

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0143c.1
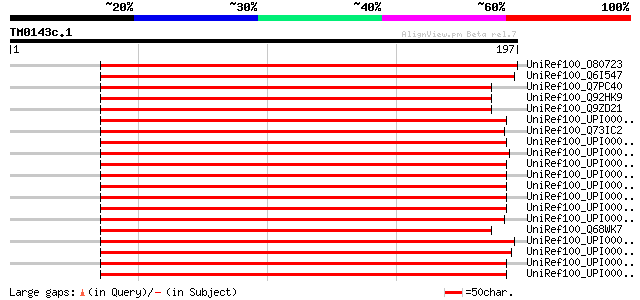
(197 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80723 Putative peptide chain release factor [Arabidop... 262 4e-69
UniRef100_Q6I547 Putative peptide chain release factor [Oryza sa... 254 7e-67
UniRef100_Q7PC40 Peptide chain release factor RF-1 [Rickettsia s... 189 3e-47
UniRef100_Q92HK9 Peptide chain release factor 1 [Rickettsia cono... 189 3e-47
UniRef100_Q9ZD21 Peptide chain release factor 1 [Rickettsia prow... 188 8e-47
UniRef100_UPI000028C765 UPI000028C765 UniRef100 entry 187 1e-46
UniRef100_Q73IC2 Peptide chain release factor 1 [Wolbachia pipie... 186 3e-46
UniRef100_UPI00003263DA UPI00003263DA UniRef100 entry 185 5e-46
UniRef100_UPI0000346082 UPI0000346082 UniRef100 entry 184 9e-46
UniRef100_UPI00002F0227 UPI00002F0227 UniRef100 entry 184 9e-46
UniRef100_UPI00002CD112 UPI00002CD112 UniRef100 entry 184 9e-46
UniRef100_UPI00002B1522 UPI00002B1522 UniRef100 entry 184 9e-46
UniRef100_UPI00002F6EDB UPI00002F6EDB UniRef100 entry 184 1e-45
UniRef100_UPI00002EC134 UPI00002EC134 UniRef100 entry 184 1e-45
UniRef100_UPI00002D4F5D UPI00002D4F5D UniRef100 entry 184 1e-45
UniRef100_Q68WK7 Peptide chain release factor 1 [Rickettsia typhi] 183 2e-45
UniRef100_UPI00003273D8 UPI00003273D8 UniRef100 entry 183 2e-45
UniRef100_UPI00002DDC72 UPI00002DDC72 UniRef100 entry 183 2e-45
UniRef100_UPI0000349FB6 UPI0000349FB6 UniRef100 entry 182 3e-45
UniRef100_UPI00003243C1 UPI00003243C1 UniRef100 entry 182 3e-45
>UniRef100_O80723 Putative peptide chain release factor [Arabidopsis thaliana]
Length = 395
Score = 262 bits (669), Expect = 4e-69
Identities = 125/162 (77%), Positives = 153/162 (94%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ SV + + +VDVQL+NEDL++DTYRSGGSGGQHANTTNSAVRI H+PTG++V+IQ
Sbjct: 233 TSAISVAILPQADEVDVQLRNEDLRIDTYRSGGSGGQHANTTNSAVRIIHLPTGMMVSIQ 292
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQHMN+AKALK+LCA+LYE+ERLR+ S+RSKLRS+QIGSGDRS RIRTYNFPQGRVT
Sbjct: 293 DERSQHMNRAKALKVLCARLYEIERLRIQSSRSKLRSDQIGSGDRSGRIRTYNFPQGRVT 352
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTSS 197
DHRVGIT+H+I+D+MQGENLD+FIDALLLR+EMDA+ASF+S+
Sbjct: 353 DHRVGITHHAIEDMMQGENLDMFIDALLLRQEMDAIASFSST 394
>UniRef100_Q6I547 Putative peptide chain release factor [Oryza sativa]
Length = 350
Score = 254 bits (650), Expect = 7e-67
Identities = 125/161 (77%), Positives = 146/161 (90%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ SV + + +VDVQL+NEDL++DTYRSGGSGGQ NTT+SAVRITH+PTG +VAIQ
Sbjct: 190 TSAVSVAILPQADEVDVQLRNEDLRIDTYRSGGSGGQSVNTTDSAVRITHVPTGTVVAIQ 249
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQH NKAKALK+L A+LYE+ER RLH +RSKLRSEQIGSGDRSERIRTYNFPQGRVT
Sbjct: 250 DERSQHQNKAKALKVLRARLYEIERHRLHMDRSKLRSEQIGSGDRSERIRTYNFPQGRVT 309
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTS 196
DHRVGIT+HSI+DVM+GE+LD+FIDALLLR EMDA+ASF S
Sbjct: 310 DHRVGITHHSIEDVMEGESLDIFIDALLLRYEMDAIASFAS 350
>UniRef100_Q7PC40 Peptide chain release factor RF-1 [Rickettsia sibirica]
Length = 355
Score = 189 bits (480), Expect = 3e-47
Identities = 88/152 (57%), Positives = 122/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER + R+ R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRKKEQERADSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD F++AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKNGQLDEFVEALIADDE 348
>UniRef100_Q92HK9 Peptide chain release factor 1 [Rickettsia conorii]
Length = 355
Score = 189 bits (480), Expect = 3e-47
Identities = 88/152 (57%), Positives = 122/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER + R+ R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRKKEQERADSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD F++AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKNGQLDEFVEALIADDE 348
>UniRef100_Q9ZD21 Peptide chain release factor 1 [Rickettsia prowazekii]
Length = 355
Score = 188 bits (477), Expect = 8e-47
Identities = 89/152 (58%), Positives = 121/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +D++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEGIDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A+LYE +R + RS R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARLYEEKRRQKEQERSDSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD FI+AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKHGQLDEFIEALIANDE 348
>UniRef100_UPI000028C765 UPI000028C765 UniRef100 entry
Length = 173
Score = 187 bits (475), Expect = 1e-46
Identities = 88/158 (55%), Positives = 123/158 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+++ + DL++D +R+GG GGQ NTT+SAVRITHIPTGI V+ Q
Sbjct: 14 TSAATVAVLPEAEEVDLKINDSDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGISVSQQ 73
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RSK R +IGSGDRSERIRTYNFPQGRVT
Sbjct: 74 DEKSQHKNKAKGMKILRARLYELERSRIEQERSKDRKTKIGSGDRSERIRTYNFPQGRVT 133
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I+AL L+ + +++++
Sbjct: 134 DHRINLTLHKLDEFLEGEVFDEMIEALTLQAQEESLSN 171
>UniRef100_Q73IC2 Peptide chain release factor 1 [Wolbachia pipientis wMel]
Length = 359
Score = 186 bits (472), Expect = 3e-46
Identities = 86/157 (54%), Positives = 120/157 (75%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E+ +VD +++ +DL++D YRS G GGQ NTT+SAVR+TH+PTGI+V Q
Sbjct: 200 TSAATVAILPEVEEVDFKIEEKDLRIDVYRSSGPGGQSVNTTDSAVRVTHLPTGIVVIQQ 259
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALK+L A+LYE+ER + RS +R QIGSGDRSERIRTYNFPQ R+T
Sbjct: 260 DEKSQHKNKAKALKVLRARLYEIERQKKEMERSTMRKSQIGSGDRSERIRTYNFPQSRIT 319
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVA 192
DHR+ +T H ++ +++ LD FI+AL+ R E + +A
Sbjct: 320 DHRINLTSHRLEQIIKEGELDEFIEALISRNEAERLA 356
>UniRef100_UPI00003263DA UPI00003263DA UniRef100 entry
Length = 266
Score = 185 bits (470), Expect = 5e-46
Identities = 85/158 (53%), Positives = 124/158 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E+ +VD+++ + DL++D +R+GG GGQ NTT+SAVRITHIPTG+ V+ Q
Sbjct: 107 TSAATVAVLPEVEEVDLKINDTDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGLTVSQQ 166
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RSK R +IG+GDRSERIRTYNFPQGRVT
Sbjct: 167 DEKSQHKNKAKGMKILRARLYELERSRIDQERSKDRKTKIGTGDRSERIRTYNFPQGRVT 226
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I++L L+ + +++++
Sbjct: 227 DHRINLTLHKLDEFLEGEVFDEMIESLTLQAQEESLSN 264
>UniRef100_UPI0000346082 UPI0000346082 UniRef100 entry
Length = 213
Score = 184 bits (468), Expect = 9e-46
Identities = 85/159 (53%), Positives = 122/159 (76%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E+ ++D+++ DL++D +R+GG GGQ NTT+SAVRITHIPTGI V+ Q
Sbjct: 54 TSAATVAVLPEVEELDLKINESDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGISVSQQ 113
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NK+K +KIL A+LYE+ER R+ RS+ R +IG+GDRSERIRTYNFPQGRVT
Sbjct: 114 DEKSQHKNKSKGMKILRARLYELERSRIEKERSQDRKSKIGTGDRSERIRTYNFPQGRVT 173
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASF 194
DHR+ +T H +++ ++GE D I+AL+LR + + + F
Sbjct: 174 DHRINLTLHKLEEFLEGEIFDEMIEALILRSQEENLKIF 212
>UniRef100_UPI00002F0227 UPI00002F0227 UniRef100 entry
Length = 196
Score = 184 bits (468), Expect = 9e-46
Identities = 87/158 (55%), Positives = 122/158 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VDV++ + DL++D +R+GG GGQ NTT+SAVRITHIPTGI V+ Q
Sbjct: 37 TSAATVAVLPEAEEVDVKINDADLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGISVSQQ 96
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RS+ R +IGSGDRSERIRTYNFPQGRVT
Sbjct: 97 DEKSQHKNKAKGMKILRARLYELERSRIDQERSQDRKTKIGSGDRSERIRTYNFPQGRVT 156
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I++L L + +++++
Sbjct: 157 DHRINLTLHKLDEFLEGEVFDEIIESLTLHAQEESLSN 194
>UniRef100_UPI00002CD112 UPI00002CD112 UniRef100 entry
Length = 230
Score = 184 bits (468), Expect = 9e-46
Identities = 84/158 (53%), Positives = 124/158 (78%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E+ +VD+++ + DL++D +R+GG GGQ NTT+SAVRITHIPTG+ V+ Q
Sbjct: 71 TSAATVAVLPEVEEVDLKINDSDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGLTVSQQ 130
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RS+ R +IG+GDRSERIRTYNFPQGRVT
Sbjct: 131 DEKSQHKNKAKGMKILRARLYELERSRIDQERSQDRKTKIGTGDRSERIRTYNFPQGRVT 190
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I++L L+ + +++++
Sbjct: 191 DHRINLTLHKLDEFLEGEAFDEMIESLTLQAQEESLSN 228
>UniRef100_UPI00002B1522 UPI00002B1522 UniRef100 entry
Length = 253
Score = 184 bits (468), Expect = 9e-46
Identities = 85/158 (53%), Positives = 123/158 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+++ DL++D +R+GG GGQ NTT+SAVRITHIPTG+ V+ Q
Sbjct: 94 TSAATVAVLPEAEEVDLKINESDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGLSVSQQ 153
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RS+ R +IGSGDRSERIRTYNFPQGRVT
Sbjct: 154 DEKSQHKNKAKGMKILRARLYELERSRIDQERSQDRKTKIGSGDRSERIRTYNFPQGRVT 213
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I++L+L+ + +++++
Sbjct: 214 DHRINLTLHKLDEFLEGEAFDEIIESLILQAQEESLSN 251
>UniRef100_UPI00002F6EDB UPI00002F6EDB UniRef100 entry
Length = 357
Score = 184 bits (466), Expect = 1e-45
Identities = 84/158 (53%), Positives = 123/158 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+++ DL++D +R+GG GGQ NTT+SAVRITHIPTG+ V+ Q
Sbjct: 198 TSAATVAVLPEAEEVDLKINESDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGLSVSQQ 257
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RSK R +IG+GDRSERIRTYNFPQGRVT
Sbjct: 258 DEKSQHKNKAKGMKILRARLYELERSRIDQERSKDRKSKIGTGDRSERIRTYNFPQGRVT 317
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +++ ++GE D I++L+L+ + +++++
Sbjct: 318 DHRINLTLHKLEEFLEGEAFDEMIESLILQAQEESLSN 355
>UniRef100_UPI00002EC134 UPI00002EC134 UniRef100 entry
Length = 266
Score = 184 bits (466), Expect = 1e-45
Identities = 85/158 (53%), Positives = 120/158 (75%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +++ ++ +DL++D +RS G GGQ NTT+SAVRITH+P+GI+V+ Q
Sbjct: 107 TSAATVAVLPEAEDLEISIEEKDLRIDVFRSSGPGGQSVNTTDSAVRITHLPSGIVVSQQ 166
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH N+AKA+KIL ++LYE ER + RS R +Q+GSGDRSERIRTYN+PQGRVT
Sbjct: 167 DEKSQHKNRAKAMKILLSRLYETERQKQQDQRSSSRKDQVGSGDRSERIRTYNYPQGRVT 226
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D V+QGE LD IDAL++ + +AS
Sbjct: 227 DHRINLTLHKLDKVIQGEALDELIDALIVEDRSLKLAS 264
>UniRef100_UPI00002D4F5D UPI00002D4F5D UniRef100 entry
Length = 359
Score = 184 bits (466), Expect = 1e-45
Identities = 88/157 (56%), Positives = 116/157 (73%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E VD+ +K+ DL++D YRSGG+GGQ NTT+SAVRITH+PT I+V Q
Sbjct: 198 TSAATVAVLPEAEDVDLDIKDTDLRIDVYRSGGAGGQSVNTTDSAVRITHLPTNIVVTQQ 257
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQH NKAKALKIL ++LYE+ER++ S R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 258 DERSQHKNKAKALKILRSRLYEVERIKKQEEESSNRKNQVGSGDRSERIRTYNFPQGRVS 317
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVA 192
DHR+ +T + +D + GE LD I AL E+ D ++
Sbjct: 318 DHRINLTLYKLDQFLTGEALDEMISALSASEQADKLS 354
>UniRef100_Q68WK7 Peptide chain release factor 1 [Rickettsia typhi]
Length = 355
Score = 183 bits (465), Expect = 2e-45
Identities = 89/152 (58%), Positives = 119/152 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E VD++++++DL++DTYRS G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEGVDIKIEDKDLRIDTYRSSGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A+LYE +R + RS R Q+GSGDRSERIRTYNF GRV+
Sbjct: 257 DEKSQHKNKAKALKILRARLYEEKRRQKEQERSDSRRWQVGSGDRSERIRTYNFLHGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD FI+AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKHGQLDEFIEALIANDE 348
>UniRef100_UPI00003273D8 UPI00003273D8 UniRef100 entry
Length = 184
Score = 183 bits (464), Expect = 2e-45
Identities = 87/161 (54%), Positives = 119/161 (73%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+++K+ DL++D YRSGG+GGQ NTT+SAVRITH+PT I+V Q
Sbjct: 22 TSAATVAVLPEAEEVDLEIKDTDLRIDVYRSGGAGGQSVNTTDSAVRITHLPTNIVVTQQ 81
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQH NKAKALKIL ++LYE+ER++ S R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 82 DERSQHKNKAKALKILRSRLYEVERIKKQEEVSSNRKNQVGSGDRSERIRTYNFPQGRVS 141
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTS 196
DHR+ +T + +D + GE LD I AL E+ + ++ +
Sbjct: 142 DHRINLTLYKLDQFLTGEALDEMISALSASEQAEKLSQLNA 182
>UniRef100_UPI00002DDC72 UPI00002DDC72 UniRef100 entry
Length = 206
Score = 183 bits (464), Expect = 2e-45
Identities = 84/160 (52%), Positives = 124/160 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E+ +VD+++ DL+VD +R+GG GGQ NTT+SAVRITHIPTG+ V+ Q
Sbjct: 47 TSAATVAVLPEVEEVDLKINESDLRVDVFRAGGPGGQSVNTTDSAVRITHIPTGLSVSQQ 106
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RS+ R +IG+GDRSERIRTYNFPQGRVT
Sbjct: 107 DEKSQHKNKAKGMKILRARLYELERTRIDQERSQDRKSKIGTGDRSERIRTYNFPQGRVT 166
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFT 195
DHR+ +T H +++ ++GE D I++L L+ + +++++ +
Sbjct: 167 DHRINLTLHKLEEFLEGEVFDEVIESLTLQAQEESLSNLS 206
>UniRef100_UPI0000349FB6 UPI0000349FB6 UniRef100 entry
Length = 247
Score = 182 bits (463), Expect = 3e-45
Identities = 84/158 (53%), Positives = 123/158 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+++ + DL++D +R+GG GGQ NTT+SAVRITHIPTG+ V+ Q
Sbjct: 88 TSAATVAVLPEAEEVDLKINDSDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGLSVSQQ 147
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RS+ R +IG+GDRSERIRTYNFPQGRVT
Sbjct: 148 DEKSQHKNKAKGMKILRARLYELERSRIDQERSQDRKTKIGTGDRSERIRTYNFPQGRVT 207
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I++L L+ + +++++
Sbjct: 208 DHRINLTLHKLDEFLEGEAFDEMIESLTLQAQEESLSN 245
>UniRef100_UPI00003243C1 UPI00003243C1 UniRef100 entry
Length = 254
Score = 182 bits (463), Expect = 3e-45
Identities = 85/158 (53%), Positives = 122/158 (76%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+++ + DL++D +R+GG GGQ NTT+SAVRITHIPTGI V+ Q
Sbjct: 95 TSAATVAVLPEAEEVDLKINDSDLRIDVFRAGGPGGQSVNTTDSAVRITHIPTGISVSQQ 154
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAK +KIL A+LYE+ER R+ RS+ R +IG GDRSERIRTYNFPQGRVT
Sbjct: 155 DEKSQHKNKAKGMKILRARLYELERSRIDQERSQDRKTKIGKGDRSERIRTYNFPQGRVT 214
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H +D+ ++GE D I++L L+ + +++++
Sbjct: 215 DHRINLTLHKLDEFLEGEAFDEVIESLTLQAQEESLSN 252
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.128 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 277,605,255
Number of Sequences: 2790947
Number of extensions: 10431322
Number of successful extensions: 37124
Number of sequences better than 10.0: 1462
Number of HSP's better than 10.0 without gapping: 1418
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 29750
Number of HSP's gapped (non-prelim): 3907
length of query: 197
length of database: 848,049,833
effective HSP length: 121
effective length of query: 76
effective length of database: 510,345,246
effective search space: 38786238696
effective search space used: 38786238696
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0143c.1


