

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0133.10
(382 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
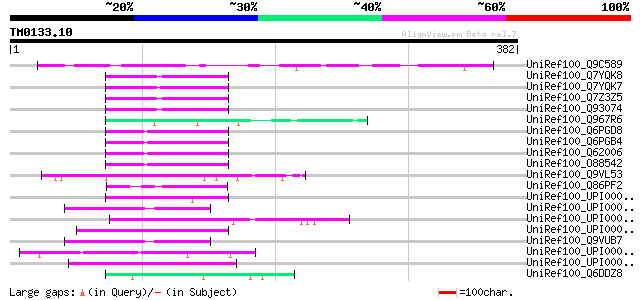
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C589 Hypothetical protein At5g21280 [Arabidopsis tha... 136 9e-31
UniRef100_Q7YQK8 OPA-containing protein [Homo sapiens] 59 3e-07
UniRef100_Q7YQK7 TNRC11 protein [Pongo pygmaeus] 59 3e-07
UniRef100_Q7Z3Z5 TNRC11 protein [Homo sapiens] 59 3e-07
UniRef100_Q93074 Thyroid hormone receptor-associated protein com... 59 3e-07
UniRef100_Q967R6 Cytoplasmic polyadenylation element-binding pro... 55 3e-06
UniRef100_Q6PGD8 Hypothetical protein [Mus musculus] 55 3e-06
UniRef100_Q6PGB4 Tnrc11 protein [Mus musculus] 55 3e-06
UniRef100_Q62006 Opa repeat [Mus musculus] 55 3e-06
UniRef100_O88542 OPA-containing protein 1 [Mus musculus] 55 3e-06
UniRef100_Q9VL53 CG4722-PA [Drosophila melanogaster] 55 3e-06
UniRef100_Q86PF2 CG12071-PB, isoform B [Drosophila melanogaster] 54 7e-06
UniRef100_UPI00001D17E8 UPI00001D17E8 UniRef100 entry 53 1e-05
UniRef100_UPI0000080BEC UPI0000080BEC UniRef100 entry 52 3e-05
UniRef100_UPI000042CEE4 UPI000042CEE4 UniRef100 entry 52 3e-05
UniRef100_UPI0000345110 UPI0000345110 UniRef100 entry 52 3e-05
UniRef100_Q9VUB7 CG32133-PA [Drosophila melanogaster] 52 3e-05
UniRef100_UPI00004375D6 UPI00004375D6 UniRef100 entry 52 4e-05
UniRef100_UPI000034C0FD UPI000034C0FD UniRef100 entry 52 4e-05
UniRef100_Q6DDZ8 MGC81192 protein [Xenopus laevis] 52 4e-05
>UniRef100_Q9C589 Hypothetical protein At5g21280 [Arabidopsis thaliana]
Length = 302
Score = 136 bits (343), Expect = 9e-31
Identities = 122/351 (34%), Positives = 156/351 (43%), Gaps = 99/351 (28%)
Query: 22 RNLEEEPPFSGAYMRSLVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQP 81
R+ ++EP S Y+RSLVKQ ++ ++ T G G Q+Q
Sbjct: 3 RSNKQEPRVSSTYIRSLVKQQ------LAYSTTMTTTTTTTTNDGSGGGKTQTQT----- 51
Query: 82 QQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQ 141
HKKQVRRRLHTSRPYQERLLNMAEARREIVTALK HRASM++A+ Q
Sbjct: 52 --HKKQVRRRLHTSRPYQERLLNMAEARREIVTALKQHRASMRQATRIPPPQ-------- 101
Query: 142 RAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVPNSYPVPVA 201
P PQ + FS PP
Sbjct: 102 -PPPPPQ-------------------------------PLNLFSPPPP------------ 117
Query: 202 SPIAPPPLMAENP--NFILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSNSFSSATSSSQ 259
P P P NP NF+LPNQPLGLNLNF DFN+ I S+T+ SS+S S+++SSS
Sbjct: 118 -PPPPDPFSWTNPSLNFLLPNQPLGLNLNFQDFNDF---IQTSSTTSSSSSSSTSSSSSS 173
Query: 260 EVPSVELSQGDGISSLVNSTESNAASQVNAGLHAAMDEEAIAEIRSLGEQYQMEWNDTMN 319
P+ + + S+ S + A +A Q N N
Sbjct: 174 IFPT---------NPHIYSSPSPPPTFTTATSDSA-------------PQLPSSSNGENN 211
Query: 320 LVKSACWFK-FLRNME-HGAPEAN----TEGDAYHNFDQPLEFPAWLNANE 364
+V SA W + L+ +E PE E D + F +EFP+WLN E
Sbjct: 212 VVTSAWWSELMLKTVEPEIKPETEEVIVVEDDVFPKFSDVMEFPSWLNQTE 262
>UniRef100_Q7YQK8 OPA-containing protein [Homo sapiens]
Length = 2027
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1908 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 1964
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1965 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1997
Score = 39.3 bits (90), Expect = 0.19
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1942 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2001
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2002 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2027
>UniRef100_Q7YQK7 TNRC11 protein [Pongo pygmaeus]
Length = 2028
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1908 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ--QQQQQQQQQQQQQQQQQQQ 1965
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1966 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1998
Score = 39.3 bits (90), Expect = 0.19
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1943 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2002
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2003 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2028
>UniRef100_Q7Z3Z5 TNRC11 protein [Homo sapiens]
Length = 2027
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1908 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 1964
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1965 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1997
Score = 39.3 bits (90), Expect = 0.19
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1942 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2001
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2002 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2027
>UniRef100_Q93074 Thyroid hormone receptor-associated protein complex 230 kDa component
[Homo sapiens]
Length = 2212
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 2093 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 2149
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 2150 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2182
Score = 39.3 bits (90), Expect = 0.19
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 2127 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2186
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2187 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2212
>UniRef100_Q967R6 Cytoplasmic polyadenylation element-binding protein isoform 1
[Aplysia californica]
Length = 687
Score = 55.5 bits (132), Expect = 3e-06
Identities = 54/216 (25%), Positives = 85/216 (39%), Gaps = 39/216 (18%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAE--ARREIVTALKFHRASMKEASEQQ 130
Q QQ TQQ QQ ++Q +++ H + Q++LL + A+++ + + K+ +QQ
Sbjct: 58 QQQQQTQQQQQQQQQQQQQQHLQQVQQQQLLKQQQQQAQQQQIQQQLLQQQQQKQQLQQQ 117
Query: 131 QQQQLQQQQQ--------------QRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP---SC 173
QQQ+ QQQQ Q+ P S + PS + P +C
Sbjct: 118 QQQEQLQQQQLQLQQQLQQQLQHIQKEPSSHTYTPGPSPELQSVLNYANVPLSKSAAFNC 177
Query: 174 NTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFN 233
N + S +G P PV SP+ P P + + N P N N
Sbjct: 178 NNSSSYSVG--------------PTPVQSPVTPSPAASA----VTVNSPSYGNFQLFGEN 219
Query: 234 NLDATIHLSNTSLSSNSFSSATSSSQEVPSVELSQG 269
D+T + S + S + S P V +S G
Sbjct: 220 AFDSTTPFQSDGTSQSHSRSLANDSD--PMVVMSPG 253
Score = 33.9 bits (76), Expect = 7.8
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 4/75 (5%)
Query: 78 TQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVT----ALKFHRASMKEASEQQQQQ 133
+Q PQ + + + QE + + E R I +L+ + S ++ QQQQQ
Sbjct: 2 SQSPQTVDQAISVKTDYEDNQQEHIPSNFEIFRRINALLDNSLEANNVSCSQSQSQQQQQ 61
Query: 134 QLQQQQQQRAPDSPQ 148
Q QQQQQQ+ Q
Sbjct: 62 QTQQQQQQQQQQQQQ 76
>UniRef100_Q6PGD8 Hypothetical protein [Mus musculus]
Length = 866
Score = 55.1 bits (131), Expect = 3e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 746 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 803
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 804 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 836
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 735 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 785
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 786 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 824
Score = 35.8 bits (81), Expect = 2.1
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 732 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 780
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 781 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 815
>UniRef100_Q6PGB4 Tnrc11 protein [Mus musculus]
Length = 2158
Score = 55.1 bits (131), Expect = 3e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 2038 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 2095
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 2096 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2128
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 2027 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 2077
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 2078 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 2116
Score = 35.8 bits (81), Expect = 2.1
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 2024 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 2072
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 2073 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 2107
>UniRef100_Q62006 Opa repeat [Mus musculus]
Length = 139
Score = 55.1 bits (131), Expect = 3e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 19 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 76
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 77 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 109
Score = 39.3 bits (90), Expect = 0.19
Identities = 26/110 (23%), Positives = 48/110 (43%), Gaps = 13/110 (11%)
Query: 68 IGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEAS 127
+G+ Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ +
Sbjct: 3 LGQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ-------------QQM 49
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNF 177
+QQQQQ QQQQQQ+ Q Q + + P+ P F
Sbjct: 50 LRQQQQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQPQF 99
>UniRef100_O88542 OPA-containing protein 1 [Mus musculus]
Length = 2074
Score = 55.1 bits (131), Expect = 3e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 1954 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 2011
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 2012 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2044
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 1943 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 1993
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 1994 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 2032
Score = 35.8 bits (81), Expect = 2.1
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 1940 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 1988
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 1989 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 2023
>UniRef100_Q9VL53 CG4722-PA [Drosophila melanogaster]
Length = 696
Score = 55.1 bits (131), Expect = 3e-06
Identities = 65/252 (25%), Positives = 105/252 (40%), Gaps = 59/252 (23%)
Query: 25 EEEPPFSGA---YMRS--LVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKA-----CQS 74
++EP + G Y RS L + Q S+ + + + + G + A ++
Sbjct: 379 QQEPLYGGTRSLYCRSPTLTRSNLNRSQSVYAKSNTAINRDIVPRPGPLVPAQSLYPMRT 438
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ QQ QQ ++QV +S + + N + R E + ++ ++ +QQQQQQ
Sbjct: 439 QQQQQQQQQQQQQVAPAPQSSHLQNQNVQNQMQQRSESIYGMRGSMRGQQQPIQQQQQQQ 498
Query: 135 LQQQQQQRAPD----------SPQLSQNPS----------------FDQDGRYK-SRRNP 167
QQQ QQ+ P+ PQ+ +P DG +K RR+P
Sbjct: 499 QQQQLQQQQPNMGVQQQQMQPPPQMMSDPQQQPQGFQPVYGTRTNPTPMDGNHKYDRRDP 558
Query: 168 -RIY------------PSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPI---APPPLMA 211
++Y S ++++ SY G + PPA P+ P P P+ APPP
Sbjct: 559 QQMYGVTGPRNRGQSAQSDDSSYGSYHGS-AVTPPARHPSVEPSPPPPPMLMYAPPP--- 614
Query: 212 ENPNFILPNQPL 223
PN P QP+
Sbjct: 615 -QPNAAHP-QPI 624
>UniRef100_Q86PF2 CG12071-PB, isoform B [Drosophila melanogaster]
Length = 578
Score = 53.9 bits (128), Expect = 7e-06
Identities = 30/91 (32%), Positives = 52/91 (56%), Gaps = 11/91 (12%)
Query: 74 SQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQ 133
S Q QQPQQH +Q + + Q++ ++A ++ A + H+ +++ +QQQQQ
Sbjct: 399 SHQQQQQPQQHPQQQQHQ-------QQQQQHVAHGKK----AARKHQQHLQQQQQQQQQQ 447
Query: 134 QLQQQQQQRAPDSPQLSQNPSFDQDGRYKSR 164
Q QQQQQQ+ +P + N S + +G +S+
Sbjct: 448 QQQQQQQQQQTSAPMYNNNSSSNHNGNNQSQ 478
>UniRef100_UPI00001D17E8 UPI00001D17E8 UniRef100 entry
Length = 160
Score = 53.1 bits (126), Expect = 1e-05
Identities = 29/101 (28%), Positives = 52/101 (50%), Gaps = 8/101 (7%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q+ + + +++I+ + + ++ +QQQQ
Sbjct: 30 QQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQQQILRQQQQQQQQQQQQQQQQQQ 89
Query: 133 QQLQ--------QQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ Q QQQQQ AP PQ P F + G ++++
Sbjct: 90 QQQQQPQQQQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 130
Score = 36.2 bits (82), Expect = 1.6
Identities = 26/90 (28%), Positives = 45/90 (49%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + ++ A + + + +F R +++ +QQQ
Sbjct: 75 QQQQQQQQQQQQQQQQQQQQPQQQQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 134
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 135 AALVRQLQQQLSNTQPQ----PSTNIFGRY 160
Score = 35.0 bits (79), Expect = 3.5
Identities = 18/38 (47%), Positives = 21/38 (54%)
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
EQQQQQQ QQQQQQ+ Q Q Q +Y R+
Sbjct: 25 EQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQ 62
Score = 34.7 bits (78), Expect = 4.6
Identities = 25/108 (23%), Positives = 45/108 (41%), Gaps = 18/108 (16%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++H ++ + +Q
Sbjct: 28 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQ---------------QYHIRQQQQQQQILRQ 72
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRN---PRIYPSCNTNF 177
QQ QQQQQQ+ Q Q Q ++ ++ P+ P F
Sbjct: 73 QQQQQQQQQQQQQQQQQQQQQQPQQQQPHQQQQQAAPPQPQPQSQPQF 120
>UniRef100_UPI0000080BEC UPI0000080BEC UniRef100 entry
Length = 2294
Score = 52.0 bits (123), Expect = 3e-05
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 6/110 (5%)
Query: 42 LRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQER 101
L +N ++ Q + + ++ Q QQ QQPQQ ++Q ++ HT P Q
Sbjct: 794 LSSNQDSLIMRQQQLKQQQQMQQQQQMAPQPQQQQMAQQPQQQQQQQPQQQHTPSPRQSP 853
Query: 102 LLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
L +++ T + + + A + QQQQQ QQQQQQ+ Q Q
Sbjct: 854 L------QQQPTTPTLQQQPNQQNAQQIQQQQQQQQQQQQQQQQQQQQQQ 897
Score = 42.4 bits (98), Expect = 0.022
Identities = 43/156 (27%), Positives = 61/156 (38%), Gaps = 16/156 (10%)
Query: 75 QQSTQQPQQHKKQVRRRL----HTSRPYQERLLNMAEARREIVTALKFHRASMKEA---S 127
QQ QQ QQ + V+++L H P L M + + + +E
Sbjct: 1657 QQQQQQQQQQQMVVQQQLPPQQHVEPPSAAGALRMMGQQHNATAGVPGPPRTQQELLMLQ 1716
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYM--GDFS 185
+QQQQQ LQQQQ +R PQ Q P ++ ++ + P+ + M G
Sbjct: 1717 QQQQQQLLQQQQMRRVVALPQQQQAPQ-----QHLVQQQIVVAPTQSPQQQQQMAVGVGV 1771
Query: 186 SYPPAFVPNSY--PVPVASPIAPPPLMAENPNFILP 219
P P+ Y P PI P NPN LP
Sbjct: 1772 GVPVQRTPHGYIQTRPGQQPIPMPQFYGHNPNLKLP 1807
Score = 40.8 bits (94), Expect = 0.064
Identities = 39/160 (24%), Positives = 65/160 (40%), Gaps = 19/160 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHT----SRPYQERLLNMAEARREIVTALKFHRASMKEASE 128
Q+ Q QQ + Q R+ H +P + +++ + ++ H+ + +
Sbjct: 659 QNAQQQHLRQQQQMQQNRQQHMMGSPQQPQPQSMMSPQQQQQMQPFQQPVHQQQRMQLQQ 718
Query: 129 QQQQQQLQQQQQQRAPD-----SPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGD 183
QQQ Q QQQQQQ++P SPQ+SQ P ++ P T
Sbjct: 719 QQQLAQQQQQQQQQSPQHISPQSPQISQTPPMQ-----AKLHQHQVVPGQATQPQQ---S 770
Query: 184 FSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPL 223
FS P + + PV VA ++ L + + I+ Q L
Sbjct: 771 FSQQKP--IDPTDPVQVAQVLSRSALSSNQDSLIMRQQQL 808
Score = 39.7 bits (91), Expect = 0.14
Identities = 44/178 (24%), Positives = 71/178 (39%), Gaps = 17/178 (9%)
Query: 41 QLRTNDQGCVVNSDASSHGQNLT---KHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRP 97
Q++ Q VVN S Q L + + K Q QQ QQ QQ ++Q +++ +
Sbjct: 1129 QMQQQPQQIVVNQQILSDPQRLQYLQQQQLMQKQQQQQQQQQQQQQQQQQQQQQQLVGQQ 1188
Query: 98 YQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQL----QQQQQQRAPDSPQLSQNP 153
Q+ L+ + T + K +++ QQQQ + QQQ QR P Q Q
Sbjct: 1189 QQQILIQQQQ------TPQQQQILQQKLSNDNQQQQIMQLITQQQLPQRTPPPQQQQQQQ 1242
Query: 154 SFDQDGRYKSRRNPRI---YPSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPP 208
Q + + +P++ + S + M + P V P+P P P P
Sbjct: 1243 LLLQQQQQSPQHHPQMQQQHWSPQSPAGGQMASSTPGTPTSVGMQSPLP-GGPTTPQP 1299
Score = 39.3 bits (90), Expect = 0.19
Identities = 31/116 (26%), Positives = 50/116 (42%), Gaps = 9/116 (7%)
Query: 42 LRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQER 101
L+ Q V A Q + ++ + +QQ Q PQQH+ Q ++ Q++
Sbjct: 938 LQKQQQLLHVQQQAQQQPQQQQQITQVQQLPPAQQQQQLPQQHQVQQQQPQQVQFTQQQQ 997
Query: 102 LLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRA----PDSPQLSQNP 153
+ A + I+ + + + QQQQQ+ QQQQQ+ P QL Q P
Sbjct: 998 IALGAGGQVRII-----QQTIQRPPTNVQQQQQVPQQQQQQIIGQFPQQQQLQQQP 1048
Score = 38.9 bits (89), Expect = 0.24
Identities = 37/144 (25%), Positives = 56/144 (38%), Gaps = 10/144 (6%)
Query: 79 QQPQQHKKQVRRRL-HTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQ 137
QQPQQ V ++ H Q+++LN + + + + ++ ++Q QQQLQQ
Sbjct: 518 QQPQQQSVPVSPQIIHQHLMRQQQILNQNQMQLQQIVQSGQALTPQQQQQQRQIQQQLQQ 577
Query: 138 -----QQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSC---NTNFSSYMGDFSSYPP 189
QQ Q Q QN Q R R+P + PS + + + PP
Sbjct: 578 VMQQQQQLQMQMQQQQQQQNLQQQQSPRMVQNRSPVMPPSTPSPSLQQQQHHATSNMLPP 637
Query: 190 AFVPNSYPVPVASPIAPPPLMAEN 213
P P PPP +N
Sbjct: 638 Q-SPRQLQSPAPQMTPPPPPSPQN 660
Score = 38.9 bits (89), Expect = 0.24
Identities = 26/88 (29%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ+TQQ Q ++Q + H + Q++ + +++ + + ++ +QQQQ
Sbjct: 199 QKQQATQQQQIQQQQQLQHQHQMQQQQQQQQQQQQQQQQ-----QQQQQQQQQQQQQQQQ 253
Query: 133 QQLQQQQ--QQRAPDSPQLSQNPSFDQD 158
QQ QQQ QQ+A + LS F +D
Sbjct: 254 QQQHQQQIIQQQATATSALSDILGFGED 281
Score = 34.7 bits (78), Expect = 4.6
Identities = 19/70 (27%), Positives = 35/70 (49%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q QQ ++ ++ +++L+ + + + + ++ +QQQQ
Sbjct: 1114 QQQQIIPQQQQQQRMQMQQQPQQIVVNQQILSDPQRLQYLQQQQLMQKQQQQQQQQQQQQ 1173
Query: 133 QQLQQQQQQR 142
QQ QQQQQQ+
Sbjct: 1174 QQQQQQQQQQ 1183
Score = 34.3 bits (77), Expect = 6.0
Identities = 28/102 (27%), Positives = 44/102 (42%), Gaps = 17/102 (16%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPY---------QERLLNMAEARREIVTALKFHRASM 123
Q QQ QQ QQ ++Q ++ L +P Q ++N + A+ + + A
Sbjct: 880 QQQQQQQQQQQQQQQQQQVLTQQQPQPGQQQQVITQRHVINTSTAQGQQIIQSHMSLALQ 939
Query: 124 KEA--------SEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
K+ ++QQ QQQ Q Q Q+ P + Q Q P Q
Sbjct: 940 KQQQLLHVQQQAQQQPQQQQQITQVQQLPPAQQQQQLPQQHQ 981
>UniRef100_UPI000042CEE4 UPI000042CEE4 UniRef100 entry
Length = 751
Score = 52.0 bits (123), Expect = 3e-05
Identities = 49/199 (24%), Positives = 82/199 (40%), Gaps = 21/199 (10%)
Query: 76 QSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREI-VTALKFHRASMKEASEQQQQQQ 134
QS P Q +L + P + N + + ++ ++ S +S Q+ QQ
Sbjct: 61 QSVNMPPQQPPGSNMQLQSISPRDQAQFNQQNIQSQRPISPRPPNQRSNNSSSSQRSFQQ 120
Query: 135 LQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP----RIYPSCNTNFSSYMGDFSSYPPA 190
+QQQQQQ+ + Q S + + DG S +P +Y S + ++ P
Sbjct: 121 IQQQQQQQHQNQQQFSPSIKYASDGYSNSPYSPMDSNNLYQQDQFKASPTL---NATPVK 177
Query: 191 FVPNSYPVPVASPIAPPPLMAENPNFIL--PNQPL-------GLNLN----FHDFNNLDA 237
P P PV +PPP+ A + ++ P+ PL LNLN +H + +
Sbjct: 178 SPPVDAPYPVYDAPSPPPVFASKESIVISPPSVPLSATSSVTNLNLNQADIYHSQSENNL 237
Query: 238 TIHLSNTSLSSNSFSSATS 256
+H N + S SS +S
Sbjct: 238 AVHTGNKFGHNRSVSSTSS 256
>UniRef100_UPI0000345110 UPI0000345110 UniRef100 entry
Length = 275
Score = 52.0 bits (123), Expect = 3e-05
Identities = 30/115 (26%), Positives = 55/115 (47%)
Query: 51 VNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARR 110
V + A+ Q + ++ + Q QQ QQ QQ ++Q RRR + Q++ + ++
Sbjct: 48 VTATAAQQRQQYQQQQQLKQQQQQQQQQQQQQQQQQQQRRRQQQQQQQQQQQQFQQQQQQ 107
Query: 111 EIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
+ + + ++ +QQQQQQ QQQQQQ+ Q Q Q + + ++
Sbjct: 108 QQFQQQQQQQQQQQQQQQQQQQQQFQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 162
Score = 45.1 bits (105), Expect = 0.003
Identities = 25/87 (28%), Positives = 42/87 (47%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q + + + Q++ + +++ + + ++ +QQQQ
Sbjct: 93 QQQQQQQQFQQQQQQQQFQQQQQQQQQQQQQQQQQQQQQFQQQQQQQQQQQQQQQQQQQQ 152
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDG 159
QQ QQQQQQ+ Q Q Q G
Sbjct: 153 QQQQQQQQQQQQQQQQQQQQQQQQQQG 179
Score = 43.1 bits (100), Expect = 0.013
Identities = 24/85 (28%), Positives = 42/85 (49%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ + +++ + + ++ +QQQQ
Sbjct: 90 QQQQQQQQQQQFQQQQQQQQFQQQQQQQQQQQQQQQQQQQQQFQQQQQQQQQQQQQQQQQ 149
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQ 157
QQ QQQQQQ+ Q Q Q
Sbjct: 150 QQQQQQQQQQQQQQQQQQQQQQQQQ 174
Score = 43.1 bits (100), Expect = 0.013
Identities = 24/85 (28%), Positives = 41/85 (48%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ +Q +++ + Q++ + +++ + + ++ +QQQQ
Sbjct: 89 QQQQQQQQQQQQFQQQQQQQQFQQQQQQQQQQQQQQQQQQQQQFQQQQQQQQQQQQQQQQ 148
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQ 157
QQ QQQQQQ+ Q Q Q
Sbjct: 149 QQQQQQQQQQQQQQQQQQQQQQQQQ 173
Score = 41.2 bits (95), Expect = 0.049
Identities = 25/92 (27%), Positives = 47/92 (50%), Gaps = 3/92 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ + +++ + + ++ +QQQQ
Sbjct: 102 QQQQQQQQFQQQQQQQQQQQQQQQQQQQQQFQQQQQQQQ---QQQQQQQQQQQQQQQQQQ 158
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSR 164
QQ QQQQQQ+ Q Q + +++SR
Sbjct: 159 QQQQQQQQQQQQQQQQQQQQGGEVFEFQFESR 190
>UniRef100_Q9VUB7 CG32133-PA [Drosophila melanogaster]
Length = 2294
Score = 52.0 bits (123), Expect = 3e-05
Identities = 32/110 (29%), Positives = 51/110 (46%), Gaps = 6/110 (5%)
Query: 42 LRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQER 101
L +N ++ Q + + ++ Q QQ QQPQQ ++Q ++ HT P Q
Sbjct: 794 LSSNQDSLIMRQQQLKQQQQMQQQQQMAPQPQQQQMAQQPQQQQQQQPQQQHTPSPRQSP 853
Query: 102 LLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
L +++ T + + + A + QQQQQ QQQQQQ+ Q Q
Sbjct: 854 L------QQQPTTPTLQQQPNQQNAQQIQQQQQQQQQQQQQQQQQQQQQQ 897
Score = 42.4 bits (98), Expect = 0.022
Identities = 43/156 (27%), Positives = 61/156 (38%), Gaps = 16/156 (10%)
Query: 75 QQSTQQPQQHKKQVRRRL----HTSRPYQERLLNMAEARREIVTALKFHRASMKEA---S 127
QQ QQ QQ + V+++L H P L M + + + +E
Sbjct: 1657 QQQQQQQQQQQMVVQQQLPPQQHVEPPSAAGALRMMGQQHNATAGVPGPPRTQQELLMLQ 1716
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYM--GDFS 185
+QQQQQ LQQQQ +R PQ Q P ++ ++ + P+ + M G
Sbjct: 1717 QQQQQQLLQQQQMRRVVALPQQQQAPQ-----QHLVQQQIVVAPTQSPQQQQQMAVGVGV 1771
Query: 186 SYPPAFVPNSY--PVPVASPIAPPPLMAENPNFILP 219
P P+ Y P PI P NPN LP
Sbjct: 1772 GVPVQRTPHGYIQTRPGQQPIPMPQFYGHNPNLKLP 1807
Score = 40.8 bits (94), Expect = 0.064
Identities = 39/160 (24%), Positives = 65/160 (40%), Gaps = 19/160 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHT----SRPYQERLLNMAEARREIVTALKFHRASMKEASE 128
Q+ Q QQ + Q R+ H +P + +++ + ++ H+ + +
Sbjct: 659 QNAQQQHLRQQQQMQQNRQQHMMGSPQQPQPQSMMSPQQQQQMQPFQQPVHQQQRMQLQQ 718
Query: 129 QQQQQQLQQQQQQRAPD-----SPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGD 183
QQQ Q QQQQQQ++P SPQ+SQ P ++ P T
Sbjct: 719 QQQLAQQQQQQQQQSPQHISPQSPQISQTPPMQ-----AKLHQHQVVPGQATQPQQ---S 770
Query: 184 FSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPL 223
FS P + + PV VA ++ L + + I+ Q L
Sbjct: 771 FSQQKP--IDPTDPVQVAQVLSRSALSSNQDSLIMRQQQL 808
Score = 39.7 bits (91), Expect = 0.14
Identities = 44/178 (24%), Positives = 71/178 (39%), Gaps = 17/178 (9%)
Query: 41 QLRTNDQGCVVNSDASSHGQNLT---KHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRP 97
Q++ Q VVN S Q L + + K Q QQ QQ QQ ++Q +++ +
Sbjct: 1129 QMQQQPQQIVVNQQILSDPQRLQYLQQQQLMQKQQQQQQQQQQQQQQQQQQQQQQLVGQQ 1188
Query: 98 YQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQL----QQQQQQRAPDSPQLSQNP 153
Q+ L+ + T + K +++ QQQQ + QQQ QR P Q Q
Sbjct: 1189 QQQILIQQQQ------TPQQQQILQQKLSNDNQQQQIMQLITQQQLPQRTPPPQQQQQQQ 1242
Query: 154 SFDQDGRYKSRRNPRI---YPSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPP 208
Q + + +P++ + S + M + P V P+P P P P
Sbjct: 1243 LLLQQQQQSPQHHPQMQQQHWSPQSPAGGQMASSTPGTPTSVGMQSPLP-GGPTTPQP 1299
Score = 39.3 bits (90), Expect = 0.19
Identities = 31/116 (26%), Positives = 50/116 (42%), Gaps = 9/116 (7%)
Query: 42 LRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQER 101
L+ Q V A Q + ++ + +QQ Q PQQH+ Q ++ Q++
Sbjct: 938 LQKQQQLLHVQQQAQQQPQQQQQITQVQQLPPAQQQQQLPQQHQVQQQQPQQVQFTQQQQ 997
Query: 102 LLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRA----PDSPQLSQNP 153
+ A + I+ + + + QQQQQ+ QQQQQ+ P QL Q P
Sbjct: 998 IALGAGGQVRII-----QQTIQRPPTNVQQQQQVPQQQQQQIIGQFPQQQQLQQQP 1048
Score = 38.9 bits (89), Expect = 0.24
Identities = 26/88 (29%), Positives = 45/88 (50%), Gaps = 7/88 (7%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ+TQQ Q ++Q + H + Q++ + +++ + + ++ +QQQQ
Sbjct: 199 QKQQATQQQQIQQQQQLQHQHQMQQQQQQQQQQQQQQQQ-----QQQQQQQQQQQQQQQQ 253
Query: 133 QQLQQQQ--QQRAPDSPQLSQNPSFDQD 158
QQ QQQ QQ+A + LS F +D
Sbjct: 254 QQQHQQQIIQQQATATSALSDILGFGED 281
Score = 38.9 bits (89), Expect = 0.24
Identities = 37/144 (25%), Positives = 56/144 (38%), Gaps = 10/144 (6%)
Query: 79 QQPQQHKKQVRRRL-HTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQ 137
QQPQQ V ++ H Q+++LN + + + + ++ ++Q QQQLQQ
Sbjct: 518 QQPQQQSVPVSPQIIHQHLMRQQQILNQNQMQLQQIVQSGQALTPQQQQQQRQIQQQLQQ 577
Query: 138 -----QQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSC---NTNFSSYMGDFSSYPP 189
QQ Q Q QN Q R R+P + PS + + + PP
Sbjct: 578 VMQQQQQLQMQMQQQQQQQNLQQQQSPRMVQNRSPVMPPSTPSPSLQQQQHHATSNMLPP 637
Query: 190 AFVPNSYPVPVASPIAPPPLMAEN 213
P P PPP +N
Sbjct: 638 Q-SPRQLQSPAPQMTPPPPPSPQN 660
Score = 35.8 bits (81), Expect = 2.1
Identities = 31/96 (32%), Positives = 46/96 (47%), Gaps = 21/96 (21%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAE---ARREIVTALKFHRASMKEASEQ 129
Q QQ QQ QQH++Q+ ++ T+ +L E A + I + ++ ++ ++EQ
Sbjct: 246 QQQQQQQQQQQHQQQIIQQQATATSALSDILGFGEDSVAAKSIASI----KSQLQASAEQ 301
Query: 130 QQQQQL-------------QQQQQQRAPDSP-QLSQ 151
QQQQL Q Q QQ A SP QL Q
Sbjct: 302 IQQQQLPVPAQQQVVFVRPQPQPQQPAQQSPHQLQQ 337
Score = 34.7 bits (78), Expect = 4.6
Identities = 19/70 (27%), Positives = 35/70 (49%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q QQ ++ ++ +++L+ + + + + ++ +QQQQ
Sbjct: 1114 QQQQIIPQQQQQQRMQMQQQPQQIVVNQQILSDPQRLQYLQQQQLMQKQQQQQQQQQQQQ 1173
Query: 133 QQLQQQQQQR 142
QQ QQQQQQ+
Sbjct: 1174 QQQQQQQQQQ 1183
Score = 34.3 bits (77), Expect = 6.0
Identities = 28/102 (27%), Positives = 44/102 (42%), Gaps = 17/102 (16%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPY---------QERLLNMAEARREIVTALKFHRASM 123
Q QQ QQ QQ ++Q ++ L +P Q ++N + A+ + + A
Sbjct: 880 QQQQQQQQQQQQQQQQQQVLTQQQPQPGQQQQVITQRHVINTSTAQGQQIIQSHMSLALQ 939
Query: 124 KEA--------SEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
K+ ++QQ QQQ Q Q Q+ P + Q Q P Q
Sbjct: 940 KQQQLLHVQQQAQQQPQQQQQITQVQQLPPAQQQQQLPQQHQ 981
>UniRef100_UPI00004375D6 UPI00004375D6 UniRef100 entry
Length = 575
Score = 51.6 bits (122), Expect = 4e-05
Identities = 48/190 (25%), Positives = 84/190 (43%), Gaps = 15/190 (7%)
Query: 8 KQESNRGMELNKLS-------RNLEEEPPFSGAYMRSLVKQLRTNDQGCVVNSDASSHGQ 60
+QE+ R EL + R L+++ L + Q NS S Q
Sbjct: 144 EQEAQRQQELQRQRQQQQEALRRLQQQQQQQQLAQMKLPSSSKWGQQSTTANS--LSQSQ 201
Query: 61 NLTKHGKIGKACQSQQ-STQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFH 119
N +I K + ++ T++ Q+ ++Q +R+ +P Q +L ++ +
Sbjct: 202 NALSLAEIQKLEEERERQTREEQRRQQQELQRVQQQQP-QTKLPGWGSVAKQPMATKSLL 260
Query: 120 RASMKEASEQQQQ--QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR--NPRIYPSCNT 175
+EA + +Q+ QQ QQQQQQ+ P PQ Q+ + Q R ++R N ++ S N
Sbjct: 261 EIQREEAQQMKQRKDQQQQQQQQQQPPPQPQPQQHSTNTQQNRTQNRAAINTSVWGSVNN 320
Query: 176 NFSSYMGDFS 185
S++M D S
Sbjct: 321 VSSNWMMDSS 330
>UniRef100_UPI000034C0FD UPI000034C0FD UniRef100 entry
Length = 189
Score = 51.6 bits (122), Expect = 4e-05
Identities = 34/127 (26%), Positives = 61/127 (47%)
Query: 45 NDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLN 104
N+ +N++ +++ N + K + Q QQ QQ QQ ++Q +++ + QE+
Sbjct: 55 NNNNNNINNNINNNINNNNNNNKQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQEQEQEEE 114
Query: 105 MAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSR 164
E +++ + + ++ +QQQQQQ QQQQQQ+ QL RY ++
Sbjct: 115 EEEEQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQLQLQLQLQHYNTTTTRYNNK 174
Query: 165 RNPRIYP 171
PR P
Sbjct: 175 PTPRQRP 181
Score = 44.3 bits (103), Expect = 0.006
Identities = 31/128 (24%), Positives = 54/128 (41%), Gaps = 13/128 (10%)
Query: 52 NSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARRE 111
N+ ++ N+ + K Q QQ QQ QQ ++Q +++ + Q+ E E
Sbjct: 59 NNINNNINNNINNNNNNNKQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQEQEQEEEEEEE 118
Query: 112 IVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
+ +QQQQQQ QQQQQQ+ Q Q Q + + + + Y
Sbjct: 119 -------------QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQLQLQLQLQHYN 165
Query: 172 SCNTNFSS 179
+ T +++
Sbjct: 166 TTTTRYNN 173
>UniRef100_Q6DDZ8 MGC81192 protein [Xenopus laevis]
Length = 1270
Score = 51.6 bits (122), Expect = 4e-05
Identities = 48/182 (26%), Positives = 72/182 (39%), Gaps = 40/182 (21%)
Query: 73 QSQQSTQQPQQHKKQVRRR-----------LHTSRPYQERLLNMAEARREIVTALKFHRA 121
Q ++ Q+ QQ K+Q RRR + Y+ + L + L+ A
Sbjct: 410 QRREREQRKQQEKEQQRRRDDIRREEERRMAEREQEYKRKQLEEQRQSERLQRQLQQEHA 469
Query: 122 SMKEASEQQQQQQLQQQQQQRAP------DSPQLSQNPSFDQDGRYKSRRNPRIYPSCNT 175
+K +QQQQQQ Q+QQQ++ P S+ P++ ++ +SR N + P T
Sbjct: 470 YLKSLQQQQQQQQQQKQQQEKKPLYHYNRGVMNPSEKPAWAREVEERSRLNKQSSPLAIT 529
Query: 176 NFSSY--MGDFSSYPP---------------------AFVPNSYPVPVASPIAPPPLMAE 212
SS G+ S PP AF P P + P+APP A
Sbjct: 530 KLSSVESAGNPSQTPPSQRPPEIQEHKPLRTTPQNRAAFRPTPRPNSLQDPLAPPVRHAP 589
Query: 213 NP 214
P
Sbjct: 590 VP 591
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.126 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 622,409,323
Number of Sequences: 2790947
Number of extensions: 26091187
Number of successful extensions: 248253
Number of sequences better than 10.0: 2406
Number of HSP's better than 10.0 without gapping: 1117
Number of HSP's successfully gapped in prelim test: 1395
Number of HSP's that attempted gapping in prelim test: 172594
Number of HSP's gapped (non-prelim): 20073
length of query: 382
length of database: 848,049,833
effective HSP length: 129
effective length of query: 253
effective length of database: 488,017,670
effective search space: 123468470510
effective search space used: 123468470510
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0133.10


