQVLVQNNSDGD-----DSDDDDVPLFETLPESVGARLKRKRRT 180
+ S +E ++E + GD + + D+ P+ L + RL+ ++
Sbjct: 175 EE--STEEEEQATEETPAPRAPEPAPGDLIDLEEVESDEEPIANKLAPGIAERLQSRKGK 232
Query: 181 T----ASEAPSVPQKKTKP-SPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDS 235
T + ++ QKK+ P +PTT + K + K K+ S+SD
Sbjct: 233 TPITRSGRIKTMAQKKSTPITPTTSRWSKVAIPSK-------------KRKEFSSSDSDD 279
Query: 236 DVEADVPDIVLAEMKKFAGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVG 295
DVE DVPDI A K +G+++P NVP APLDN+SFHS N +WK+VYQRRLA ERE+G
Sbjct: 280 DVELDVPDIKRA---KKSGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALERELG 336
Query: 296 SDVLECKEVMALIEKAGLIKTVLQVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREK 355
D L+CKE+M LI+ AGL+KTV ++G C+E LV+EF+VN+ ++ +SDEY+KV+VR K
Sbjct: 337 RDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVFVRGK 396
Query: 356 CVKFSPDVINKALGRSATAVTDEEPSLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAIL 415
CV+FSP VINK LGR V D S +AKE+TA QVQ WPKK LS LSVKYAIL
Sbjct: 397 CVRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAIL 456
Query: 416 NRIGAVNWVPTQHTSAISATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFP 475
+RIGA NWVPT HTS ++ L + +Y +GT +FG ++F+QT+KH+E+ AVKLPI FP
Sbjct: 457 HRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGKYIFDQTVKHSESFAVKLPIAFP 516
Query: 476 SLITEIILQQHPKIVRDDEEAMPKGSPITLDHRFFYGPHVPDIDVPSNKTFASVPVSKSG 535
+++ I+L QHP I+ + + M + S ++L ++ F G HVPDI S K AS VSK
Sbjct: 517 TVLCGIMLSQHPNILNNIDSVMKRESALSLHYKLFEGTHVPDIVSTSGKAAASGAVSKD- 575
Query: 536 RRAIIDELQAVSKALQETITASTARKVKVDELLLKLQE 573
A+I EL+ K L+ TI A+T +K++++ L+ +L +
Sbjct: 576 --ALIAELKDTCKVLEATIKATTEKKMELERLIKRLSD 611
>UniRef100_Q84VI3 Envelope-like protein [Glycine max]
Length = 680
Score = 367 bits (941), Expect = e-100
Identities = 211/505 (41%), Positives = 310/505 (60%), Gaps = 35/505 (6%)
Query: 81 VPDAEASAPQDTNVSQENPIPENPDVE-DVIAEETVSTET-PSSSKKG--DKGSVSSKET 136
VP+A+ P + NP PDV+ DV + E PS S++G ++ +++ET
Sbjct: 156 VPEADEDVP-----TSSNPNVSVPDVKKDVPTSSAPNAEALPSPSEEGSTEEDDQAAEET 210
Query: 137 HSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT----ASEAPSVPQKK 192
+ + + D ++ + D+ P+ L + RL+ ++ T + ++ QKK
Sbjct: 211 PAPRAPEPAPGDLIDLEEVESDEEPIANRLAPGIAERLQSRKGKTPIKRSGRIKTMAQKK 270
Query: 193 TKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKF 252
+ +P TP + ++ K KK+ + S+SD DVE DV ++ K
Sbjct: 271 S--TPITPATSRRS----------KVAIPSKKRKEISSSDSDKDVELDVST---SKKAKT 315
Query: 253 AGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVGSDVLECKEVMALIEKAG 312
+G+++P NVP APLDN+SFHS N KWKYVYQRRLA ERE+G D L+CKE+M LI+ AG
Sbjct: 316 SGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDALDCKEIMDLIKAAG 375
Query: 313 LIKTVLQVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREKCVKFSPDVINKALGRSA 372
L+KTV ++G C+E LV+EF+VN+ ++ +SDEY+KV+VR KCVKFSP VINK LGR
Sbjct: 376 LLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKFSPAVINKYLGRPT 435
Query: 373 TAVTDEEPSLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTSAI 432
V D + S +AKE+TA +VQ WPKK LS LSVKYAIL+RIGA NWVPT HTS +
Sbjct: 436 DGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTV 495
Query: 433 SATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFPSLITEIILQQHPKIVRD 492
+ L + +Y +GT +FGN++F+QT+KH+E+ A+KLPI FP+++ I+L QHP ++
Sbjct: 496 ATGLGKFLYAVGTKSKFNFGNYIFDQTVKHSESFAIKLPIAFPTVLCGIMLSQHPNMLNY 555
Query: 493 DEEAMPKGSPITLDHRFFYGPHVPDI----DVPSNKTFASVPVSKSGRRAIIDELQAVSK 548
+ M + SP++L ++ F G HVPDI S K AS VSK A+I EL+ K
Sbjct: 556 TDSVMKRESPLSLHYKLFEGTHVPDIVSTSVSTSGKAAASGAVSKD---ALIAELKDTCK 612
Query: 549 ALQETITASTARKVKVDELLLKLQE 573
L+ TI A+T +K++++ L+ +L E
Sbjct: 613 VLEATIKATTEKKMELELLIKRLSE 637
>UniRef100_Q84VH9 Envelope-like protein [Glycine max]
Length = 656
Score = 366 bits (939), Expect = 1e-99
Identities = 221/576 (38%), Positives = 329/576 (56%), Gaps = 32/576 (5%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
S K TSS S+ + P+ + + + ++ E+ P
Sbjct: 67 SPKETSSPVSPSVPSPPSSTKAPSNQ--EQPEFNIQPIQMIPGPAPVPEKLVPKRQQGVK 124
Query: 67 INEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVE-DVIAEETVSTETPSSSKK 125
I+E +L S R+V D E + VS PD + DV T + E SSS K
Sbjct: 125 ISENPSLATSPREV--DTEMDKKIRSIVSSILKNASVPDADKDVPTSSTPNAEVLSSSSK 182
Query: 126 G---DKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT- 181
++ +++ET + + + D ++ + D+ P+ + L + RL+ ++ T
Sbjct: 183 EKSTEEEDQATEETPAPRAPEPAPGDLIDLEEVESDEEPIVKKLALGIAERLQSRKGKTP 242
Query: 182 ---ASEAPSVPQKKTKP-SPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDV 237
+ ++ QKK+ P +PTT + K + K K+ + S+SD DV
Sbjct: 243 ITRSGRIKTIAQKKSTPITPTTSRWSKVAIPSK-------------KRKEISSSDSDDDV 289
Query: 238 EADVPDIVLAEMKKFAGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVGSD 297
E DVPDI A K +G+++P NVP APLDN+SFHS N +WK+VYQRRLA ERE+G D
Sbjct: 290 ELDVPDIKRA---KKSGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALERELGRD 346
Query: 298 VLECKEVMALIEKAGLIKTVLQVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREKCV 357
L+CKE+M LI+ AGL+KTV ++G C+E LV+EF+VN+ ++ +SDEY+ V+VR K +
Sbjct: 347 ALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQTVFVRGKGI 406
Query: 358 KFSPDVINKALGRSATAVTDEEPSLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILNR 417
+FSP VINK LGR V D S +AKE+TA QVQ WPKK LS LSVKYAIL+R
Sbjct: 407 RFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVKYAILHR 466
Query: 418 IGAVNWVPTQHTSAISATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFPSL 477
IG NWVPT HTS ++ L + +Y +GT +FGN++F+QT+KH+E+ AVKLPI FP++
Sbjct: 467 IGTANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFDQTVKHSESFAVKLPIAFPTV 526
Query: 478 ITEIILQQHPKIVRDDEEAMPKGSPITLDHRFFYGPHVPDIDVPSNKTFASVPVSKSGRR 537
+ I+L QHP I+ + + + S ++L ++ F G HVPDI S K AS V+K
Sbjct: 527 LCGIMLSQHPNILNNIDSVKKRESALSLHYKLFEGTHVPDIVSTSGKAAASGAVTKD--- 583
Query: 538 AIIDELQAVSKALQETITASTARKVKVDELLLKLQE 573
A+I EL+ K L+ TI A+T +K++++ L+ +L +
Sbjct: 584 ALIAELKDTCKVLEATIKATTEKKMELERLIKRLSD 619
>UniRef100_Q84VI1 Envelope-like protein [Glycine max]
Length = 682
Score = 363 bits (933), Expect = 6e-99
Identities = 210/507 (41%), Positives = 304/507 (59%), Gaps = 39/507 (7%)
Query: 81 VPDAEASAPQDTNVSQENPIPENPDVE-DVIAEETVSTETPSSSKKGDKGSVSSKETHSD 139
VP+A+ P + NP PDV+ DV + E S G++GS + ++
Sbjct: 158 VPEADEDVP-----TSSNPNVSVPDVKKDVPTSSAPNAEALPSP--GEEGSTEEDDQAAE 210
Query: 140 EPIQVLVQNNSDGD-----DSDDDDVPLFETLPESVGARLKRKRRTT----ASEAPSVPQ 190
E + GD + + D+ P+ L + RL+ ++ T + ++ Q
Sbjct: 211 ETPAPRAPEPAPGDLIDLEEVESDEEPIANRLAPGIAERLQSRKGKTPIKRSGRIKTMAQ 270
Query: 191 KKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMK 250
KK+ +P TP + ++ K KK+ + S+SD DVE DV ++
Sbjct: 271 KKS--TPITPATSRRS----------KVAIPSKKRKEISSSDSDKDVELDVST---SKKA 315
Query: 251 KFAGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVGSDVLECKEVMALIEK 310
K +G+++P NVP APLDN+SFHS N KWKYVYQRRLA ERE+G D L+CKE+M LI+
Sbjct: 316 KTSGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDALDCKEIMDLIKA 375
Query: 311 AGLIKTVLQVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREKCVKFSPDVINKALGR 370
GL+KTV ++G C+E LV+EF+VN+ ++ +SDEY+KV+VR KCVKFSP VINK LGR
Sbjct: 376 GGLLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDEYQKVFVRGKCVKFSPAVINKYLGR 435
Query: 371 SATAVTDEEPSLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTS 430
V D + S +AKE+TA +VQ WPKK LS LSVKYAIL+RIGA NWVPT HTS
Sbjct: 436 PTDGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTS 495
Query: 431 AISATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFPSLITEIILQQHPKIV 490
++ L + +Y +GT +FGN++F+QT+KH+E+ A+KLPI FP+++ I+L QHP ++
Sbjct: 496 TVATGLGKFLYAVGTKSKFNFGNYIFDQTVKHSESFAIKLPIAFPTVLCGIMLSQHPNML 555
Query: 491 RDDEEAMPKGSPITLDHRFFYGPHVPDI----DVPSNKTFASVPVSKSGRRAIIDELQAV 546
+ M + SP++L ++ F G HVPDI S K AS VSK A+I EL+
Sbjct: 556 NYTDSVMKRESPLSLHYKLFEGTHVPDIVSTSVSTSGKAAASGAVSKD---ALIAELKDT 612
Query: 547 SKALQETITASTARKVKVDELLLKLQE 573
K L+ TI A+T +K++++ L+ +L E
Sbjct: 613 CKVLEATIKATTEKKMELELLIKRLSE 639
>UniRef100_Q84VH7 Envelope-like protein [Glycine max]
Length = 680
Score = 363 bits (932), Expect = 7e-99
Identities = 208/505 (41%), Positives = 310/505 (61%), Gaps = 35/505 (6%)
Query: 81 VPDAEASAPQDTNVSQENPIPENPDVE-DVIAEETVSTET-PSSSKKG--DKGSVSSKET 136
VP+A+ P + NP PDV+ DV + E PS S++G ++ +++ET
Sbjct: 156 VPEADEDVP-----TSSNPDVSVPDVKKDVPTSSAPNAEALPSPSEEGSTEEDDQAAEET 210
Query: 137 HSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT----ASEAPSVPQKK 192
+ + + D ++ + D+ P+ L + RL+ ++ T + ++ QKK
Sbjct: 211 PAPRAPEPAPGDLIDLEEVESDEEPIANRLAPGIAERLQSRKGKTPIKRSGRIKTMAQKK 270
Query: 193 TKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKF 252
+ +P TP + ++ K KK+ + S+SD DVE DV ++ K
Sbjct: 271 S--TPITPATSRRS----------KVAIPSKKRKEISSSDSDKDVELDVST---SKKAKT 315
Query: 253 AGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVGSDVLECKEVMALIEKAG 312
+G+++P NVP APLDN+SFHS N KWKYVYQRRLA ERE+G D L+CKE+M LI+ AG
Sbjct: 316 SGKKVPGNVPDAPLDNISFHSIGNVEKWKYVYQRRLAVERELGRDALDCKEIMDLIKAAG 375
Query: 313 LIKTVLQVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREKCVKFSPDVINKALGRSA 372
L+KTV ++G C+E LV+EF+VN+ ++ +SD+Y++V+VR KCV+FSP VINK LGR
Sbjct: 376 LLKTVSKLGDCYEGLVREFIVNIPSDISNRKSDDYQRVFVRGKCVRFSPAVINKYLGRPT 435
Query: 373 TAVTDEEPSLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTSAI 432
V D + S +AKE+TA +VQ WPKK LS LSVKYAIL+RIGA NWVPT HTS +
Sbjct: 436 DGVIDIDVSEHQIAKEITAKRVQHWPKKGKLSAGKLSVKYAILHRIGAANWVPTNHTSTV 495
Query: 433 SATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFPSLITEIILQQHPKIVRD 492
+ L + +Y +GT +FGN++F+QT+KH+E+ A+KLPI FP+++ I+L QHP ++
Sbjct: 496 ATGLGKFLYAVGTKSKFNFGNYIFDQTVKHSESFAIKLPIAFPTVLCGIMLSQHPNMLNY 555
Query: 493 DEEAMPKGSPITLDHRFFYGPHVPDI----DVPSNKTFASVPVSKSGRRAIIDELQAVSK 548
+ M + SP++L ++ F G HVPDI S K AS VSK A+I EL+ K
Sbjct: 556 TDSVMKRESPLSLHYKLFEGTHVPDIVSTSVSTSGKAAASGAVSKD---ALIAELKDTCK 612
Query: 549 ALQETITASTARKVKVDELLLKLQE 573
L+ TI A+T +K++++ L+ +L E
Sbjct: 613 VLEATIKATTEKKMELELLIKRLSE 637
>UniRef100_Q9FG83 Gb|AAF18631.1 [Arabidopsis thaliana]
Length = 515
Score = 81.6 bits (200), Expect = 6e-14
Identities = 92/375 (24%), Positives = 153/375 (40%), Gaps = 54/375 (14%)
Query: 149 NSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTT-----PKSK 203
+ D D S DD VP + V K+ A+EAP P SP PK+
Sbjct: 58 SDDSDSSSDDAVPDPKAARSPVAVPSKQ-----AAEAPPPPSHAPDDSPAPVSDLPPKTS 112
Query: 204 KKDLKGKGKATAEKSMSEKKKKAPVIVSESDS-----DVEADVPDIVLAEMKKFAGRRIP 258
++ + + P S+ S E D D +
Sbjct: 113 TAPPINDPAPDLLRNAFSSRGRLPYDPSDRCSIHRHHQAERDAADPSIW----------- 161
Query: 259 KNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVGSDVLECKEVMALIEKAGLIKTVL 318
P PL + F + + ++K R +R V E +++M +IE L+ TV
Sbjct: 162 -TPPYEPL-TIQFLTSQGVERYKDFKLRDPYEQRYVPRGPPEMRDMMKVIEDNHLLDTVA 219
Query: 319 QVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREKCVKFSPDVINKALGRSAT---AV 375
++ + ++ EF N + E + KV+VR K +FSP +INK A A
Sbjct: 220 EIDPFVKEVIWEFYANFHI---FNEETKKSKVFVRGKMYEFSPQMINKTFRSPAIKWHAK 276
Query: 376 TDEEP---SLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTSAI 432
+ E S +AK L+ G+V W K L T + A L I +NW+P+++ S +
Sbjct: 277 AEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLAGLYLICCINWIPSKNNSNV 333
Query: 433 SATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCA-VKLPIYFPSLITEIILQQ------ 485
+ A+++Y + +P +FG V++Q K A + ++ + FP+LI +++ Q
Sbjct: 334 TVDRAKMLYLLNERIPFNFGKLVYDQVEKAARSPGQIQNILPFPNLIYRVLMFQRVITLE 393
Query: 486 -------HPKIVRDD 493
HPK+ D
Sbjct: 394 NYEKPGPHPKLFNPD 408
>UniRef100_Q9C5V0 Putative envelope protein [Arabidopsis thaliana]
Length = 461
Score = 81.6 bits (200), Expect = 6e-14
Identities = 92/375 (24%), Positives = 153/375 (40%), Gaps = 54/375 (14%)
Query: 149 NSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTT-----PKSK 203
+ D D S DD VP + V K+ A+EAP P SP PK+
Sbjct: 58 SDDSDSSSDDAVPDPKAARSPVAVPSKQ-----AAEAPPPPSHAPDDSPAPVSDLPPKTS 112
Query: 204 KKDLKGKGKATAEKSMSEKKKKAPVIVSESDS-----DVEADVPDIVLAEMKKFAGRRIP 258
++ + + P S+ S E D D +
Sbjct: 113 TAPPINDPAPDLLRNAFSSRGRLPYDPSDRCSIHRHHQAERDAADPSIW----------- 161
Query: 259 KNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVGSDVLECKEVMALIEKAGLIKTVL 318
P PL + F + + ++K R +R V E +++M +IE L+ TV
Sbjct: 162 -TPPYEPL-TIQFLTSQGVERYKDFKLRDPYEQRYVPRGPPEMRDMMKVIEDNHLLDTVA 219
Query: 319 QVGRCFERLVKEFVVNLSVEVGLPESDEYRKVYVREKCVKFSPDVINKALGRSAT---AV 375
++ + ++ EF N + E + KV+VR K +FSP +INK A A
Sbjct: 220 EIDPFVKEVIWEFYANFHI---FNEETKKSKVFVRGKMYEFSPQMINKTFRSPAIKWHAK 276
Query: 376 TDEEP---SLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTSAI 432
+ E S +AK L+ G+V W K L T + A L I +NW+P+++ S +
Sbjct: 277 AEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLAGLYLICCINWIPSKNNSNV 333
Query: 433 SATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCA-VKLPIYFPSLITEIILQQ------ 485
+ A+++Y + +P +FG V++Q K A + ++ + FP+LI +++ Q
Sbjct: 334 TVDRAKMLYLLNERIPFNFGKLVYDQVEKAARSPGQIQNILPFPNLIYRVLMFQRVITLE 393
Query: 486 -------HPKIVRDD 493
HPK+ D
Sbjct: 394 NYEKPGPHPKLFNPD 408
>UniRef100_Q9SKW8 F5J5.2 [Arabidopsis thaliana]
Length = 463
Score = 80.9 bits (198), Expect = 1e-13
Identities = 85/345 (24%), Positives = 138/345 (39%), Gaps = 45/345 (13%)
Query: 186 PSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIV 245
P P K P SKK K ++ + K D DV P +
Sbjct: 66 PVAPMFIPKTEPGICMSKKTKSKVSTLTQRKRQCTAASAKFRSKAHLQDGDVTEIAPPLF 125
Query: 246 LA-----------EMKKFAGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREV 294
L+ +K+ + P P + F +EE ++K + R R +
Sbjct: 126 LSCYRENRRLLAVRDRKYPELKFPSETPYSSC----FLTEEGLGRYKVIGNRCFNDMRFL 181
Query: 295 GSDVLECKEVMALIEKAGLIKTVLQVGRCFERLVKEFVVNLSVEVGLPESDE----YRKV 350
D L+ GL+ TV ++ +V EF NL P+ +E V
Sbjct: 182 PLDGNNTASTQQLLFNGGLLPTVTEIDSYVHEVVMEFYANL------PDGEEGDNLAYSV 235
Query: 351 YVREKCVKFSPDVINKAL----------GRSATAVTDEEPSLDVVAKELTAGQVQKWPKK 400
+VR +FS +IN+ G S V + S+D VA L+ G+ W
Sbjct: 236 FVRRNMYEFSLAIINQMFQLPNPSYPLDGMSEIQVPE---SMDEVAIALSNGKANSW--- 289
Query: 401 KLLSTCNLSVKYAILNRIGAVNWVPTQHTSAISATLARLIYKIGTMVPVDFGNFVFEQTL 460
K+L++ LS + A+LN+I NW PT + S + L+Y + +P +FG +F++ L
Sbjct: 290 KMLTSRLLSPELALLNKICCHNWSPTVNRSVLKPERMTLLYMVAKALPFNFGKLIFDENL 349
Query: 461 KHAETC--AVKLPIYFPSLITEIILQQHPKIVRDDEEAMPKGSPI 503
E + L + P+LI +++ Q +IVR DE +P+
Sbjct: 350 GVFEFANPSSTLCLVLPNLIDQMLRFQ--RIVRSDEGDTTSSAPL 392
>UniRef100_Q8W0W1 Putative envelope protein [Vicia faba]
Length = 59
Score = 77.8 bits (190), Expect = 8e-13
Identities = 36/59 (61%), Positives = 42/59 (71%)
Query: 418 IGAVNWVPTQHTSAISATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFPS 476
IGA NWVPT H S IS L RLIY +GT D+G ++F+QTLKHA + AVK PI FPS
Sbjct: 1 IGAANWVPTNHKSTISTGLGRLIYAVGTRTEFDYGRYIFDQTLKHAGSYAVKGPIAFPS 59
>UniRef100_Q9M2B7 Hypothetical protein F23N14_70 [Arabidopsis thaliana]
Length = 539
Score = 70.5 bits (171), Expect = 1e-10
Identities = 84/364 (23%), Positives = 148/364 (40%), Gaps = 51/364 (14%)
Query: 180 TTASEAPSVP-------------QKKTKPSP-TTPKSKKKDLKGKGKATAEKSMSEKKKK 225
TT+S+ PSV K +P+ T P + L K +++
Sbjct: 127 TTSSQPPSVSIPPPSAPPLVLSDSKDAEPAGLTNPSAPPSPLAPKNITPVASPVADVPMP 186
Query: 226 APVIVSESDSDVEADVPDIVLAEMKKFAGRR--------IPKNV--PAAPLDNVSFHSEE 275
P+I +++ A VPD ++ + A R + + + P P F S
Sbjct: 187 DPLISPTAETAEGASVPDAAVSYAARAAAHRQVFAERDELDRTLRRPLVPPHTKRFLSAA 246
Query: 276 NALKWKYVYQRRLATEREVGSDVLECKEVMALIEKAGLIKTVLQVGRCFERLVKEFVVNL 335
A ++K++ +R ++ + D +E +G+ +TV+ V + +V+EF NL
Sbjct: 247 AAERYKHIAKRDFIFQKTLPLDPEVLTATKYFLEHSGMAQTVVAVEQFVPEVVREFYANL 306
Query: 336 SVEVGLPESDEYRK-----VYVREKCVKFSPDVINKALGRSATAVTDEEP------SLDV 384
PE EYR+ VYVR K +FSP +IN +A+ E P S D
Sbjct: 307 ------PEM-EYRECGLDLVYVRGKMYEFSPALINHMFSIDDSALDPEAPVTLSTASRDD 359
Query: 385 VAKELTAGQVQKWPKKKLLSTCNLSVKYAILNRIGAVNWVPTQHTSAISATLARLIYKIG 444
+A +T G ++W + L + +L+++ NW PT +TS + RLI
Sbjct: 360 LALMMTGGTTRRWLR---LQPADHLDTMKMLHKVCCGNWFPTTNTSTLRVDRLRLIDMGT 416
Query: 445 TMVPVDFGNFVFEQTLKHAETCAVKL-PIYFPSLITEII-----LQQHPKIVRDDEEAMP 498
+ G V T+ A + + +P+LI +++ ++ P+ DE +
Sbjct: 417 HGKSFNLGKLVVTHTMSLARLGPLSSHRLAYPNLIYQLLTFQRDVRSRPRDTLSDEPGVF 476
Query: 499 KGSP 502
P
Sbjct: 477 VNDP 480
>UniRef100_UPI00004307AD UPI00004307AD UniRef100 entry
Length = 679
Score = 61.6 bits (148), Expect = 6e-08
Identities = 55/239 (23%), Positives = 105/239 (43%), Gaps = 11/239 (4%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKY-YKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
K S K SS + S + + K P + + +K+ V ED S EEE P
Sbjct: 270 KISSKKEESSSEDSSEEEPQVKKVQPVAPFSFITKNVPVKKEESSSED--SSEEEPPVKK 327
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A+I S V + E+S+ +D++ +E+P ++ + V + TV +T SS
Sbjct: 328 VAASIAPVTPKKMS----VKNEESSSSEDSSEEEESPTKKSVSAK-VTSANTVIKKTESS 382
Query: 123 SKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDD--DDVPLFETLPESVGARLKRKRRT 180
++ D + K + ++ S +DS + D+ P+ P + K+ +
Sbjct: 383 TEDSDDSDENEKSVTKSTTKSIGKKDESSSEDSSESEDEKPVPTKTPVPTKTKADTKKES 442
Query: 181 TASEAPSVPQKKTKPSPTTPKSKKKDLK-GKGKATAEKSMSEKKKKAPVIVSESDSDVE 238
++ ++ S ++ KP KSK ++K G+ + + +EK+K + + D D+E
Sbjct: 443 SSEDSDSDSSEEEKPKTIVSKSKTTEIKQDTGEKSQKTPKAEKRKHKEIEQEKDDEDIE 501
Score = 58.9 bits (141), Expect = 4e-07
Identities = 56/235 (23%), Positives = 104/235 (43%), Gaps = 19/235 (8%)
Query: 11 TSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEE 70
T+ T++ S S + + A+K +S V+S ++ +S E+ +
Sbjct: 237 TTPMTKEESSSSEESSEEEAKKTNILQSTSVASKISSKKEESSSEDSSEEEPQVKKVQPV 296
Query: 71 ETLMGSDRDVVPDAEASAPQDTNVSQENPI--------PENPDVEDVIAEETVSTETPSS 122
++V E S+ +D++ +E P+ P P V EE+ S+E S
Sbjct: 297 APFSFITKNVPVKKEESSSEDSS-EEEPPVKKVAASIAPVTPKKMSVKNEESSSSEDSSE 355
Query: 123 SKKGD-KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
++ K SVS+K T ++ I+ + D DDSD+++ + ++ +S+G ++
Sbjct: 356 EEESPTKKSVSAKVTSANTVIKKTESSTEDSDDSDENEKSVTKSTTKSIG------KKDE 409
Query: 182 ASEAPSVPQKKTKPSPT---TPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSES 233
+S S + KP PT P K D K + + S S +++K IVS+S
Sbjct: 410 SSSEDSSESEDEKPVPTKTPVPTKTKADTKKESSSEDSDSDSSEEEKPKTIVSKS 464
Score = 45.8 bits (107), Expect = 0.003
Identities = 52/246 (21%), Positives = 100/246 (40%), Gaps = 33/246 (13%)
Query: 3 LKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
+K+ + K S S ++ TP + + +S +D S+ EEE +
Sbjct: 64 VKTSKASPVKKKESTDESSSEEEEETPKTQPKAVSTPKISKEQKSSDDSDSESEEESKTN 123
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A N + + + E+S+ + E+P V+ V+ + T T S+
Sbjct: 124 ITAKTNTKSIITSKTKVGANQKESSSEDSSESEDESP------VKPVLKKATPIPPTKST 177
Query: 123 SKKGDKGSVSSKETHSDE------PIQ---------VLVQNNSDGDDSDD--DDVPLFET 165
+KK + S S ++ +E PI+ + S DD+DD ++ + +T
Sbjct: 178 AKKEESSSDDSDDSSEEENAKPQLPIKAKSNIKSANITKPQESSSDDTDDSSEEATVTKT 237
Query: 166 LPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEK--- 222
P + ++ +++ E+ KKT +T + K K K ++++E S E+
Sbjct: 238 TP------MTKEESSSSEESSEEEAKKTNILQSTSVASKISSK-KEESSSEDSSEEEPQV 290
Query: 223 KKKAPV 228
KK PV
Sbjct: 291 KKVQPV 296
>UniRef100_UPI00004307AC UPI00004307AC UniRef100 entry
Length = 707
Score = 61.6 bits (148), Expect = 6e-08
Identities = 55/239 (23%), Positives = 105/239 (43%), Gaps = 11/239 (4%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKY-YKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
K S K SS + S + + K P + + +K+ V ED S EEE P
Sbjct: 323 KISSKKEESSSEDSSEEEPQVKKVQPVAPFSFITKNVPVKKEESSSED--SSEEEPPVKK 380
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A+I S V + E+S+ +D++ +E+P ++ + V + TV +T SS
Sbjct: 381 VAASIAPVTPKKMS----VKNEESSSSEDSSEEEESPTKKSVSAK-VTSANTVIKKTESS 435
Query: 123 SKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDD--DDVPLFETLPESVGARLKRKRRT 180
++ D + K + ++ S +DS + D+ P+ P + K+ +
Sbjct: 436 TEDSDDSDENEKSVTKSTTKSIGKKDESSSEDSSESEDEKPVPTKTPVPTKTKADTKKES 495
Query: 181 TASEAPSVPQKKTKPSPTTPKSKKKDLK-GKGKATAEKSMSEKKKKAPVIVSESDSDVE 238
++ ++ S ++ KP KSK ++K G+ + + +EK+K + + D D+E
Sbjct: 496 SSEDSDSDSSEEEKPKTIVSKSKTTEIKQDTGEKSQKTPKAEKRKHKEIEQEKDDEDIE 554
Score = 58.9 bits (141), Expect = 4e-07
Identities = 56/235 (23%), Positives = 104/235 (43%), Gaps = 19/235 (8%)
Query: 11 TSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEE 70
T+ T++ S S + + A+K +S V+S ++ +S E+ +
Sbjct: 290 TTPMTKEESSSSEESSEEEAKKTNILQSTSVASKISSKKEESSSEDSSEEEPQVKKVQPV 349
Query: 71 ETLMGSDRDVVPDAEASAPQDTNVSQENPI--------PENPDVEDVIAEETVSTETPSS 122
++V E S+ +D++ +E P+ P P V EE+ S+E S
Sbjct: 350 APFSFITKNVPVKKEESSSEDSS-EEEPPVKKVAASIAPVTPKKMSVKNEESSSSEDSSE 408
Query: 123 SKKGD-KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
++ K SVS+K T ++ I+ + D DDSD+++ + ++ +S+G ++
Sbjct: 409 EEESPTKKSVSAKVTSANTVIKKTESSTEDSDDSDENEKSVTKSTTKSIG------KKDE 462
Query: 182 ASEAPSVPQKKTKPSPT---TPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSES 233
+S S + KP PT P K D K + + S S +++K IVS+S
Sbjct: 463 SSSEDSSESEDEKPVPTKTPVPTKTKADTKKESSSEDSDSDSSEEEKPKTIVSKS 517
Score = 45.8 bits (107), Expect = 0.003
Identities = 52/246 (21%), Positives = 100/246 (40%), Gaps = 33/246 (13%)
Query: 3 LKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
+K+ + K S S ++ TP + + +S +D S+ EEE +
Sbjct: 117 VKTSKASPVKKKESTDESSSEEEEETPKTQPKAVSTPKISKEQKSSDDSDSESEEESKTN 176
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A N + + + E+S+ + E+P V+ V+ + T T S+
Sbjct: 177 ITAKTNTKSIITSKTKVGANQKESSSEDSSESEDESP------VKPVLKKATPIPPTKST 230
Query: 123 SKKGDKGSVSSKETHSDE------PIQ---------VLVQNNSDGDDSDD--DDVPLFET 165
+KK + S S ++ +E PI+ + S DD+DD ++ + +T
Sbjct: 231 AKKEESSSDDSDDSSEEENAKPQLPIKAKSNIKSANITKPQESSSDDTDDSSEEATVTKT 290
Query: 166 LPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEK--- 222
P + ++ +++ E+ KKT +T + K K K ++++E S E+
Sbjct: 291 TP------MTKEESSSSEESSEEEAKKTNILQSTSVASKISSK-KEESSSEDSSEEEPQV 343
Query: 223 KKKAPV 228
KK PV
Sbjct: 344 KKVQPV 349
>UniRef100_UPI00004307AB UPI00004307AB UniRef100 entry
Length = 605
Score = 61.6 bits (148), Expect = 6e-08
Identities = 55/239 (23%), Positives = 105/239 (43%), Gaps = 11/239 (4%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKY-YKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
K S K SS + S + + K P + + +K+ V ED S EEE P
Sbjct: 270 KISSKKEESSSEDSSEEEPQVKKVQPVAPFSFITKNVPVKKEESSSED--SSEEEPPVKK 327
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A+I S V + E+S+ +D++ +E+P ++ + V + TV +T SS
Sbjct: 328 VAASIAPVTPKKMS----VKNEESSSSEDSSEEEESPTKKSVSAK-VTSANTVIKKTESS 382
Query: 123 SKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDD--DDVPLFETLPESVGARLKRKRRT 180
++ D + K + ++ S +DS + D+ P+ P + K+ +
Sbjct: 383 TEDSDDSDENEKSVTKSTTKSIGKKDESSSEDSSESEDEKPVPTKTPVPTKTKADTKKES 442
Query: 181 TASEAPSVPQKKTKPSPTTPKSKKKDLK-GKGKATAEKSMSEKKKKAPVIVSESDSDVE 238
++ ++ S ++ KP KSK ++K G+ + + +EK+K + + D D+E
Sbjct: 443 SSEDSDSDSSEEEKPKTIVSKSKTTEIKQDTGEKSQKTPKAEKRKHKEIEQEKDDEDIE 501
Score = 58.9 bits (141), Expect = 4e-07
Identities = 56/235 (23%), Positives = 104/235 (43%), Gaps = 19/235 (8%)
Query: 11 TSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEE 70
T+ T++ S S + + A+K +S V+S ++ +S E+ +
Sbjct: 237 TTPMTKEESSSSEESSEEEAKKTNILQSTSVASKISSKKEESSSEDSSEEEPQVKKVQPV 296
Query: 71 ETLMGSDRDVVPDAEASAPQDTNVSQENPI--------PENPDVEDVIAEETVSTETPSS 122
++V E S+ +D++ +E P+ P P V EE+ S+E S
Sbjct: 297 APFSFITKNVPVKKEESSSEDSS-EEEPPVKKVAASIAPVTPKKMSVKNEESSSSEDSSE 355
Query: 123 SKKGD-KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
++ K SVS+K T ++ I+ + D DDSD+++ + ++ +S+G ++
Sbjct: 356 EEESPTKKSVSAKVTSANTVIKKTESSTEDSDDSDENEKSVTKSTTKSIG------KKDE 409
Query: 182 ASEAPSVPQKKTKPSPT---TPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSES 233
+S S + KP PT P K D K + + S S +++K IVS+S
Sbjct: 410 SSSEDSSESEDEKPVPTKTPVPTKTKADTKKESSSEDSDSDSSEEEKPKTIVSKS 464
Score = 45.8 bits (107), Expect = 0.003
Identities = 52/246 (21%), Positives = 100/246 (40%), Gaps = 33/246 (13%)
Query: 3 LKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
+K+ + K S S ++ TP + + +S +D S+ EEE +
Sbjct: 64 VKTSKASPVKKKESTDESSSEEEEETPKTQPKAVSTPKISKEQKSSDDSDSESEEESKTN 123
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A N + + + E+S+ + E+P V+ V+ + T T S+
Sbjct: 124 ITAKTNTKSIITSKTKVGANQKESSSEDSSESEDESP------VKPVLKKATPIPPTKST 177
Query: 123 SKKGDKGSVSSKETHSDE------PIQ---------VLVQNNSDGDDSDD--DDVPLFET 165
+KK + S S ++ +E PI+ + S DD+DD ++ + +T
Sbjct: 178 AKKEESSSDDSDDSSEEENAKPQLPIKAKSNIKSANITKPQESSSDDTDDSSEEATVTKT 237
Query: 166 LPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEK--- 222
P + ++ +++ E+ KKT +T + K K K ++++E S E+
Sbjct: 238 TP------MTKEESSSSEESSEEEAKKTNILQSTSVASKISSK-KEESSSEDSSEEEPQV 290
Query: 223 KKKAPV 228
KK PV
Sbjct: 291 KKVQPV 296
>UniRef100_Q9S9S8 F28J9.1 [Arabidopsis thaliana]
Length = 505
Score = 59.7 bits (143), Expect = 2e-07
Identities = 44/133 (33%), Positives = 62/133 (46%), Gaps = 9/133 (6%)
Query: 358 KFSPDVINKALGRSATA-VTDEEPSLDVVAKELTAGQVQKWPKKKLLSTCNLSVKYAILN 416
KFS + + TA + DEE VA+ LT G+VQ ++ S L
Sbjct: 152 KFSSPLAEQRTQCIQTAGLVDEE-----VARYLTDGRVQNLQN---IAMNKFSENCKSLF 203
Query: 417 RIGAVNWVPTQHTSAISATLARLIYKIGTMVPVDFGNFVFEQTLKHAETCAVKLPIYFPS 476
+I NW P + S RL+Y+I +P+DFG V++ L+ A K + FPS
Sbjct: 204 KISCRNWSPQTNEGYASIDRGRLVYRIAHKMPIDFGEMVYDHVLQLALMSVSKFFLMFPS 263
Query: 477 LITEIILQQHPKI 489
LI +I QHP I
Sbjct: 264 LIYRLIQTQHPDI 276
>UniRef100_UPI00004307A7 UPI00004307A7 UniRef100 entry
Length = 386
Score = 58.9 bits (141), Expect = 4e-07
Identities = 53/230 (23%), Positives = 99/230 (43%), Gaps = 23/230 (10%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
SS+ TSS+ EK+L K T A+ K+ ++ +S+EEEE P
Sbjct: 48 SSEDTSSENEKAL-----KVVTKAKTAVKTSKASPVKKKESTDESSSEEEEETPKTQPKA 102
Query: 67 INEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKG 126
++ + D+E+ TN++ + + + +I T T+ ++ K+
Sbjct: 103 VSTPKISKEQKSSDDSDSESEEESKTNITAKT------NTKSII---TSKTKVGANQKES 153
Query: 127 DKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAP 186
S E ++ I+ + D DDSD+++ + ++ +S+G ++ +S
Sbjct: 154 SSEDSSESEDETNTVIKKTESSTEDSDDSDENEKSVTKSTTKSIG------KKDESSSED 207
Query: 187 SVPQKKTKPSPT---TPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSES 233
S + KP PT P K D K + + S S +++K IVS+S
Sbjct: 208 SSESEDEKPVPTKTPVPTKTKADTKKESSSEDSDSDSSEEEKPKTIVSKS 257
Score = 51.6 bits (122), Expect = 6e-05
Identities = 48/222 (21%), Positives = 97/222 (43%), Gaps = 15/222 (6%)
Query: 3 LKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
+K+ + K S S ++ TP + + +S +D S+ EEE +
Sbjct: 70 VKTSKASPVKKKESTDESSSEEEEETPKTQPKAVSTPKISKEQKSSDDSDSESEEESKTN 129
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
A N + +++ S V + + S+ +D++ S++ + VI + STE
Sbjct: 130 ITAKTNTK-SIITSKTKVGANQKESSSEDSSESED-------ETNTVIKKTESSTEDSDD 181
Query: 123 SKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTA 182
S + +K SV+ T S I +++S+ +D+ P+ P + K+ +++
Sbjct: 182 SDENEK-SVTKSTTKS---IGKKDESSSEDSSESEDEKPVPTKTPVPTKTKADTKKESSS 237
Query: 183 SEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKK 224
++ S ++ KP KSK ++K + T EK S K++
Sbjct: 238 EDSDSDSSEEEKPKTIVSKSKTTEIK---QDTGEKKDSRKRE 276
>UniRef100_Q79LN3 Biofilm-associated surface protein [Staphylococcus aureus]
Length = 2276
Score = 58.2 bits (139), Expect = 7e-07
Identities = 66/313 (21%), Positives = 131/313 (41%), Gaps = 26/313 (8%)
Query: 42 SSSHVLLEDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIP 101
S V ED + E + + N +T + S D V E + +Q++
Sbjct: 71 SEKDVTTEDDNNAEVQNSAQTVDKSENSNDTAVESTNDSVKTDETKETSENKSAQDD--- 127
Query: 102 ENPDVEDVIAEETVSTETPSSS-KKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDV 160
+N + EE+ +T + SS + K + + ET DE QNN D D + DDD
Sbjct: 128 DNIKEDSNTQEESTNTSSQSSEVPQTKKDTNETSETAIDEDASTKEQNNKDNDTAQDDD- 186
Query: 161 PLFETLPESVGARLKRKRRTTASEAPSVPQ-KKTKPSPTTPKSKKKDLKGKGKATAEKSM 219
+ E + + + T+S++ VPQ KK +P ++ K+ D + + A EK++
Sbjct: 187 ----NIKED--SNTQEESTNTSSQSSEVPQTKKEQPDKSSNSIKEPDKQQEEVAKEEKAI 240
Query: 220 SE---KKKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRRIPKNVPAAPLDNVSFHSEEN 276
+E K K+ + +++D + E+++ + + K K+ + L++ SE
Sbjct: 241 TEIADKNKELELKNNKTDKNEESELESNLSSSENK-------KDTVESFLNSQLSDSETK 293
Query: 277 ALKWKYVYQRRLATEREVGSDVLECKEVMALIEKAGLIKTVLQVGRCFERLVKEFVVNLS 336
+ AT+ E+ +++L +LIE A K + + + +
Sbjct: 294 KIMENANIDYDKATDEEINTEILRA----SLIEMANNKKKTETLATPQRTMFRAMATPTA 349
Query: 337 VEVGLPESDEYRK 349
+ + + +E +K
Sbjct: 350 LRAAVNQDEELQK 362
>UniRef100_Q6P1U1 LOC395027 protein [Xenopus tropicalis]
Length = 916
Score = 57.8 bits (138), Expect = 9e-07
Identities = 65/274 (23%), Positives = 105/274 (37%), Gaps = 46/274 (16%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKA--RTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFH 61
+S S K T K +Q+ K A +TP +K +S S SSS ED S ++++
Sbjct: 126 ESDSEKETKKPPVKKPAQTPKGAALKTPTQKKAESSSSESSSS----EDEASKKKKQSVT 181
Query: 62 DAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPS 121
P + + +D D+ +S D+ +++ P+ P + A + + +T
Sbjct: 182 KVPPKQTVVKAGLANDNQKAADSSSSEDSDSPPEKKSAAPKTPPAKPATAAKPQAKKTAG 241
Query: 122 SSKKGDKGS--------------------------VSSKETHSDEPIQVLVQNNSDGDDS 155
+ S ++SK+ S +P +SD DS
Sbjct: 242 KKSSSEDSSDSSDEEAKPAKSKPKPGVYSAVPPPTMNSKKKASSQPGTKTKAESSDSSDS 301
Query: 156 DDDDVP------LFETLPESVGARLKRKRRTTASEAPSVPQKKTKPS-PTTPK--SKKKD 206
DD+ P + + SV L +K T++ + KPS PK + K D
Sbjct: 302 SDDEQPPKKAKIVPAKITASVPKPLAKKAETSSDSESDSSSEDEKPSVKVAPKKAAAKPD 361
Query: 207 LKGKGKATAEKSMSEKKKKAPVIVSESDSDVEAD 240
K A A+KS KK S SDSD +D
Sbjct: 362 AKSTPAAAAKKSAPAKKAS-----SSSDSDSSSD 390
Score = 48.9 bits (115), Expect = 4e-04
Identities = 83/330 (25%), Positives = 124/330 (37%), Gaps = 70/330 (21%)
Query: 8 SKSTSSKTEKS--LSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPA 65
+KS ++ T K+ S++ ++T K K+ SS D +SDE+E+ PA
Sbjct: 404 AKSAATPTSKTPTTSKATPTSKTLPAKQGTPKTSKKDSSSSDSSDSSSDEDEKKTPAKPA 463
Query: 66 NINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKK 125
P A+ + + +E+ + D D +E+ E SS+K
Sbjct: 464 AKTTPAKPAAKTTPAKPAAKTTPGKKAPTKKES---SSSDSSDSSSED----EKKSSAKP 516
Query: 126 GDK---GSVSSKETHSDEPIQVLVQNNSDGDDSDDDDV---------------PLFETLP 167
G K G +SK S V + S DSD D P ++LP
Sbjct: 517 GVKTTHGKAASKTDASAAAKSVPAKKASSSSDSDSDSSEEETTKTTKPLNKPSPAVKSLP 576
Query: 168 ESVGARLKRKRRTTASEAPSVPQKKTKPS--------PTTPKSKKKDLK----------- 208
K + + +S + S +KKTKP+ P TPK + ++ K
Sbjct: 577 SK-----KAESSSDSSSSDSDSKKKTKPAKPPAKATTPVTPKVQVQNKKPSKANSTDSSS 631
Query: 209 ---GKGKATAEKSMSEK----KKKAPVIV----------SESDSDVEADVPDIVLAEMKK 251
GK K T KS + K KK+PV V S+S SD E P A
Sbjct: 632 EEEGKSKQTTGKSPAAKATVPPKKSPVAVNKAKASSSSSSDSSSDDEKQKPKQPPAAKDV 691
Query: 252 FAGRRIPKNVPAAPLDNVS--FHSEENALK 279
G + K P D+ S SE++ LK
Sbjct: 692 KQGAKATKPAPKKAADSSSEDSSSEDDDLK 721
Score = 43.9 bits (102), Expect = 0.013
Identities = 61/266 (22%), Positives = 98/266 (35%), Gaps = 41/266 (15%)
Query: 5 SGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAP 64
S SS S S +E+ +++ K P+ S SS D +S + + P
Sbjct: 544 SSSSDSDSDSSEEETTKTTKPLNKPSPAVKSLPSKKAESS----SDSSSSDSDSKKKTKP 599
Query: 65 ANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSK 124
A P A+A+ P V +N P + D +EE ++ +
Sbjct: 600 AK---------------PPAKATTPVTPKVQVQNKKPSKANSTDSSSEEEGKSKQTTGKS 644
Query: 125 KGDKGSVSSKETHSDEPIQV---LVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
K +V K++ P+ V ++S D S DD+ + P+ A K+
Sbjct: 645 PAAKATVPPKKS----PVAVNKAKASSSSSSDSSSDDE----KQKPKQPPAAKDVKQGAK 696
Query: 182 ASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA--EKSMSEKKKKAPVIVSESDSDVEA 239
A++ P+K S S+ DLK TA + ++ K K AP S S+S
Sbjct: 697 ATKP--APKKAADSSSEDSSSEDDDLKATKTNTAISKSPVTPKAKPAPKKESSSESSDSE 754
Query: 240 DVPDIVLAEMKKFAGRRIPKNVPAAP 265
D E +K PK + P
Sbjct: 755 D-------EKQKGKNISTPKTANSTP 773
Score = 41.2 bits (95), Expect = 0.084
Identities = 49/194 (25%), Positives = 77/194 (39%), Gaps = 26/194 (13%)
Query: 82 PDAEASAPQDTNVSQENPIPENPDVEDVIAEE------TVSTETPSSSKKGDKGSVSSKE 135
PDA+ P + ++ P E+ ED +EE ++T++ K KG + K
Sbjct: 57 PDAKKRKPSANGLPKKKPTNESSSSEDSSSEEDEPPAKKIATQSAEGKKPVVKGVQAKKA 116
Query: 136 THSDEPIQVLVQNNSDGDDSDDD--DVPLFETLPESVGARLK----RKRRTTASEAPS-- 187
S E ++SD DS+ + P+ + GA LK +K +++SE+ S
Sbjct: 117 KSSSE-------DSSDESDSEKETKKPPVKKPAQTPKGAALKTPTQKKAESSSSESSSSE 169
Query: 188 -VPQKKTKPSPTTPKSK----KKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVP 242
KK K S T K K L + A+ S SE P S + A
Sbjct: 170 DEASKKKKQSVTKVPPKQTVVKAGLANDNQKAADSSSSEDSDSPPEKKSAAPKTPPAKPA 229
Query: 243 DIVLAEMKKFAGRR 256
+ KK AG++
Sbjct: 230 TAAKPQAKKTAGKK 243
Score = 40.4 bits (93), Expect = 0.14
Identities = 68/315 (21%), Positives = 113/315 (35%), Gaps = 46/315 (14%)
Query: 4 KSGSSKSTSSKTEKSLS-------QSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSD-- 54
K+ S T +K E S S Q KKA+ K S ++ D SD
Sbjct: 282 KASSQPGTKTKAESSDSSDSSDDEQPPKKAKIVPAKITASVPKPLAKKAETSSDSESDSS 341
Query: 55 -EEEEPF-----HDAPANINEEETLMGSDRDVVP--------DAEASAPQDTNVSQENPI 100
E+E+P A A + + T + + P D+++S+ ++T I
Sbjct: 342 SEDEKPSVKVAPKKAAAKPDAKSTPAAAAKKSAPAKKASSSSDSDSSSDEETTPKPSTKI 401
Query: 101 -PENPDVEDVIAEETVSTETPSS----SKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDS 155
P T S TP+S +K+G + + SD ++SD D+
Sbjct: 402 TPAKSAATPTSKTPTTSKATPTSKTLPAKQGTPKTSKKDSSSSDSS-----DSSSDEDEK 456
Query: 156 DDDDVPLFETLPESVGAR-LKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGK---- 210
P +T P A+ K + P KK S + S +D K
Sbjct: 457 KTPAKPAAKTTPAKPAAKTTPAKPAAKTTPGKKAPTKKESSSSDSSDSSSEDEKKSSAKP 516
Query: 211 ------GKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRRIP--KNVP 262
GKA ++ S K P + S SD ++D + + K + P K++P
Sbjct: 517 GVKTTHGKAASKTDASAAAKSVPAKKASSSSDSDSDSSEEETTKTTKPLNKPSPAVKSLP 576
Query: 263 AAPLDNVSFHSEENA 277
+ ++ S S ++
Sbjct: 577 SKKAESSSDSSSSDS 591
>UniRef100_Q7XTT4 OSJNBa0058K23.21 protein [Oryza sativa]
Length = 707
Score = 57.8 bits (138), Expect = 9e-07
Identities = 66/281 (23%), Positives = 113/281 (39%), Gaps = 51/281 (18%)
Query: 5 SGSSKSTSSKTEKSLSQSR--KKARTPARKYYKSKSHGVSSSHVLLEDVTSD---EEEEP 59
S +S S+ +++SLS KK P +K ++G S V + +SD +EE
Sbjct: 136 STNSSSSDDSSDESLSDDEPVKKPAAPLKKPVALATNG--SKKVETDSSSSDSSSDEESD 193
Query: 60 FHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTET 119
D ++ + + + ++++S + S E+ + P V E + S+++
Sbjct: 194 EDDKKTAAPVKKPSVAAIQKKTQESDSSDSDSDSESDEDVPTKAPAVAKKKEESSESSDS 253
Query: 120 PSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRR 179
S S D+ + KE S + ++SD + D D P T+P K+
Sbjct: 254 ESDSDSDDEAAAVKKEEESSD------SSDSDSESESDSDEPAKPTIPAKRPLTKDTKKG 307
Query: 180 TTASEAPSV------------PQKKTKPSPTT------PKSKKKDLKGKGKATAEKSMSE 221
+ E+ PQKK K S T+ PK+ KK++ ++ + S E
Sbjct: 308 QSKDESEDSSDESSEESGDEPPQKKIKDSTTSGTTKPSPKATKKEISSDDESDEDDSSDE 367
Query: 222 K------------KKKAPVI--------VSESDSDVEADVP 242
KK+APV SE DSD+E+D P
Sbjct: 368 SSDEDVKQKQTQAKKQAPVAQESSSSDESSEEDSDMESDEP 408
Score = 45.1 bits (105), Expect = 0.006
Identities = 51/233 (21%), Positives = 87/233 (36%), Gaps = 22/233 (9%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
++ + S S S+S + T A K K SS D SD + + +A A
Sbjct: 215 TQESDSSDSDSDSESDEDVPTKAPAVAKKKEESSESS-----DSESDSDSDD--EAAAVK 267
Query: 68 NEEETLMGSDRDVVPDAEASAPQDTNVSQENPIP----------ENPDVEDVIAEETVST 117
EEE+ SD D ++++ P + + P+ E+ D D +EE+
Sbjct: 268 KEEESSDSSDSDSESESDSDEPAKPTIPAKRPLTKDTKKGQSKDESEDSSDESSEES-GD 326
Query: 118 ETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDS----DDDDVPLFETLPESVGAR 173
E P K S ++K + ++ + SD DDS D+DV +T +
Sbjct: 327 EPPQKKIKDSTTSGTTKPSPKATKKEISSDDESDEDDSSDESSDEDVKQKQTQAKKQAPV 386
Query: 174 LKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKA 226
+ + S + +P+ T K + G K+ + E K A
Sbjct: 387 AQESSSSDESSEEDSDMESDEPAKTPQKKETAVSVGSNKSATKPGQEEPKTPA 439
Score = 37.7 bits (86), Expect = 0.93
Identities = 54/266 (20%), Positives = 98/266 (36%), Gaps = 38/266 (14%)
Query: 1 MGLKSGSSKSTSSKTEKSLSQSR--KKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEE 58
MG S S + T S+S+ + KK + A + V + V S EE +
Sbjct: 1 MGKSSKKSAVEVAPTSVSVSEGKSGKKGKRNAEDEIEKAVSAKKQKTVREKVVPSKEEAK 60
Query: 59 PFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTE 118
P E + D + + P+ T ++ P + D ++++ S +
Sbjct: 61 KVKKQPPPKKVESSSSEEDSSESEEEVKAQPKKTVQPKKAAQPAKEESSDDSSDDSSSDD 120
Query: 119 TPSSSK--KGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDV-----------PLFET 165
P+ + +K ++S+ N+S DDS D+ + PL +
Sbjct: 121 EPAKKPVARPNKAALST--------------NSSSSDDSSDESLSDDEPVKKPAAPLKKP 166
Query: 166 LPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKK 225
+ + K + +++S++ S + T KK + K T E S+
Sbjct: 167 VALATNGSKKVETDSSSSDSSSDEESDEDDKKTAAPVKKPSVAAIQKKTQESDSSD---- 222
Query: 226 APVIVSESDSDVEADVPDIVLAEMKK 251
S+SDS+ + DVP A KK
Sbjct: 223 -----SDSDSESDEDVPTKAPAVAKK 243
>UniRef100_Q5TVN3 ENSANGP00000027660 [Anopheles gambiae str. PEST]
Length = 511
Score = 57.8 bits (138), Expect = 9e-07
Identities = 65/252 (25%), Positives = 104/252 (40%), Gaps = 24/252 (9%)
Query: 9 KSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSS--SHVLLEDVTSDEEEEPFHDAPAN 66
+S S E S S +K + P K K SS S +DV+S++ E+ + +
Sbjct: 176 QSLKSAKEPSKSHPKKGSEKPDSKNKKPSEDSKSSEDSENQQDDVSSEKPEDEAAEESDD 235
Query: 67 INE-----EETLMGSDRDVV--PDAEASAPQDTNVSQENPIPENPDV-----EDVIAEET 114
+ +E+ + V PD++ P + + S E P E DV ED +AEE
Sbjct: 236 SKQSQKSGKESSKSQPKKVSDKPDSKNKKPSEDSKSSEGPEIEQDDVSSENPEDKVAEE- 294
Query: 115 VSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARL 174
S + SSK + S S + S++P +N +DS + P E + V +
Sbjct: 295 -SDDPKQSSKSAKEPSKSQPKKESEKPDS---KNKKPSEDSKSSEDP--ENQQDDVSSEK 348
Query: 175 KRKRRTTASEAPSVPQKKTK-PSPTTPK--SKKKDLKGKGKATAEKSMSEKKKKAPVIVS 231
S+ P K K S + PK S+K D+K K + KS + + + + S
Sbjct: 349 PEDEAAEESDDPKQSPKSAKESSKSQPKKGSEKPDIKNKKPSKDSKSSEDPENQQDDVSS 408
Query: 232 ESDSDVEADVPD 243
E D A+ D
Sbjct: 409 EKPEDEAAEESD 420
Score = 55.8 bits (133), Expect = 3e-06
Identities = 64/259 (24%), Positives = 101/259 (38%), Gaps = 34/259 (13%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLE--DVTSD-------EEE 57
S +S S E S SQ +K + P K K SS +E DV+S+ EE
Sbjct: 236 SKQSQKSGKESSKSQPKKVSDKPDSKNKKPSEDSKSSEGPEIEQDDVSSENPEDKVAEES 295
Query: 58 EPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDV-----EDVIAE 112
+ + + E PD++ P + + S E+P + DV ED AE
Sbjct: 296 DDPKQSSKSAKEPSKSQPKKESEKPDSKNKKPSEDSKSSEDPENQQDDVSSEKPEDEAAE 355
Query: 113 ETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQN------NSDGDDSDDDDVPLFETL 166
E+ + P S K K S S+ E + + +S+ ++ DDV
Sbjct: 356 ES---DDPKQSPKSAKESSKSQPKKGSEKPDIKNKKPSKDSKSSEDPENQQDDVS--SEK 410
Query: 167 PESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPK--SKKKDLKGKGKATAEKSMSEKKK 224
PE A + + P+ +PS + PK S+K D+K K + KS + +
Sbjct: 411 PEDEAAE-------ESDDPKQSPKSGKEPSKSQPKKGSEKPDIKNKNPSADSKSSEDPEN 463
Query: 225 KAPVIVSESDSDVEADVPD 243
+ + SE D A+ D
Sbjct: 464 QQDDVSSEKPEDEAAEESD 482
Score = 51.2 bits (121), Expect = 8e-05
Identities = 58/231 (25%), Positives = 92/231 (39%), Gaps = 35/231 (15%)
Query: 9 KSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSS----------SHVLLEDVTSDEEEE 58
+S+ S E S SQ +K++ P K K SS S ED ++E ++
Sbjct: 300 QSSKSAKEPSKSQPKKESEKPDSKNKKPSEDSKSSEDPENQQDDVSSEKPEDEAAEESDD 359
Query: 59 PFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDV-----EDVIAEE 113
P +P + E PD + P + S E+P + DV ED AEE
Sbjct: 360 P-KQSPKSAKESSKSQPKKGSEKPDIKNKKPSKDSKSSEDPENQQDDVSSEKPEDEAAEE 418
Query: 114 TVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGAR 173
S + S K G + S S + S++P ++N + DS + P
Sbjct: 419 --SDDPKQSPKSGKEPSKSQPKKGSEKPD---IKNKNPSADSKSSEDP------------ 461
Query: 174 LKRKRRTTASEAPSVPQKKTKPSP-TTPKSKKKDLKGKGKATAEKSMSEKK 223
+ ++ +SE P + P +PKS K+ K + K +EK S+ K
Sbjct: 462 -ENQQDDVSSEKPEDEAAEESDDPKQSPKSAKEPSKSQLKKGSEKPDSKNK 511
Score = 41.2 bits (95), Expect = 0.084
Identities = 61/253 (24%), Positives = 95/253 (37%), Gaps = 37/253 (14%)
Query: 21 QSRKKARTPARKYYKSKSHGVSSSH-VLLEDVTSDEEEEPFHDAPANINEEETLMGSDRD 79
QS K + P++ K S S + ED S E+ E D ++N E+
Sbjct: 35 QSLKSGKEPSKSQPKKGSDKPDSKNKKTSEDSKSSEDPENQQD---DVNSEK-------- 83
Query: 80 VVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGDK--GSVSSKETH 137
P+ EA+ D +P A+E ++ S K D+ S S +
Sbjct: 84 --PEDEAAEESD-----------DPKQSSKSAKEPSKSQLKKESDKPDRKEPSKSQPKKG 130
Query: 138 SDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTK-PS 196
S++P +N +DS + P E + V + S+ P K K PS
Sbjct: 131 SEKPDS---KNKKPSEDSKSSEDP--ENQQDDVSSEKPEDEAAEESDDPKQSLKSAKEPS 185
Query: 197 PTTPK--SKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMK--KF 252
+ PK S+K D K K + KS + + + + SE D A+ D K K
Sbjct: 186 KSHPKKGSEKPDSKNKKPSEDSKSSEDSENQQDDVSSEKPEDEAAEESDDSKQSQKSGKE 245
Query: 253 AGRRIPKNVPAAP 265
+ + PK V P
Sbjct: 246 SSKSQPKKVSDKP 258
Score = 39.7 bits (91), Expect = 0.24
Identities = 50/228 (21%), Positives = 87/228 (37%), Gaps = 52/228 (22%)
Query: 21 QSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEETLMGSDRDV 80
QS K A+ P++ K +S + +EP P +E+
Sbjct: 97 QSSKSAKEPSKSQLKKESD-------------KPDRKEPSKSQPKKGSEK---------- 133
Query: 81 VPDAEASAPQDTNVSQENPIPENPDV-----EDVIAEETVSTETPSSSKKGDKGSVSSKE 135
PD++ P + + S E+P + DV ED AEE S + S K + S S +
Sbjct: 134 -PDSKNKKPSEDSKSSEDPENQQDDVSSEKPEDEAAEE--SDDPKQSLKSAKEPSKSHPK 190
Query: 136 THSDEPIQVLVQNNSDG---DDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKK 192
S++P + + D +DS++ + PE A ++
Sbjct: 191 KGSEKPDSKNKKPSEDSKSSEDSENQQDDVSSEKPEDEAA------------------EE 232
Query: 193 TKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEAD 240
+ S + KS K+ K + K ++K S+ KK + S ++E D
Sbjct: 233 SDDSKQSQKSGKESSKSQPKKVSDKPDSKNKKPSEDSKSSEGPEIEQD 280
Score = 35.4 bits (80), Expect = 4.6
Identities = 39/169 (23%), Positives = 68/169 (40%), Gaps = 24/169 (14%)
Query: 90 QDTNVSQENPIPENPDVEDVIAEETVSTETPSSS-------------KKGDKGSVSSKET 136
Q +VS E P E + D + S + PS S KK + S SS++
Sbjct: 13 QQDDVSSEKPEDEAAEESDDPKQSLKSGKEPSKSQPKKGSDKPDSKNKKTSEDSKSSEDP 72
Query: 137 HSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPS 196
+ + + + + DD ++ E ++LK++ S+ P + +PS
Sbjct: 73 ENQQDDVNSEKPEDEAAEESDDPKQSSKSAKEPSKSQLKKE-----SDKPD----RKEPS 123
Query: 197 PTTPK--SKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPD 243
+ PK S+K D K K + KS + + + + SE D A+ D
Sbjct: 124 KSQPKKGSEKPDSKNKKPSEDSKSSEDPENQQDDVSSEKPEDEAAEESD 172
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 937,740,921
Number of Sequences: 2790947
Number of extensions: 42037467
Number of successful extensions: 203606
Number of sequences better than 10.0: 3427
Number of HSP's better than 10.0 without gapping: 176
Number of HSP's successfully gapped in prelim test: 3429
Number of HSP's that attempted gapping in prelim test: 183971
Number of HSP's gapped (non-prelim): 12071
length of query: 577
length of database: 848,049,833
effective HSP length: 133
effective length of query: 444
effective length of database: 476,853,882
effective search space: 211723123608
effective search space used: 211723123608
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0280b.3


