

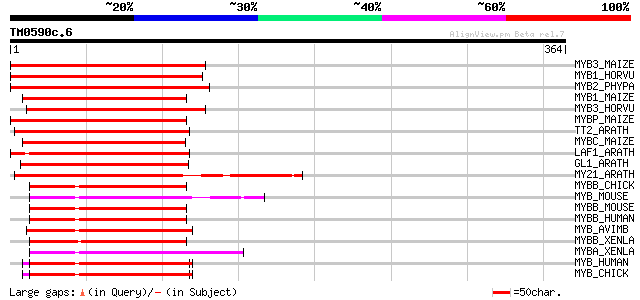
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0590c.6
(364 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MYB3_MAIZE (P20025) Myb-related protein Zm38 190 6e-48
MYB1_HORVU (P20026) Myb-related protein Hv1 187 4e-47
MYB2_PHYPA (P80073) Myb-related protein Pp2 179 1e-44
MYB1_MAIZE (P20024) Myb-related protein Zm1 177 3e-44
MYB3_HORVU (P20027) Myb-related protein Hv33 170 5e-42
MYBP_MAIZE (P27898) Myb-related protein P 169 8e-42
TT2_ARATH (Q9FJA2) TRANSPARENT TESTA 2 protein (Myb-related prot... 158 2e-38
MYBC_MAIZE (P10290) Anthocyanin regulatory C1 protein 156 9e-38
LAF1_ARATH (Q9M0K4) Transcription factor LAF1 (Long after far-re... 154 4e-37
GL1_ARATH (P27900) Trichome differentiation protein GL1 (GLABROU... 150 4e-36
MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related prot... 147 5e-35
MYBB_CHICK (Q03237) Myb-related protein B (B-Myb) 115 2e-25
MYB_MOUSE (P06876) Myb proto-oncogene protein (C-myb) 114 4e-25
MYBB_MOUSE (P48972) Myb-related protein B (B-Myb) 114 4e-25
MYBB_HUMAN (P10244) Myb-related protein B (B-Myb) 114 4e-25
MYB_AVIMB (P01104) Transforming protein Myb 113 7e-25
MYBB_XENLA (P52551) Myb-related protein B (B-Myb) (Myb-related p... 113 7e-25
MYBA_XENLA (Q05935) Myb-related protein A (A-Myb) (XAMYB) (MYB-r... 113 7e-25
MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb) 112 1e-24
MYB_CHICK (P01103) Myb proto-oncogene protein (C-myb) 112 1e-24
>MYB3_MAIZE (P20025) Myb-related protein Zm38
Length = 255
Score = 190 bits (482), Expect = 6e-48
Identities = 84/128 (65%), Positives = 102/128 (79%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK +G WT EED++LV YI+ HG G WR+LPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKAHTNRGAWTKEEDERLVAYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG F+ +E++ I++LHS+LGNKWS IAA L GRTDNEIKNYWNTH+++KLL G
Sbjct: 61 LRPDLKRGNFTADEDDLIVKLHSLLGNKWSLIAARLPGRTDNEIKNYWNTHVRRKLLGRG 120
Query: 121 LDPVTHTP 128
+DPVTH P
Sbjct: 121 IDPVTHRP 128
>MYB1_HORVU (P20026) Myb-related protein Hv1
Length = 267
Score = 187 bits (475), Expect = 4e-47
Identities = 86/126 (68%), Positives = 97/126 (76%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK KG WT EED +L YI+ HG G WR+LPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKAHTNKGAWTKEEDDRLTAYIKAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG FS EE+E II+LHS+LGNKWS IA L GRTDNEIKNYWNTHI++KL G
Sbjct: 61 LRPDLKRGNFSHEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKLTSRG 120
Query: 121 LDPVTH 126
+DPVTH
Sbjct: 121 IDPVTH 126
>MYB2_PHYPA (P80073) Myb-related protein Pp2
Length = 421
Score = 179 bits (453), Expect = 1e-44
Identities = 83/131 (63%), Positives = 96/131 (72%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + EK G+++GPWT EEDQKLV +I +G WR +PK AGL RCGKSCRLRW NY
Sbjct: 1 MGRKPCCEKVGLRRGPWTSEEDQKLVSHITNNGLSCWRAIPKLAGLLRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG FS EE I+ LH+ LGN+WS IAA L GRTDNEIKNYWNT +KK+L G
Sbjct: 61 LRPDLKRGIFSEAEENLILDLHATLGNRWSRIAAQLPGRTDNEIKNYWNTRLKKRLRSQG 120
Query: 121 LDPVTHTPRLD 131
LDP TH P D
Sbjct: 121 LDPNTHLPLED 131
>MYB1_MAIZE (P20024) Myb-related protein Zm1
Length = 340
Score = 177 bits (450), Expect = 3e-44
Identities = 79/108 (73%), Positives = 90/108 (83%)
Query: 9 KNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRG 68
K G+ +G WTP+ED +L+ YIQKHGH WR LPK AGL RCGKSCRLRW NYLRPD+KRG
Sbjct: 11 KVGLNRGSWTPQEDMRLIAYIQKHGHTNWRALPKQAGLLRCGKSCRLRWINYLRPDLKRG 70
Query: 69 KFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKL 116
F+ EEEEAII+LH +LGNKWS IAA L GRTDNEIKN WNTH+KKK+
Sbjct: 71 NFTDEEEEAIIRLHGLLGNKWSKIAACLPGRTDNEIKNVWNTHLKKKV 118
>MYB3_HORVU (P20027) Myb-related protein Hv33
Length = 302
Score = 170 bits (431), Expect = 5e-42
Identities = 74/117 (63%), Positives = 95/117 (80%)
Query: 12 VKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRGKFS 71
V+KG W+PEED+KL ++I +HG G W ++P+ A L RCGKSCRLRW NYLRPD+KRG FS
Sbjct: 14 VRKGLWSPEEDEKLYNHIIRHGVGCWSSVPRLAALNRCGKSCRLRWINYLRPDLKRGCFS 73
Query: 72 LEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHTP 128
+EE+ I+ LH +LGN+WS IA++L GRTDNEIKN+WN+ IKKKL + G+DP TH P
Sbjct: 74 QQEEDHIVALHQILGNRWSQIASHLPGRTDNEIKNFWNSCIKKKLRQQGIDPATHKP 130
>MYBP_MAIZE (P27898) Myb-related protein P
Length = 399
Score = 169 bits (429), Expect = 8e-42
Identities = 74/116 (63%), Positives = 93/116 (79%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ EK G+K+G WT EEDQ L +YI +HG G WR+LPKNAGL RCGKSCRLRW NY
Sbjct: 1 MGRTPCCEKVGLKRGRWTAEEDQLLANYIAEHGEGSWRSLPKNAGLLRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKL 116
LR D+KRG S EEE+ II+LH+ LGN+WS IA++L GRTDNEIKNYWN+H+ +++
Sbjct: 61 LRADVKRGNISKEEEDIIIKLHATLGNRWSLIASHLPGRTDNEIKNYWNSHLSRQI 116
>TT2_ARATH (Q9FJA2) TRANSPARENT TESTA 2 protein (Myb-related protein
123) (AtMYB123) (Myb-related transcription factor
LBM2-like)
Length = 258
Score = 158 bits (400), Expect = 2e-38
Identities = 71/115 (61%), Positives = 87/115 (74%)
Query: 4 SNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRP 63
+ S + + +G WT ED+ L DYI HG GKW TLP AGLKRCGKSCRLRW NYLRP
Sbjct: 6 TTSVRREELNRGAWTDHEDKILRDYITTHGEGKWSTLPNQAGLKRCGKSCRLRWKNYLRP 65
Query: 64 DIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLK 118
IKRG S +EEE II+LH++LGN+WS IA L GRTDNEIKN+WN++++K+L K
Sbjct: 66 GIKRGNISSDEEELIIRLHNLLGNRWSLIAGRLPGRTDNEIKNHWNSNLRKRLPK 120
>MYBC_MAIZE (P10290) Anthocyanin regulatory C1 protein
Length = 273
Score = 156 bits (394), Expect = 9e-38
Identities = 66/107 (61%), Positives = 84/107 (77%)
Query: 9 KNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRG 68
K GVK+G WT +ED L Y++ HG GKWR +P+ AGL+RCGKSCRLRW NYLRP+I+RG
Sbjct: 9 KEGVKRGAWTSKEDDALAAYVKAHGEGKWREVPQKAGLRRCGKSCRLRWLNYLRPNIRRG 68
Query: 69 KFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKK 115
S +EE+ II+LH +LGN+WS IA L GRTDNEIKNYWN+ + ++
Sbjct: 69 NISYDEEDLIIRLHRLLGNRWSLIAGRLPGRTDNEIKNYWNSTLGRR 115
>LAF1_ARATH (Q9M0K4) Transcription factor LAF1 (Long after far-red
light protein 1) (Myb-related protein 18) (AtMYB18)
Length = 283
Score = 154 bits (388), Expect = 4e-37
Identities = 73/118 (61%), Positives = 88/118 (73%), Gaps = 2/118 (1%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
MAK+ GE++ +KG W+PEED+KL +I +GH W T+P AGL+R GKSCRLRW NY
Sbjct: 1 MAKTKYGERH--RKGLWSPEEDEKLRSFILSYGHSCWTTVPIKAGLQRNGKSCRLRWINY 58
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLK 118
LRP +KR S EEEE I+ HS LGNKWS IA L GRTDNEIKNYW++H+KKK LK
Sbjct: 59 LRPGLKRDMISAEEEETILTFHSSLGNKWSQIAKFLPGRTDNEIKNYWHSHLKKKWLK 116
>GL1_ARATH (P27900) Trichome differentiation protein GL1 (GLABROUS1
protein)
Length = 228
Score = 150 bits (380), Expect = 4e-36
Identities = 63/110 (57%), Positives = 82/110 (74%)
Query: 8 EKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKR 67
E KKG WT EED L+DY+ HG G+W + + GLKRCGKSCRLRW NYL P++ +
Sbjct: 10 ENQEYKKGLWTVEEDNILMDYVLNHGTGQWNRIVRKTGLKRCGKSCRLRWMNYLSPNVNK 69
Query: 68 GKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLL 117
G F+ +EE+ II+LH +LGN+WS IA + GRTDN++KNYWNTH+ KKL+
Sbjct: 70 GNFTEQEEDLIIRLHKLLGNRWSLIAKRVPGRTDNQVKNYWNTHLSKKLV 119
>MY21_ARATH (Q9LK95) Transcription factor MYB21 (Myb-related protein
21) (AtMYB21) (Myb homolog 3) (AtMyb3)
Length = 226
Score = 147 bits (370), Expect = 5e-35
Identities = 82/190 (43%), Positives = 115/190 (60%), Gaps = 17/190 (8%)
Query: 4 SNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRP 63
S S + V+KGPWT EED L++YI HG G W +L K+AGLKR GKSCRLRW NYLRP
Sbjct: 12 SGSSAEAEVRKGPWTMEEDLILINYIANHGDGVWNSLAKSAGLKRTGKSCRLRWLNYLRP 71
Query: 64 DIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDP 123
D++RG + EE+ I++LH+ GN+WS IA +L GRTDNEIKN+W T I+K
Sbjct: 72 DVRRGNITPEEQLIIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQK--------- 122
Query: 124 VTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVT-TLLSCQNT 182
+ + DV +S+ + ++S++ N+ V +Q Q + + T S Q+T
Sbjct: 123 --YIKQSDVTTTSSVGS----HHSSEINDQAASTSSHNVFCTQDQAMETYSPTPTSYQHT 176
Query: 183 NQDVWNTNNY 192
N + +N NY
Sbjct: 177 NME-FNYGNY 185
>MYBB_CHICK (Q03237) Myb-related protein B (B-Myb)
Length = 686
Score = 115 bits (288), Expect = 2e-25
Identities = 51/104 (49%), Positives = 75/104 (72%), Gaps = 3/104 (2%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQK+++ ++K+G +W + K+ LK R GK CR RW N+L P++K+ ++
Sbjct: 83 KGPWTKEEDQKVIELVKKYGTKQWTLIAKH--LKGRLGKQCRERWHNHLNPEVKKSSWTE 140
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKL 116
EE+ I + H VLGN+W+ IA L GRTDN +KN+WN+ IK+K+
Sbjct: 141 EEDRIIFEAHKVLGNRWAEIAKLLPGRTDNAVKNHWNSTIKRKV 184
>MYB_MOUSE (P06876) Myb proto-oncogene protein (C-myb)
Length = 636
Score = 114 bits (285), Expect = 4e-25
Identities = 64/155 (41%), Positives = 91/155 (58%), Gaps = 15/155 (9%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQ++++ +QK+G +W + K+ LK R GK CR RW N+L P++K+ ++
Sbjct: 92 KGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTSWTE 149
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHTPRLDV 132
EE+ I Q H LGN+W+ IA L GRTDN IKN+WN+ +++K+ + G
Sbjct: 150 EEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEG-----------Y 198
Query: 133 LQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQL 167
LQ S + + S Q NN L G PSQL
Sbjct: 199 LQEPSKASQTPVATSFQKNN-HLMGFGHASPPSQL 232
>MYBB_MOUSE (P48972) Myb-related protein B (B-Myb)
Length = 704
Score = 114 bits (285), Expect = 4e-25
Identities = 51/104 (49%), Positives = 75/104 (72%), Gaps = 3/104 (2%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQK+++ ++K+G +W + K+ LK R GK CR RW N+L P++K+ ++
Sbjct: 83 KGPWTKEEDQKVIELVKKYGTKQWTLIAKH--LKGRLGKQCRERWHNHLNPEVKKSCWTE 140
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKL 116
EE+ I + H VLGN+W+ IA L GRTDN +KN+WN+ IK+K+
Sbjct: 141 EEDRIICEAHKVLGNRWAEIAKMLPGRTDNAVKNHWNSTIKRKV 184
>MYBB_HUMAN (P10244) Myb-related protein B (B-Myb)
Length = 700
Score = 114 bits (285), Expect = 4e-25
Identities = 51/104 (49%), Positives = 75/104 (72%), Gaps = 3/104 (2%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQK+++ ++K+G +W + K+ LK R GK CR RW N+L P++K+ ++
Sbjct: 83 KGPWTKEEDQKVIELVKKYGTKQWTLIAKH--LKGRLGKQCRERWHNHLNPEVKKSCWTE 140
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKL 116
EE+ I + H VLGN+W+ IA L GRTDN +KN+WN+ IK+K+
Sbjct: 141 EEDRIICEAHKVLGNRWAEIAKMLPGRTDNAVKNHWNSTIKRKV 184
>MYB_AVIMB (P01104) Transforming protein Myb
Length = 382
Score = 113 bits (283), Expect = 7e-25
Identities = 50/110 (45%), Positives = 78/110 (70%), Gaps = 3/110 (2%)
Query: 12 VKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKF 70
+ KGPWT EEDQ++++++QK+G +W + K+ LK R GK CR RW N+L P++K+ +
Sbjct: 19 LNKGPWTKEEDQRVIEHVQKYGPKRWSDIAKH--LKGRIGKQCRERWHNHLNPEVKKTSW 76
Query: 71 SLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
+ EE+ I Q H LGN+W+ IA L GRTDN +KN+WN+ +++K+ + G
Sbjct: 77 TEEEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAVKNHWNSTMRRKVEQEG 126
>MYBB_XENLA (P52551) Myb-related protein B (B-Myb) (Myb-related
protein 1) (XMYB1)
Length = 743
Score = 113 bits (283), Expect = 7e-25
Identities = 49/103 (47%), Positives = 71/103 (68%), Gaps = 1/103 (0%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRGKFSLE 73
KGPWT EED+K+++ ++K+G W + K R GK CR RW N+L P++K+ ++ E
Sbjct: 83 KGPWTKEEDEKVIELVKKYGTKHWTLIAKQLR-GRMGKQCRERWHNHLNPEVKKSSWTEE 141
Query: 74 EEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKL 116
E+ I Q H VLGN+W+ IA L GRTDN +KN+WN+ IK+K+
Sbjct: 142 EDRIICQAHKVLGNRWAEIAKLLPGRTDNAVKNHWNSTIKRKV 184
>MYBA_XENLA (Q05935) Myb-related protein A (A-Myb) (XAMYB)
(MYB-related protein 2) (XMYB2)
Length = 728
Score = 113 bits (283), Expect = 7e-25
Identities = 59/142 (41%), Positives = 85/142 (59%), Gaps = 4/142 (2%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQ++++ + K+G KW + K+ LK R GK CR RW N+L PD+K+ ++
Sbjct: 86 KGPWTKEEDQRVIELVHKYGPKKWSIIAKH--LKGRIGKQCRERWHNHLNPDVKKSSWTE 143
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG-LDPVTHTPRLD 131
EE+ I H +GN+W+ IA L GRTDN IKN+WN+ +K+K+ + G L + + R
Sbjct: 144 EEDRIIYSAHKRMGNRWAEIAKLLPGRTDNSIKNHWNSTMKRKVEQEGYLQDLMNCDRPS 203
Query: 132 VLQLASILNSSLYNNSAQFNNP 153
LQ S + QF P
Sbjct: 204 KLQAKSCAAPNHLQAQNQFYIP 225
>MYB_HUMAN (P10242) Myb proto-oncogene protein (C-myb)
Length = 640
Score = 112 bits (281), Expect = 1e-24
Identities = 51/108 (47%), Positives = 76/108 (70%), Gaps = 3/108 (2%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQ++++ +QK+G +W + K+ LK R GK CR RW N+L P++K+ ++
Sbjct: 92 KGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTSWTE 149
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
EE+ I Q H LGN+W+ IA L GRTDN IKN+WN+ +++K+ + G
Sbjct: 150 EEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEG 197
Score = 70.1 bits (170), Expect = 8e-12
Identities = 35/111 (31%), Positives = 61/111 (54%), Gaps = 2/111 (1%)
Query: 9 KNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRG 68
K + K WT EED+KL ++++G W+ + N R C+ RW L P++ +G
Sbjct: 35 KRHLGKTRWTREEDEKLKKLVEQNGTDDWKVIA-NYLPNRTDVQCQHRWQKVLNPELIKG 93
Query: 69 KFSLEEEEAIIQLHSVLGNK-WSTIAANLQGRTDNEIKNYWNTHIKKKLLK 118
++ EE++ +I+L G K WS IA +L+GR + + W+ H+ ++ K
Sbjct: 94 PWTKEEDQRVIELVQKYGPKRWSVIAKHLKGRIGKQCRERWHNHLNPEVKK 144
>MYB_CHICK (P01103) Myb proto-oncogene protein (C-myb)
Length = 641
Score = 112 bits (281), Expect = 1e-24
Identities = 51/108 (47%), Positives = 76/108 (70%), Gaps = 3/108 (2%)
Query: 14 KGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLK-RCGKSCRLRWANYLRPDIKRGKFSL 72
KGPWT EEDQ++++ +QK+G +W + K+ LK R GK CR RW N+L P++K+ ++
Sbjct: 92 KGPWTKEEDQRVIELVQKYGPKRWSVIAKH--LKGRIGKQCRERWHNHLNPEVKKTSWTE 149
Query: 73 EEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
EE+ I Q H LGN+W+ IA L GRTDN IKN+WN+ +++K+ + G
Sbjct: 150 EEDRIIYQAHKRLGNRWAEIAKLLPGRTDNAIKNHWNSTMRRKVEQEG 197
Score = 68.6 bits (166), Expect = 2e-11
Identities = 34/111 (30%), Positives = 60/111 (53%), Gaps = 2/111 (1%)
Query: 9 KNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRG 68
K + K WT EED+KL ++++G W+ + R C+ RW L P++ +G
Sbjct: 35 KRHLGKTRWTREEDEKLKKLVEQNGTEDWKVIASFLP-NRTDVQCQHRWQKVLNPELIKG 93
Query: 69 KFSLEEEEAIIQLHSVLGNK-WSTIAANLQGRTDNEIKNYWNTHIKKKLLK 118
++ EE++ +I+L G K WS IA +L+GR + + W+ H+ ++ K
Sbjct: 94 PWTKEEDQRVIELVQKYGPKRWSVIAKHLKGRIGKQCRERWHNHLNPEVKK 144
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.311 0.128 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,475,637
Number of Sequences: 164201
Number of extensions: 2034313
Number of successful extensions: 6487
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 58
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 5858
Number of HSP's gapped (non-prelim): 413
length of query: 364
length of database: 59,974,054
effective HSP length: 112
effective length of query: 252
effective length of database: 41,583,542
effective search space: 10479052584
effective search space used: 10479052584
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 66 (30.0 bits)
Lotus: description of TM0590c.6


