

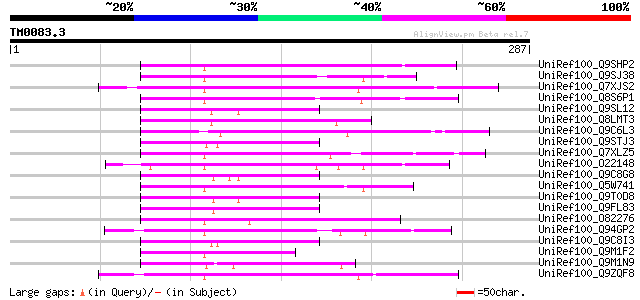
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0083.3
(287 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcrip... 77 7e-13
UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcrip... 74 6e-12
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 72 2e-11
UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa] 68 3e-10
UniRef100_Q9SL12 Putative non-LTR retroelement reverse transcrip... 67 4e-10
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 64 3e-09
UniRef100_Q9C6L3 Hypothetical protein F2J7.11 [Arabidopsis thali... 64 6e-09
UniRef100_Q9STJ3 Putative reverse transcriptase [Arabidopsis tha... 63 1e-08
UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa] 63 1e-08
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 62 1e-08
UniRef100_Q9C8G8 Hypothetical protein T4K22.11 [Arabidopsis thal... 62 2e-08
UniRef100_Q5W741 Hypothetical protein OJ1005_D04.5 [Oryza sativa] 62 2e-08
UniRef100_Q9T0D8 Hypothetical protein AT4g11710 [Arabidopsis tha... 62 2e-08
UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like... 61 3e-08
UniRef100_O82276 Putative non-LTR retroelement reverse transcrip... 61 3e-08
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 61 4e-08
UniRef100_Q9C8I3 Hypothetical protein F19C24.27 [Arabidopsis tha... 60 5e-08
UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis tha... 60 7e-08
UniRef100_Q9M1N9 Hypothetical protein T18B22.50 [Arabidopsis tha... 60 9e-08
UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcrip... 60 9e-08
>UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 773
Score = 76.6 bits (187), Expect = 7e-13
Identities = 50/184 (27%), Positives = 82/184 (44%), Gaps = 10/184 (5%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
V W ++ PK GGL +R ++ N++LL K +W+ L +R+F KY +
Sbjct: 294 VAWSKLNDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDA 353
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGCT-SFWYSDWTGLGKLCALVPFVHISDTA 184
+ T SSY WK ++ ++ GL + +G W +W L V +
Sbjct: 354 KATSQSSYAWKSILHGTKLISRGLKYIAGNGNNIQLWKDNWLPLNPPRPPVGTCDSIYSQ 413
Query: 185 LTISDVWVEGSWNLSMLYTELPDPYKPTITSMQGPSHETVDDCWVW-NSNPAGYSCSDAY 243
L +SD+ +EG WN +L + P I +++ PS +D W ++ YS Y
Sbjct: 414 LKVSDLLIEGRWNEDLLCKLIHQNDIPHIRAIR-PSITGANDAITWIYTHDGNYSVKSGY 472
Query: 244 QWLR 247
LR
Sbjct: 473 HLLR 476
>UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1229
Score = 73.6 bits (179), Expect = 6e-12
Identities = 51/168 (30%), Positives = 73/168 (43%), Gaps = 21/168 (12%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
V W ++ PKN GGLG R N SLL KL W+ L SRI + KY SS
Sbjct: 734 VAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSSSFMEC 793
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDG-CTSFWYSDWTGLGKLCALVPFVHISDTA 184
+L S+ W+ ++ +L++GL + I +G S W W + K P V I
Sbjct: 794 KLPSQPSHGWRSIIAGREILKEGLGWLITNGEKVSIWNDPWLSISK-----PLVPIGPAL 848
Query: 185 LTISDVWVEG-------SWNLSMLYTELPDPYKPTITSMQGPSHETVD 225
D+ V W+ + + LP+ Y+ I + PS VD
Sbjct: 849 REHQDLRVSALINQNTLQWDWNKIAVILPN-YENLIKQLPAPSSRGVD 895
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 72.0 bits (175), Expect = 2e-11
Identities = 58/231 (25%), Positives = 98/231 (42%), Gaps = 16/231 (6%)
Query: 50 VLFDYWRRSDEGRSHLGTVVRDCVNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK--- 106
V+ D+W S + + + ++W+++ LPK+QGG G + + N +LL K AW+
Sbjct: 799 VMMDFWWNSMQQKRKI-----HWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQ 853
Query: 107 ----LGSRIFITKYLKGSSIFSYELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGCTSF-W 161
L SR+F ++Y S S SY W+ ++ +L GL I +G +F W
Sbjct: 854 EKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSILFGRELLMQGLRTVIGNGQKTFVW 913
Query: 162 YSDWTGLGKLCALVPFVHISDTALTISDVW--VEGSWNLSMLYTELPDPYKPTITSMQGP 219
W G + + L +S + +WNL+ML P I +
Sbjct: 914 TDKWLHDGSNRRPLNRRRFINVDLKVSQLIDPTSRNWNLNMLRDLFPWKDVEIILKQRPL 973
Query: 220 SHETVDDCWVWNSNPAGYSCSDAYQWLRDWTVFRVLSVLGSMSSIISLFEE 270
+ CW+ + N YS Y++L R+ S+ SLF++
Sbjct: 974 FFKEDSFCWLHSHNGL-YSVKTGYEFLSKQVHHRLYQEAKVKPSVNSLFDK 1023
>UniRef100_Q8S6P1 Putative reverse transcriptase [Oryza sativa]
Length = 1509
Score = 67.8 bits (164), Expect = 3e-10
Identities = 48/188 (25%), Positives = 85/188 (44%), Gaps = 16/188 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
++W ++ LPKN GGLG R N+++L K W+ L SR+ KY F
Sbjct: 1040 LSWNKLTLPKNMGGLGFRDIYIFNLAMLAKQGWRLIQDPDSLCSRVLRAKYFPLGDCFRP 1099
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGC-TSFWYSDWTGLGKLCALVPFVHISDTA 184
+ T + SY W+ + K VL++G+ + + DG + W W G + P
Sbjct: 1100 KQTSNVSYTWRSIQKGLRVLQNGMIWRVGDGSKINIWADPWIPRG--WSRKPMTPRGANL 1157
Query: 185 LTISDVWVE---GSWNLSMLYTELPDPYKPTITSMQGPSHETVDDCWVWNSNPAG-YSCS 240
+T + ++ G+W+ +L + I S+ P H ++D W+ + G ++
Sbjct: 1158 VTKVEELIDPYTGTWDEDLLSQTFWEEDVAAIKSI--PVHVEMEDVLAWHFDARGCFTVK 1215
Query: 241 DAYQWLRD 248
AY+ R+
Sbjct: 1216 SAYKVQRE 1223
>UniRef100_Q9SL12 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 977
Score = 67.4 bits (163), Expect = 4e-10
Identities = 36/108 (33%), Positives = 60/108 (55%), Gaps = 9/108 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSR------IFITKYLKGSSIFSY- 125
V+W + PK +GGLG+RS +E+N K+ WK+ S +I K+L + F +
Sbjct: 817 VSWDDVCKPKKEGGLGLRSLKEANDVCCLKVVWKIVSHGNSLWVKWIEKFLLKNETFWFV 876
Query: 126 -ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDG-CTSFWYSDWTGLGKL 171
E T S++W+ L+K ++ ++ G CTSFWY DW+ +G++
Sbjct: 877 KENTTLGSWMWRKLIKFREKAKNFCKVEVNKGNCTSFWYDDWSNMGQM 924
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 64.3 bits (155), Expect = 3e-09
Identities = 41/138 (29%), Positives = 65/138 (46%), Gaps = 10/138 (7%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSR-------IFITKYLKGSSIFSY 125
+ W+ + PK GGLG TR+ N++LL K ++L S + KY+K F
Sbjct: 376 IKWEALCRPKEFGGLGFIDTRKMNIALLCKWIYRLESGKEDPCCVLLRNKYMKDGGGFFQ 435
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDG-CTSFWYSDWTGLGKLCALVPFVH--ISD 182
SS WKGL + + G S+ + +G T+FW W G L P ++ +D
Sbjct: 436 SKAEESSQFWKGLHEVKKWMDLGSSYKVGNGKATNFWSDVWIGETPLKTQYPNIYRMCAD 495
Query: 183 TALTISDVWVEGSWNLSM 200
T+S + +EG W + +
Sbjct: 496 KEKTVSQMCLEGDWYIEL 513
>UniRef100_Q9C6L3 Hypothetical protein F2J7.11 [Arabidopsis thaliana]
Length = 1213
Score = 63.5 bits (153), Expect = 6e-09
Identities = 52/210 (24%), Positives = 91/210 (42%), Gaps = 24/210 (11%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSRIFITK-----------YLKGSS 121
V+W + LPK++GGLG+R E N +L +L W R+F+ K +L S
Sbjct: 853 VSWAALCLPKSEGGLGLRRLLEWNKTLSMRLIW----RLFVAKDSLWADWQHLHHLSRGS 908
Query: 122 IFSYELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGC-TSFWYSDWTGLGKLCALVPFVHI 180
++ E S S+ WK L+ + L + +G +WY +WT LG L ++ +
Sbjct: 909 FWAVEGGQSDSWTWKRLLSLRPLAHQFLVCKVGNGLKADYWYDNWTSLGPLFRIIGDIGP 968
Query: 181 SDTAL----TISDVWVEGSWNLSMLYTELPDPYKPTITSMQGPSHETVD-DCWVWNSNPA 235
S + ++ + E W L + + + ++ PS D D + W+ N
Sbjct: 969 SSLRVPLLAKVASAFSEDGWRLPVSRSAPAKGIHDHLCTVPVPSTAQEDVDRYEWSVN-- 1026
Query: 236 GYSCSDAYQWLRDWTVFRVLSVLGSMSSII 265
G+ C + + W R + + S +S I
Sbjct: 1027 GFLC-QGFSAAKTWEAIRPKATVKSWASSI 1055
>UniRef100_Q9STJ3 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 662
Score = 62.8 bits (151), Expect = 1e-08
Identities = 34/107 (31%), Positives = 58/107 (53%), Gaps = 8/107 (7%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKL---GSRIFI----TKYLKGSSIFSY 125
+ W+ + PK +GGLG++S +E+N KL W++ G +++ T LK ++ +S+
Sbjct: 378 IAWETVCRPKREGGLGLQSIKEANDVCCLKLIWRIVSQGDSLWVQWIRTYLLKRNTFWSF 437
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGCT-SFWYSDWTGLGKL 171
S++WK L+K + I +G T SFWY DW+ G+L
Sbjct: 438 RSASQGSWMWKKLLKYRDTAKAFSKVDIRNGETASFWYDDWSSKGRL 484
>UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa]
Length = 1205
Score = 62.8 bits (151), Expect = 1e-08
Identities = 52/206 (25%), Positives = 88/206 (42%), Gaps = 23/206 (11%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
+ W+++ PK GGLG R R N +LL + AW+ L +R+ KY +I
Sbjct: 827 IAWEKLTSPKLLGGLGFRDIRCFNQALLARQAWRLIESPDSLCARVLKAKYYPNGTITDT 886
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGC-TSFWYSDWTGLGKLCALVP------FV 178
SS WKG+V +L+ GL + I DG T W + W G+ ++ +
Sbjct: 887 AFPSVSSPTWKGIVHGLELLKKGLIWRIGDGSKTKIWRNHWVAHGENLKILEKKTWNRVI 946
Query: 179 HISDTALTISDVWVEGSWNLSMLYTELPDPYKPTITSMQGPSHETVDDCWVWNSNPAG-Y 237
++ + +T + +WN ++ + + I ++ P E +D W+ G +
Sbjct: 947 YVRELIVTDTK-----TWNEPLIRHIIREEDADEILKIRIPQREE-EDFPAWHYEKTGIF 1000
Query: 238 SCSDAYQWLRDWTVFRVLSVLGSMSS 263
S Y+ W + R S S SS
Sbjct: 1001 SVRSVYRLA--WNLARKTSEQASSSS 1024
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 62.4 bits (150), Expect = 1e-08
Identities = 50/211 (23%), Positives = 92/211 (42%), Gaps = 31/211 (14%)
Query: 54 YWRRSDEGRSHLGTVVRDCVNWK---QIVLPKNQGGLGVRSTRESNVSLLGKLAWK---- 106
+W+ EGR ++WK + PK GGLG + N++LLGK W+
Sbjct: 826 WWKNKKEGRG---------LHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITE 876
Query: 107 ---LGSRIFITKYLKGSSIFSYELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGCT-SFWY 162
L +++F ++Y S + L S+ WK + +A +++ G+ I +G T + W
Sbjct: 877 KDSLMAKVFKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQVLIKQGIRAVIGNGETINVWT 936
Query: 163 SDWTGL--GKLCALVPFVHI-----SDTALTISDVWVEG--SWNLSMLYTELPDPYKPTI 213
W G K V H+ +++ + D+ + WN +++ PD + I
Sbjct: 937 DPWIGAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWNWNLVSLLFPDNTQENI 996
Query: 214 TSMQGPSHETVDDCWVWNSNPAG-YSCSDAY 243
+++ P + D + W + +G YS Y
Sbjct: 997 LALR-PGGKETRDRFTWEYSRSGHYSVKSGY 1026
>UniRef100_Q9C8G8 Hypothetical protein T4K22.11 [Arabidopsis thaliana]
Length = 504
Score = 61.6 bits (148), Expect = 2e-08
Identities = 36/108 (33%), Positives = 59/108 (54%), Gaps = 9/108 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSRI--FITKYLKGS---SIFSY-- 125
+ W + PK +GGLG+RS +E+N KL W++ S K+++ S +F +
Sbjct: 190 IAWAFVCKPKEEGGLGLRSLKEANDVCCLKLIWRIISHADSLWVKWIQSSLLKKVFFWAV 249
Query: 126 -ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGC-TSFWYSDWTGLGKL 171
E T S++W+ ++K + R I++G TSFWY DW+ LG+L
Sbjct: 250 RENTSLGSWMWRKILKFRDIARTLCKVEINNGAQTSFWYDDWSDLGRL 297
>UniRef100_Q5W741 Hypothetical protein OJ1005_D04.5 [Oryza sativa]
Length = 697
Score = 61.6 bits (148), Expect = 2e-08
Identities = 40/161 (24%), Positives = 74/161 (45%), Gaps = 11/161 (6%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
++W+++ PK GGLG R TR N +LL + AW+ L +R+ KY S+
Sbjct: 273 ISWEKLTSPKLHGGLGFRDTRSFNQALLARQAWRLIAEPNSLCARLLKAKYYPNGSLTDT 332
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHD-GCTSFWYSDWTGLGKLCALVPFVHISDTA 184
++S WKG+ +L+ GL + I D T W + W G+ ++ + +
Sbjct: 333 AFPSNTSPTWKGIEHGLELLKKGLIWRIGDRRTTKIWRNRWVAHGEKVEVIQKKNWNRLT 392
Query: 185 LTISDVWVEG--SWNLSMLYTELPDPYKPTITSMQGPSHET 223
+ ++ + G +WN +++ + I + P+ ET
Sbjct: 393 Y-VHELLIPGTNAWNEALIRHVATEEDANAILKIHVPNQET 432
>UniRef100_Q9T0D8 Hypothetical protein AT4g11710 [Arabidopsis thaliana]
Length = 473
Score = 61.6 bits (148), Expect = 2e-08
Identities = 35/108 (32%), Positives = 57/108 (52%), Gaps = 9/108 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSRI--FITKYLKGSSIFSY----- 125
+ W + PK +GGLG+RS +E+N KL W++ S K+++ S +
Sbjct: 109 ITWAFVCKPKEEGGLGLRSLKEANDVCCLKLIWRIISHADSLWVKWIQSSLLKKVSFWAV 168
Query: 126 -ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGC-TSFWYSDWTGLGKL 171
E T S++W+ ++K + R I++G TSFWY DW+ LG+L
Sbjct: 169 RENTSLGSWMWRKILKFRDIARTLCKVEINNGARTSFWYDDWSDLGRL 216
>UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1223
Score = 61.2 bits (147), Expect = 3e-08
Identities = 36/108 (33%), Positives = 62/108 (57%), Gaps = 9/108 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSRI------FITKY-LKGSSIFSY 125
++W + PK++GGLG+RS +E+N KL WK+ S ++ ++ L+ +S +
Sbjct: 860 ISWHMVCKPKDEGGLGLRSLKEANDVCCLKLVWKIVSHSNSLWVKWVDQHLLRNASFWEV 919
Query: 126 ELTGS-SSYVWKGLVKAFSVLRDGLSFAIHDG-CTSFWYSDWTGLGKL 171
+ T S S++WK L+K V + + +G TSFWY +W+ LG+L
Sbjct: 920 KQTVSQGSWIWKKLLKYREVAKTLSKVEVGNGKQTSFWYDNWSDLGQL 967
>UniRef100_O82276 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 61.2 bits (147), Expect = 3e-08
Identities = 46/157 (29%), Positives = 74/157 (46%), Gaps = 13/157 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
++W++I PK +GG+G+RS R+ N +L+ K+ W+ L +R+ KY G +
Sbjct: 702 LSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRKKYKVGGVQDTS 761
Query: 126 ELTGSS--SYVWKGL-VKAFSVLRDGLSFAIHDGCT-SFWYSDWTGLGKLCAL-VPFVHI 180
L S W+ + V V+ G+ + DGCT FW W L L +
Sbjct: 762 WLKPQPRWSSTWRSVAVGLREVVVKGVGWVPGDGCTIRFWLDRWLLQEPLVELGTDMIPE 821
Query: 181 SDTALTISDVWVEGS-WNLSMLYTELPDPYKPTITSM 216
+ +D W+ GS WNL +L LP+ K + S+
Sbjct: 822 GERIKVAADYWLPGSGWNLEILGLYLPETVKRRLLSV 858
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 60.8 bits (146), Expect = 4e-08
Identities = 48/210 (22%), Positives = 94/210 (43%), Gaps = 32/210 (15%)
Query: 53 DYWRRSDEGRSHLGTVVRDCVNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK------ 106
++W D G + + W++I+ PK+ GGLG R + N +LL + AW+
Sbjct: 736 NFWWGHDNGERKVHWIA-----WEKILYPKSHGGLGFRDMKCFNQALLARQAWRLLTSPD 790
Query: 107 -LGSRIFITKYLKGSSIFSYELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGCT-SFWYSD 164
L +R+F KY ++ SS VWKG+ +L+ G + + +G W
Sbjct: 791 SLCARVFKAKYYPNGNLVDTAFPTVSSSVWKGIQHGLDLLKQGTIWRVGNGHNIKIWRHK 850
Query: 165 WTGLGKLCALVPFVHIS-------DTALTISDVWVEGS--WNLSMLYTELPDPYKPTITS 215
W G+ HI+ + + ++++ E + WN +++ L + +
Sbjct: 851 WVPHGE--------HINVLEKKGRNRLIYVNELINEENRCWNETLVRHVLKEEDANEVLK 902
Query: 216 MQGPSHETVDDCWVWNSNPAG-YSCSDAYQ 244
++ P+H+ +DD W+ +G ++ AY+
Sbjct: 903 IRLPNHQ-MDDFPAWHHEKSGLFTVKSAYK 931
>UniRef100_Q9C8I3 Hypothetical protein F19C24.27 [Arabidopsis thaliana]
Length = 629
Score = 60.5 bits (145), Expect = 5e-08
Identities = 36/108 (33%), Positives = 59/108 (54%), Gaps = 9/108 (8%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKLGSR---IFI----TKYLKGSSIFSY 125
V+W I PK +GGLG+RS E+NV + KL W++ S +++ LK S +S
Sbjct: 265 VSWDDICKPKQEGGLGLRSLTEANVVSVLKLIWRVTSNDDSLWVKWSKMNLLKQESFWSL 324
Query: 126 ELTGS-SSYVWKGLVKAFSVLRDGLSFAIHDGC-TSFWYSDWTGLGKL 171
S S++WK ++K + +++G TSFW+ +W+G+G L
Sbjct: 325 TPNSSLGSWMWKKMLKYRETAKPFSRVEVNNGARTSFWFDNWSGMGHL 372
>UniRef100_Q9M1F2 Hypothetical protein F9K21.130 [Arabidopsis thaliana]
Length = 851
Score = 60.1 bits (144), Expect = 7e-08
Identities = 32/93 (34%), Positives = 47/93 (50%), Gaps = 7/93 (7%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK-------LGSRIFITKYLKGSSIFSY 125
V WK++ K +GGLG R + N +LL K AW+ L +R+ +Y K +SI
Sbjct: 460 VAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRIIQYPNSLFARVMKARYFKDNSIIDA 519
Query: 126 ELTGSSSYVWKGLVKAFSVLRDGLSFAIHDGCT 158
+ SY W L+ ++LR G + I DG T
Sbjct: 520 KTRSQQSYGWSSLLSGIALLRKGTRYVIGDGKT 552
>UniRef100_Q9M1N9 Hypothetical protein T18B22.50 [Arabidopsis thaliana]
Length = 762
Score = 59.7 bits (143), Expect = 9e-08
Identities = 36/133 (27%), Positives = 69/133 (51%), Gaps = 15/133 (11%)
Query: 73 VNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWKL---GSRIFITKYLKGSSI------F 123
++W Q+ PK++GGLG+RS +E+N KL W++ G +++ K+++ + +
Sbjct: 413 ISWNQVCKPKSEGGLGLRSLKEANDVCCLKLVWRIISHGDSLWV-KWVEHNLLKREIFWI 471
Query: 124 SYELTGSSSYVWKGLVKAFSVLRDGLSFAIHDG-CTSFWYSDWTGLGKLCALVPFVHISD 182
E S++WK ++K V + + +G TSFW+ DW+ LG+L + D
Sbjct: 472 VKENANLGSWIWKKILKYRGVAKRFCKAEVGNGESTSFWFDDWSLLGRLIDVAGIRGTID 531
Query: 183 ----TALTISDVW 191
++++D W
Sbjct: 532 MGISRTMSVADAW 544
>UniRef100_Q9ZQF8 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1225
Score = 59.7 bits (143), Expect = 9e-08
Identities = 54/210 (25%), Positives = 86/210 (40%), Gaps = 17/210 (8%)
Query: 50 VLFDYWRRSDEGRSHLGTVVRDCVNWKQIVLPKNQGGLGVRSTRESNVSLLGKLAWK--- 106
V+ ++W ++ E + + + W ++ P + GGLG R+ E N+ LL K W+
Sbjct: 701 VVANFWWKTREESNGIHWIA-----WDKLCTPFSDGGLGFRTLEEFNLVLLAKQLWRLIR 755
Query: 107 ----LGSRIFITKYLKGSSIFSYELTGSSSYVWKGLVKAFSVLRDGLSFAIHDG-CTSFW 161
L SR+ +Y + S S+ W+ ++ A +L GL I G T W
Sbjct: 756 FPNSLLSRVLRGRYFRYSDPIQIGKANRPSFGWRSIMAAKPLLLSGLRRTIGSGMLTRVW 815
Query: 162 YSDWTGLGKLCALVPFVHISDTALTISDVW--VEGSWNLSMLYTELPDPYKPTITSMQGP 219
W ++I DT L ++D+ V W L L EL DP + P
Sbjct: 816 EDPWIPSFPPRPAKSILNIRDTHLYVNDLIDPVTKQWKLGRL-QELVDPSDIPLILGIRP 874
Query: 220 SHETVDDCWVWNSNPAG-YSCSDAYQWLRD 248
S D + W+ +G Y+ Y RD
Sbjct: 875 SRTYKSDDFSWSFTKSGNYTVKSGYWAARD 904
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 517,882,910
Number of Sequences: 2790947
Number of extensions: 22142892
Number of successful extensions: 42449
Number of sequences better than 10.0: 137
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 63
Number of HSP's that attempted gapping in prelim test: 42279
Number of HSP's gapped (non-prelim): 156
length of query: 287
length of database: 848,049,833
effective HSP length: 126
effective length of query: 161
effective length of database: 496,390,511
effective search space: 79918872271
effective search space used: 79918872271
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0083.3


