

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371a.1
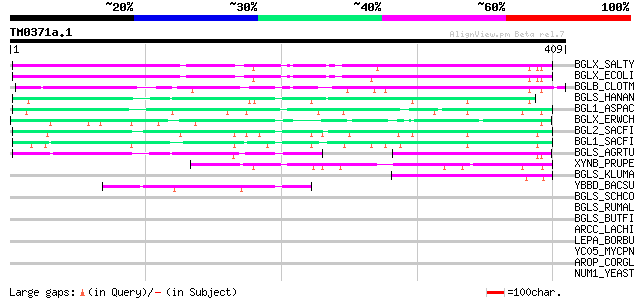
(409 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BGLX_SALTY (Q56078) Periplasmic beta-glucosidase precursor (EC 3... 154 4e-37
BGLX_ECOLI (P33363) Periplasmic beta-glucosidase precursor (EC 3... 150 7e-36
BGLB_CLOTM (P14002) Thermostable beta-glucosidase B (EC 3.2.1.21... 90 9e-18
BGLS_HANAN (P06835) Beta-glucosidase precursor (EC 3.2.1.21) (Ge... 85 3e-16
BGL1_ASPAC (P48825) Beta-glucosidase 1 precursor (EC 3.2.1.21) (... 77 6e-14
BGLX_ERWCH (Q46684) Periplasmic beta-glucosidase/beta-xylosidase... 74 5e-13
BGL2_SACFI (P22507) Beta-glucosidase 2 precursor (EC 3.2.1.21) (... 72 2e-12
BGL1_SACFI (P22506) Beta-glucosidase 1 precursor (EC 3.2.1.21) (... 71 6e-12
BGLS_AGRTU (P27034) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)... 59 2e-08
XYNB_PRUPE (P83344) Putative beta-D-xylosidase (EC 3.2.1.-) (PpA... 57 9e-08
BGLS_KLUMA (P07337) Beta-glucosidase precursor (EC 3.2.1.21) (Ge... 57 1e-07
YBBD_BACSU (P40406) Hypothetical lipoprotein ybbD precursor (ORF1) 51 5e-06
BGLS_SCHCO (P29091) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)... 43 0.002
BGLS_RUMAL (P15885) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)... 39 0.024
BGLS_BUTFI (P16084) Beta-glucosidase A (EC 3.2.1.21) (Gentiobias... 35 0.45
ARCC_LACHI (Q8G997) Carbamate kinase (EC 2.7.2.2) 33 1.3
LEPA_BORBU (O51115) GTP-binding protein lepA 32 2.3
YC05_MYCPN (P75571) Hypothetical protein MPN205 (GT9_orf438V) 32 2.9
AROP_CORGL (Q46065) Aromatic amino acid transport protein aroP (... 31 5.0
NUM1_YEAST (Q00402) Nuclear migration protein NUM1 31 6.5
>BGLX_SALTY (Q56078) Periplasmic beta-glucosidase precursor (EC
3.2.1.21) (Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase) (T-cell inhibitor)
Length = 765
Score = 154 bits (389), Expect = 4e-37
Identities = 131/465 (28%), Positives = 212/465 (45%), Gaps = 87/465 (18%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGIK 62
V+ KH+ G G + N + L +MP Y + + G +MV+ +S NG
Sbjct: 203 VMTSVKHFAAYGAVEGGKEYNTVDMSSQRLFNDYMPPYKAGLDAGSGAVMVALNSLNGTP 262
Query: 63 MHTNHDMITGFLKNTLHFKGFVISDFEGI-DRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
++ ++ L++ FKG +SD I + I A+ +V + AG+DM M +
Sbjct: 263 ATSDSWLLKDVLRDEWGFKGITVSDHGAIKELIKHGTAADPEDAVRVALKAGVDMSMADE 322
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQK--- 178
+++++ L L+K+ + M+ +DDA + +L VK+ MG+F +P++ +LG ++
Sbjct: 323 YYSKY---LPGLIKSGKVTMAELDDATRHVLNVKYDMGLFNDPYS------HLGPKESDP 373
Query: 179 ---------HRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCG 229
HRK ARE R+S+VLLKN + LPL KK I V G AD+ G
Sbjct: 374 VDTNAESRLHRKEAREVARESVVLLKNRLET----LPL-KKSGTIAVVGPLADSQRDVMG 428
Query: 230 GWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENP-------------------- 269
W+ GV+ + T+L I+N V ++Y +
Sbjct: 429 SWSA--AGVAN----QSVTVLAGIQNAVGDGAKILYAKGANITNDKGIVDFLNLYEEAVK 482
Query: 270 ----------DTEFVESNGFSYAIVVVGEHPYAEMEGDS-MNLTIPSPGPETITNV-CEA 317
D + + VVGE E S N+TIP + IT +
Sbjct: 483 IDPRSPQAMIDEAVQAAKQADVVVAVVGESQGMAHEASSRTNITIPQSQRDLITALKATG 542
Query: 318 ITCVVIIISGRPLVIEPYLALIDALVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTWFKS 376
V+++++GRPL + DA++ W G+E G +ADVL+GDY +GKLP ++ +S
Sbjct: 543 KPLVLVLMNGRPLALVKEDQQADAILETWFAGTEGGNAIADVLFGDYNPSGKLPISFPRS 602
Query: 377 VDQLP-----MNVGDP------------HYD----PLFPFGFGLS 400
V Q+P +N G P ++D PL+PFG+GLS
Sbjct: 603 VGQIPVYYSHLNTGRPYNPEKPNKYTSRYFDEANGPLYPFGYGLS 647
>BGLX_ECOLI (P33363) Periplasmic beta-glucosidase precursor (EC
3.2.1.21) (Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 765
Score = 150 bits (378), Expect = 7e-36
Identities = 132/465 (28%), Positives = 212/465 (45%), Gaps = 87/465 (18%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGIK 62
V+ KH+ G G + N + L +MP Y + + G +MV+ +S NG
Sbjct: 203 VMTSVKHFAAYGAVEGGKEYNTVDMSPQRLFNDYMPPYKAGLDAGSGAVMVALNSLNGTP 262
Query: 63 MHTNHDMITGFLKNTLHFKGFVISDFEGI-DRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
++ ++ L++ FKG +SD I + I A+ +V + +GI+M M +
Sbjct: 263 ATSDSWLLKDVLRDQWGFKGITVSDHGAIKELIKHGTAADPEDAVRVALKSGINMSMSDE 322
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQK--- 178
+++++ L L+K+ + M+ +DDA + +L VK+ MG+F +P++ +LG ++
Sbjct: 323 YYSKY---LPGLIKSGKVTMAELDDAARHVLNVKYDMGLFNDPYS------HLGPKESDP 373
Query: 179 ---------HRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCG 229
HRK ARE R+SLVLLKN + LPL KK I V G AD+ G
Sbjct: 374 VDTNAESRLHRKEAREVARESLVLLKNRLET----LPL-KKSATIAVVGPLADSKRDVMG 428
Query: 230 GWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIY------------------------ 265
W+ GV+ + T+LT IKN V V+Y
Sbjct: 429 SWSA--AGVAD----QSVTVLTGIKNAVGENGKVLYAKGANVTSDKGIIDFLNQYEEAVK 482
Query: 266 ------KENPDTEFVESNGFSYAIVVVGEHPYAEMEGDS-MNLTIPSPGPETITNV-CEA 317
+E D + + VVGE E S ++TIP + I +
Sbjct: 483 VDPRSPQEMIDEAVQTAKQSDVVVAVVGEAQGMAHEASSRTDITIPQSQRDLIAALKATG 542
Query: 318 ITCVVIIISGRPLVIEPYLALIDALVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTWFKS 376
V+++++GRPL + DA++ W G+E G +ADVL+GDY +GKLP ++ +S
Sbjct: 543 KPLVLVLMNGRPLALVKEDQQADAILETWFAGTEGGNAIADVLFGDYNPSGKLPMSFPRS 602
Query: 377 VDQLP-----MNVGDP------------HYD----PLFPFGFGLS 400
V Q+P +N G P ++D L+PFG+GLS
Sbjct: 603 VGQIPVYYSHLNTGRPYNADKPNKYTSRYFDEANGALYPFGYGLS 647
>BGLB_CLOTM (P14002) Thermostable beta-glucosidase B (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 754
Score = 90.1 bits (222), Expect = 9e-18
Identities = 116/447 (25%), Positives = 186/447 (40%), Gaps = 82/447 (18%)
Query: 5 ACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVA-TIMVSYSSWNGIKM 63
AC KH+ + + + T++D L ++ + +++ K +M +Y+ NG
Sbjct: 149 ACLKHFAANNQEHRRMTVD-TIVDERTLREIYFASFENAVKKARPWVVMCAYNKLNGEYC 207
Query: 64 HTNHDMITGFLKNTLHFKGFVISDFEGI-DRITSPPHANFTYSVEAGVSAGIDMFMIPKF 122
N ++T LKN GFV+SD+ + DR+ +G+ AG+D+ M
Sbjct: 208 SENRYLLTEVLKNEWMHDGFVVSDWGAVNDRV-------------SGLDAGLDLEMPT-- 252
Query: 123 FTEFINDLTIL--VKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHR 180
+ I D I+ VK+ + + ++ AV+RIL V M + A Y H
Sbjct: 253 -SHGITDKKIVEAVKSGKLSENILNRAVERILKVIIMALENKKENAQYEQ------DAHH 305
Query: 181 KLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSG 240
+LAR+A +S+VLLKN E +LPL KK I + G YQ SG
Sbjct: 306 RLARQAAAESMVLLKN----EDDVLPL-KKSGTIALIGAFVKKPRYQ----------GSG 350
Query: 241 NNHLEGT---TILTAIKNTVDPETTVIYKE-------NPDTEFVE-----SNGFSYAIVV 285
++H+ T I IK + ++Y E D E + ++ A+V
Sbjct: 351 SSHITPTRLDDIYEEIKKAGADKVNLVYSEGYRLENDGIDEELINEAKKAASSSDVAVVF 410
Query: 286 VGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVA 344
G E EG D +++IP I V E + +V+++ V P++ + +++
Sbjct: 411 AGLPDEYESEGFDRTHMSIPENQNRLIEAVAEVQSNIVVVLLNGSPVEMPWIDKVKSVLE 470
Query: 345 GWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLP-----------------MNVGDP 387
+L G G + Y GKL T+ + P + VG
Sbjct: 471 AYLGGQALGGRWRMCYSVKSIVGKLAETFPVKLSHNPSYLNFPGEDDRVEYKEGLFVGYR 530
Query: 388 HYD-----PLFPFGFGLSTQPTKAIYS 409
+YD PLFPFG GLS TK YS
Sbjct: 531 YYDTKGIEPLFPFGHGLSY--TKFEYS 555
>BGLS_HANAN (P06835) Beta-glucosidase precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 825
Score = 85.1 bits (209), Expect = 3e-16
Identities = 113/455 (24%), Positives = 180/455 (38%), Gaps = 88/455 (19%)
Query: 3 VIACAKHYVG------------------DGGTINGIDGNNTVIDRDGLMRVHMPGYFSSI 44
V++ AKH +G D G N ++ ID + +++ + ++
Sbjct: 197 VVSTAKHLIGNEQEHFRFAKKDKHAGKIDPGMFNTSSSLSSEIDDRAMHEIYLWPFAEAV 256
Query: 45 SKGVATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTY 104
GV++IM SY+ NG N ++ LK L F+GFV++D+ + Y
Sbjct: 257 RGGVSSIMCSYNKLNGSHACQNSYLLNYLLKEELGFQGFVMTDWGAL------------Y 304
Query: 105 SVEAGVSAGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENP 164
S +AG+DM M P F +LT V N +P R+DD RIL G+
Sbjct: 305 SGIDAANAGLDMDM-PCEAQYFGGNLTTAVLNGTLPQDRLDDMATRILSALIYSGVHNPD 363
Query: 165 FADYSLVGYLG---------------IQKH--------RKLAREAVRKSLVLLKNGISAE 201
+Y+ +L + KH R +A + + +VLLKN E
Sbjct: 364 GPNYNAQTFLTEGHEYFKQQEGDIVVLNKHVDVRSDINRAVALRSAVEGVVLLKN----E 419
Query: 202 KPILPL-TKKVPKILVAGTHA--DNLGYQCGGWTIEWQGVSGNNHLEGTTILTAIKNTVD 258
LPL +KV +I + G A D+ G C G G + G + D
Sbjct: 420 HETLPLGREKVKRISILGQAAGDDSKGTSCSLRGC-GSGAIGTGYGSGAGTFSYFVTPAD 478
Query: 259 PETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEME-----------------GDSMNL 301
+E EF+ + A + + A +E GD NL
Sbjct: 479 GIGARAQQEKISYEFIGDSWNQAAAMDSALYADAAIEVANSVAGEEIGDVDGNYGDLNNL 538
Query: 302 TIPSPGPETITNVCEA-ITCVVIIISGRPLVIEPYL---ALIDALVAGWLPGSEGQGVAD 357
T+ I N+ +VI+ SG+ + +EP++ + + + +L G +A
Sbjct: 539 TLWHNAVPLIKNISSINNNTIVIVTSGQQIDLEPFIDNENVTAVIYSSYLGQDFGTVLAK 598
Query: 358 VLYGDYGFTGKLPRTWFKSV-DQLP----MNVGDP 387
VL+GD +GKLP T K V D +P ++V DP
Sbjct: 599 VLFGDENPSGKLPFTIAKDVNDYIPVIEKVDVPDP 633
>BGL1_ASPAC (P48825) Beta-glucosidase 1 precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 860
Score = 77.4 bits (189), Expect = 6e-14
Identities = 120/495 (24%), Positives = 187/495 (37%), Gaps = 122/495 (24%)
Query: 3 VIACAKHYV------------GDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVAT 50
V+A AKHY+ G N D ++ +D + +++ + ++ GV
Sbjct: 184 VVATAKHYILNEQEHFRQVAEAAGYGFNISDTISSNVDDKTIHEMYLWPFADAVRAGVGA 243
Query: 51 IMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGV 110
IM SY+ N N + LK L F+GFV+SD+ +S
Sbjct: 244 IMCSYNQINNSYGCQNSYTLNKLLKAELGFQGFVMSDW------------GAHHSGVGSA 291
Query: 111 SAGIDMFM-----IPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMG---IFE 162
AG+DM M + + +LTI V N +P R+DD RI+ + +G +++
Sbjct: 292 LAGLDMSMPGDITFDSATSFWGTNLTIAVLNGTVPQWRVDDMAVRIMAAYYKVGRDRLYQ 351
Query: 163 NP-FADYSLVGY------------------LGIQK-HRKLAREAVRKSLVLLKNGISAEK 202
P F+ ++ Y + +Q+ H ++ R+ S VLLKN +
Sbjct: 352 PPNFSSWTRDEYGFKYFYPQEGPYEKVNHFVNVQRNHSEVIRKLGADSTVLLKNNNA--- 408
Query: 203 PILPLTKKVPKILVAGTHADNLGY--------QCGGWTIEWQGVSGNNHL---------- 244
LPLT K K+ + G A + Y C T+ SG
Sbjct: 409 --LPLTGKERKVAILGEDAGSNSYGANGCSDRGCDNGTLAMAWGSGTAEFPYLVTPEQAI 466
Query: 245 -------EGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGD 297
+G+ + T+ + + FV S+ I V G EGD
Sbjct: 467 QAEVLKHKGSVYAITDNWALSQVETLAKQASVSLVFVNSDAGEGYISVDGN------EGD 520
Query: 298 SMNLTIPSPGPETI---TNVCEAITCVVIIISGRPLVIEPYL--ALIDALVAGWLPGSE- 351
NLT+ G I N C +V+I S P++++ + + A++ LPG E
Sbjct: 521 RNNLTLWKNGDNLIKAAANNCN--NTIVVIHSVGPVLVDEWYDHPNVTAILWAGLPGQES 578
Query: 352 GQGVADVLYGDYGFTGKLPRTWFKS--------VDQLPMNVGDPHYD------------- 390
G +ADVLYG K P TW K+ V +L G P D
Sbjct: 579 GNSLADVLYGRVNPGAKSPFTWGKTREAYGDYLVRELNNGNGAPQDDFSEGVFIDYRGFD 638
Query: 391 -----PLFPFGFGLS 400
P++ FG GLS
Sbjct: 639 KRNETPIYEFGHGLS 653
>BGLX_ERWCH (Q46684) Periplasmic beta-glucosidase/beta-xylosidase
precursor [Includes: Beta-glucosidase (EC 3.2.1.21)
(Gentiobiase) (Cellobiase); Beta-xylosidase (EC
3.2.1.37) (1,4-beta-D-xylan xylohydrolase) (Xylan
1,4-beta-xylosidase)]
Length = 654
Score = 74.3 bits (181), Expect = 5e-13
Identities = 104/440 (23%), Positives = 172/440 (38%), Gaps = 88/440 (20%)
Query: 1 EKVIACAKHYVGDGGTINGIDGNNTVID----RDGLMRVHMPGYFSSISKGVATIMVSYS 56
+ VI+ KH+VG G +G D +N R ++ H+ + + A IM +YS
Sbjct: 260 QSVISIVKHWVGYGAAKDGWDSHNVYGKYAQFRQNNLQWHIDPFTGAFEAHAAGIMPTYS 319
Query: 57 -----SWNGIKMHT-----NHDMITGFLKNTLHFKGFVISDF-----------EGIDRIT 95
SW+G + N ++T L+ F G ++SD+ G+
Sbjct: 320 ILRNASWHGKPIEQVGAGFNRFLLTDLLRGQYGFDGVILSDWLITNDCKGDCLTGVKPGE 379
Query: 96 SPPHANFTYSVEA---------GVSAGIDMFMIPKFFTEFINDLTILVK---NKLIPMSR 143
P + VE V+AG+D F + D +LV+ + + +R
Sbjct: 380 KPVPRGMPWGVEKLTPAERFVKAVNAGVDQF-------GGVTDSALLVQAVQDGKLTEAR 432
Query: 144 IDDAVKRILWVKFMMGIFENPFADYSLVG-YLGIQKHRKLAREAVRKSLVLLKNGISAEK 202
+D +V RIL KF G+FE P+ + + +G ++LA + +SLVLL+N
Sbjct: 433 LDTSVNRILKQKFQTGLFERPYVNATQANDIVGRADWQQLADDTQARSLVLLQN-----N 487
Query: 203 PILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETT 262
+LPL K G + G++ N E I + NT +
Sbjct: 488 NLLPLRK--------------------GSRVWLHGIAANAAQEVGFI---VVNTPEQADV 524
Query: 263 VIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCVV 322
+ + + E N F G + EG S+ +P + I + +V
Sbjct: 525 ALIRTHTPYEQPHKNFF------FGSRHH---EG-SLAFRNDNPDYQAIVRASAKVPTLV 574
Query: 323 IIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPM 382
+ RP ++ + A+VA + G + + L +T KLP S+ +
Sbjct: 575 TVYMERPAILTNVVDKTRAVVANF--GVSDSVLLNRLMSGAAYTAKLPFELPSSMSAVRN 632
Query: 383 NVGDPHYD---PLFPFGFGL 399
D YD PLFPFG+GL
Sbjct: 633 QQPDLPYDSAKPLFPFGYGL 652
>BGL2_SACFI (P22507) Beta-glucosidase 2 precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 880
Score = 72.4 bits (176), Expect = 2e-12
Identities = 120/494 (24%), Positives = 193/494 (38%), Gaps = 111/494 (22%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTV---------------IDRDGLMRVHMPGYFSSISKG 47
V+AC KH++G+ I N+ V I + +++ + SI G
Sbjct: 200 VMACVKHFIGNEQDIYRQPSNSKVDPEYDPATKESISANIPDRAMHELYLWPFADSIRAG 259
Query: 48 VATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVE 107
V ++M SY+ N N MI LK L F+GFV+SD+ + YS
Sbjct: 260 VGSVMCSYNRVNNTYSCENSYMINHLLKEELGFQGFVVSDWAA--------QMSGAYSAI 311
Query: 108 AGVSAGIDMFMIPKFFT---EFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENP 164
+G+ + ++ + T + +LT V N+ +P+ R+DD RIL + F
Sbjct: 312 SGLDMSMPGELLGGWNTGKSYWGQNLTKAVYNETVPIERLDDMATRILAALYATNSFPTK 371
Query: 165 -----FADYSLVGY---LGIQKHRKL-----------------AREAVRKSLVLLKNGIS 199
F+ ++ Y + K + A + +S+VLLKN
Sbjct: 372 DRLPNFSSFTTKEYGNEFFVDKTSPVVKVNHFVDPSNDFTEDTALKVAEESIVLLKN--- 428
Query: 200 AEKPILPLT-KKVPKILVAGTHA--DNLGYQCG-----------GWTIEWQGVSGNNHLE 245
EK LP++ KV K+L++G A D GY+C GW G G +
Sbjct: 429 -EKNTLPISPNKVRKLLLSGIAAGPDPKGYECSDQSCVDGALFEGW---GSGSVGYPKYQ 484
Query: 246 GTTI----LTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVV--VGEHPYAEME---G 296
T A KN + + + V S+ +VV V Y ++ G
Sbjct: 485 VTPFEEISANARKNKMQFDYIRESFDLTQVSTVASDAHMSIVVVSAVSGEGYLIIDGNRG 544
Query: 297 DSMNLTIPSPGPETITNVCE-AITCVVIIISGRPLVIEPYL--ALIDALV-AGWLPGSEG 352
D N+T+ I V E VV+I S + +E + + A+V AG L G
Sbjct: 545 DKNNVTLWHNSDNLIKAVAENCANTVVVITSTGQVDVESFADHPNVTAIVWAGPLGDRSG 604
Query: 353 QGVADVLYGDYGFTGKLPRTWFKSVDQ-LPMNVGDP------------------------ 387
+A++L+G+ +G LP T KS D +P+ +P
Sbjct: 605 TAIANILFGNANPSGHLPFTVAKSNDDYIPIVTYNPPNGEPEDNTLAEHDLLVDYRYFEE 664
Query: 388 -HYDPLFPFGFGLS 400
+ +P + FG+GLS
Sbjct: 665 KNIEPRYAFGYGLS 678
>BGL1_SACFI (P22506) Beta-glucosidase 1 precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 876
Score = 70.9 bits (172), Expect = 6e-12
Identities = 117/497 (23%), Positives = 196/497 (38%), Gaps = 119/497 (23%)
Query: 3 VIACAKHYVGDG-------GTINGIDGNNT-------VIDRDGLMRVHMPGYFSSISKGV 48
V+AC KH++G+ IN T + DR + +++ + S+ GV
Sbjct: 198 VMACVKHFIGNEQEKYRQPDDINPATNQTTKEAISANIPDR-AMHALYLWPFADSVRAGV 256
Query: 49 ATIMVSYSSWNGIKMHTNHDMITGFLKNTLHFKGFVISDF----EGIDRITSPPHANFTY 104
++M SY+ N N M+ LK L F+GFV+SD+ G+ S +
Sbjct: 257 GSVMCSYNRVNNTYACENSYMMNHLLKEELGFQGFVVSDWGAQLSGVYSAISGLDMSMPG 316
Query: 105 SVEAGVSAGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENP 164
V G + G + +LT + N+ +P+ R+DD RIL + F
Sbjct: 317 EVYGGWNTGTSFWG---------QNLTKAIYNETVPIERLDDMATRILAALYATNSFPTE 367
Query: 165 -----FADYSLVGYLGIQKHRKLAREAVR---------------------KSLVLLKNGI 198
F+ ++ Y G + + E V+ +S+VLLKN
Sbjct: 368 DHLPNFSSWTTKEY-GNKYYADNTTEIVKVNYNVDPSNDFTEDTALKVAEESIVLLKN-- 424
Query: 199 SAEKPILPLT-KKVPKILVAGTHA--DNLGYQCGGWTIEWQGVSGNNHLE--GTTILTAI 253
E LP++ +K ++L++G A D +GYQC E Q + + G+ + +
Sbjct: 425 --ENNTLPISPEKAKRLLLSGIAAGPDPIGYQC-----EDQSCTNGALFQGWGSGSVGSP 477
Query: 254 KNTVDPETTVIY---KENPDTEFVE-----------SNGFSYAIVVV------GEHPYAE 293
K V P + Y K +++ ++ +IVVV G
Sbjct: 478 KYQVTPFEEISYLARKNKMQFDYIRESYDLAQVTKVASDAHLSIVVVSAASGEGYITVDG 537
Query: 294 MEGDSMNLTIPSPGPETITNVCE-AITCVVIIISGRPLVIEPYL--ALIDALV-AGWLPG 349
+GD NLT+ + G + I V E VV++ S + E + + A+V AG L
Sbjct: 538 NQGDRKNLTLWNNGDKLIETVAENCANTVVVVTSTGQINFEGFADHPNVTAIVWAGPLGD 597
Query: 350 SEGQGVADVLYGDYGFTGKLPRTWFKSVDQ-LPMNVGDP--------------------- 387
G +A++L+G +G LP T K+ D +P+ P
Sbjct: 598 RSGTAIANILFGKANPSGHLPFTIAKTDDDYIPIETYSPSSGEPEDNHLVENDLLVDYRY 657
Query: 388 ----HYDPLFPFGFGLS 400
+ +P + FG+GLS
Sbjct: 658 FEEKNIEPRYAFGYGLS 674
>BGLS_AGRTU (P27034) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
Length = 818
Score = 59.3 bits (142), Expect = 2e-08
Identities = 44/145 (30%), Positives = 67/145 (45%), Gaps = 28/145 (19%)
Query: 283 IVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDA 341
+++VG + EG D ++ +P E I V E VV+++ + P+L + A
Sbjct: 547 LLLVGREGEWDTEGLDLPDMRLPGRQEELIEAVAETNPNVVVVLQTGGPIEMPWLGKVRA 606
Query: 342 LVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVGDP------------- 387
++ W PG E G +ADVL+GD G+LP+T+ K++ DP
Sbjct: 607 VLQMWYPGQELGNALADVLFGDVEPAGRLPQTFPKALTDNSAITDDPSIYPGQDGHVRYA 666
Query: 388 --------HYD-----PLFPFGFGL 399
H+D PLFPFGFGL
Sbjct: 667 EGIFVGYRHHDTREIEPLFPFGFGL 691
Score = 57.8 bits (138), Expect = 5e-08
Identities = 61/232 (26%), Positives = 105/232 (44%), Gaps = 27/232 (11%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISK-GVATIMVSYSSWNGI 61
V A KH+V + I ++ V +R L +++P + ++ K GV +M SY+ NG
Sbjct: 138 VAATIKHFVANESEIERQTMSSDVDERT-LREIYLPPFEEAVKKAGVKAVMSSYNKLNGT 196
Query: 62 KMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
N ++T L+ F G V+SD+ G ++S ++AG+D+ M P
Sbjct: 197 YTSENPWLLTKVLREEWGFDGVVMSDWFG------------SHSTAETINAGLDLEM-PG 243
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFEN--PFADYSLVGYLGIQKH 179
+ + L V+ + + + +RIL + +G FE A+++ L + +
Sbjct: 244 PWRDRGEKLVAAVREGKVKAETVRASARRILLLLERVGAFEKAPDLAEHA----LDLPED 299
Query: 180 RKLAREAVRKSLVLLKNGISAEKPILPLTK-KVPKILVAGTHADNLGYQCGG 230
R L R+ + VLLKN +LPL K +I V G +A + GG
Sbjct: 300 RALIRQLGAEGAVLLKN-----DGVLPLAKSSFDQIAVIGPNAASARVMGGG 346
>XYNB_PRUPE (P83344) Putative beta-D-xylosidase (EC 3.2.1.-)
(PpAz152) (Fragment)
Length = 461
Score = 57.0 bits (136), Expect = 9e-08
Identities = 78/309 (25%), Positives = 132/309 (42%), Gaps = 60/309 (19%)
Query: 134 VKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQK-----HRKLAREAVR 188
V+ L+ I+ A+ + V+ +G+F+ + + G LG + H++LA EA R
Sbjct: 24 VRRGLVSQLEINWALANTMTVQMRLGMFDGEPSAHQY-GNLGPRDVCTPAHQQLALEAAR 82
Query: 189 KSLVLLKNGISAEKPILPL-TKKVPKILVAGTHAD----NLGYQCG---GWTIEWQGVSG 240
+ +VLL+N + LPL T++ + V G ++D +G G G+T QG+
Sbjct: 83 QGIVLLENRGRS----LPLSTRRHRTVAVIGPNSDVTVTMIGNYAGVACGYTTPLQGIGR 138
Query: 241 NN---HLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEG- 296
H G T + N + ++ T ++V+G E E
Sbjct: 139 YTRTIHQAGCTDVHCNGNQLFGAAEAAARQADAT-----------VLVMGLDQSIEAEFV 187
Query: 297 DSMNLTIPSPGPETITNVCEAIT--CVVIIISGRPLVI-----EPYLALIDALVAGWLPG 349
D L +P E ++ V A +++++SG P+ + +P ++ I + G+
Sbjct: 188 DRAGLLLPGHQQELVSRVARASRGPTILVLMSGGPIDVTFAKNDPRISAI--IWVGYPGQ 245
Query: 350 SEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNVGDPHYDP---------------- 391
+ G +A+VL+G GKLP TW+ V LPM DP
Sbjct: 246 AGGTAIANVLFGTANPGGKLPMTWYPQNYVTHLPMTDMAMRADPARGYPGRTYRFYIGPV 305
Query: 392 LFPFGFGLS 400
+FPFG GLS
Sbjct: 306 VFPFGLGLS 314
>BGLS_KLUMA (P07337) Beta-glucosidase precursor (EC 3.2.1.21)
(Gentiobiase) (Cellobiase) (Beta-D-glucoside
glucohydrolase)
Length = 845
Score = 56.6 bits (135), Expect = 1e-07
Identities = 48/143 (33%), Positives = 67/143 (46%), Gaps = 24/143 (16%)
Query: 282 AIVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALID 340
A++++G + E EG D N+ +P E + V +A VI+ V P+L +
Sbjct: 577 AVLIIGLNGEWETEGYDRENMDLPKRTNELVRAVLKANPNTVIVNQSGTPVEFPWLEEAN 636
Query: 341 ALVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTW-FKSVDQ----------------LPM 382
ALV W G+E G +ADVLYGD GKL +W FK D +
Sbjct: 637 ALVQAWYGGNELGNAIADVLYGDVVPNGKLSLSWPFKLQDNPAFLNFKTEFGRVVYGEDI 696
Query: 383 NVGDPHYDPL-----FPFGFGLS 400
VG +Y+ L FPFG+GLS
Sbjct: 697 FVGYRYYEKLQRKVAFPFGYGLS 719
>YBBD_BACSU (P40406) Hypothetical lipoprotein ybbD precursor (ORF1)
Length = 642
Score = 51.2 bits (121), Expect = 5e-06
Identities = 43/170 (25%), Positives = 81/170 (47%), Gaps = 22/170 (12%)
Query: 69 MITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIP-------- 120
++TG L+ + F G +++D + I H +V V AG+D+ ++P
Sbjct: 300 VMTGLLRQEMGFNGVIVTDALNMKAIAD--HFGQEEAVVMAVKAGVDIALMPASVTSLKE 357
Query: 121 -KFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYS------LVGY 173
+ F I L VKN IP +I+++V+RI+ +K G++ +D +
Sbjct: 358 EQKFARVIQALKEAVKNGDIPEQQINNSVERIISLKIKRGMYPARNSDSTKEKIAKAKKI 417
Query: 174 LGIQKHRKLAREAVRKSLVLLKNGISAEKPILPL-TKKVPKILVAGTHAD 222
+G ++H K ++ K++ +LKN E+ LP KK +IL+ + +
Sbjct: 418 VGSKQHLKAEKKLAEKAVTVLKN----EQHTLPFKPKKGSRILIVAPYEE 463
>BGLS_SCHCO (P29091) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
(Fragment)
Length = 192
Score = 42.7 bits (99), Expect = 0.002
Identities = 23/47 (48%), Positives = 28/47 (58%), Gaps = 1/47 (2%)
Query: 339 IDALVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNV 384
+ A+V LPG E G VAD+LYG Y +G+LP T KS D P V
Sbjct: 111 VKAVVWSGLPGQEAGNSVADILYGAYNPSGRLPYTIAKSADDYPAQV 157
>BGLS_RUMAL (P15885) Beta-glucosidase (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
Length = 947
Score = 38.9 bits (89), Expect = 0.024
Identities = 34/123 (27%), Positives = 55/123 (44%), Gaps = 16/123 (13%)
Query: 9 HYVGDGGTINGIDGNN---------TVIDRDGLMRVHMPGYFSSISKGVA-TIMVSYSSW 58
H VG GTI NN +V L +++ G+ ++ K A ++M +Y
Sbjct: 608 HSVGVEGTIKHFCANNQETNRHFIDSVASERALREIYLKGFEIAVRKSKARSVMTTYGKV 667
Query: 59 NGIKMHTNHDMITGFLKNTLHFKGFVISDFEG--IDRITSPPHANFTYSVEAGVSAGIDM 116
NG+ + D+ T L+ F GF ++D+ DR +P NF A V A D+
Sbjct: 668 NGLWTAGSFDLNTMILRKQWGFDGFTMTDWWANINDRGCAPDKNNFA----AMVRAQNDV 723
Query: 117 FMI 119
+M+
Sbjct: 724 YMV 726
Score = 30.8 bits (68), Expect = 6.5
Identities = 26/84 (30%), Positives = 35/84 (40%), Gaps = 23/84 (27%)
Query: 340 DALVAGWLPG-SEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMN--------------- 383
DA++ W G + G G A VL G+ GKLP T + P +
Sbjct: 190 DAVMYVWQGGMTGGTGTARVLLGEVSPCGKLPDTIAYDITDYPSDKNFHNRDVDIYAEDI 249
Query: 384 -VGDPHYDPL------FPFGFGLS 400
VG ++D FPFG+GLS
Sbjct: 250 FVGYRYFDTFAKDRVRFPFGYGLS 273
>BGLS_BUTFI (P16084) Beta-glucosidase A (EC 3.2.1.21) (Gentiobiase)
(Cellobiase) (Beta-D-glucoside glucohydrolase)
Length = 830
Score = 34.7 bits (78), Expect = 0.45
Identities = 20/89 (22%), Positives = 44/89 (48%), Gaps = 2/89 (2%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISK-GVATIMVSYSSWNGI 61
+IA KH+ + N ++ +R + +++ + + + + Y+ NG
Sbjct: 685 IIATPKHFALNNKESNRKGSDSRASER-AIREIYLKAFEIIVKEQSPGASCLQYNIVNGQ 743
Query: 62 KMHTNHDMITGFLKNTLHFKGFVISDFEG 90
+ +HD++TG L++ F+G V+SD+ G
Sbjct: 744 RSSESHDLLTGILRDEWGFEGVVVSDWWG 772
>ARCC_LACHI (Q8G997) Carbamate kinase (EC 2.7.2.2)
Length = 318
Score = 33.1 bits (74), Expect = 1.3
Identities = 17/44 (38%), Positives = 22/44 (49%)
Query: 260 ETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTI 303
E + KENPD FVE G Y VV P +E D++N +
Sbjct: 138 EADLAKKENPDFTFVEDAGRGYRRVVPSPKPIGVVETDAVNTLV 181
>LEPA_BORBU (O51115) GTP-binding protein lepA
Length = 606
Score = 32.3 bits (72), Expect = 2.3
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 14/79 (17%)
Query: 87 DFEGIDRITSPPHANFTYSVEAGVSA------------GIDMFMIPKFFTEFINDLTIL- 133
DF ++ + +P H +F+Y V +S+ GI + F+ F +DL I+
Sbjct: 77 DFYELNFVDTPGHVDFSYEVSRAISSCEGALLLIDASQGIQAQTVSNFYMAFEHDLEIIP 136
Query: 134 VKNKL-IPMSRIDDAVKRI 151
V NK+ +P + +D K+I
Sbjct: 137 VINKIDLPNANVDFVKKQI 155
>YC05_MYCPN (P75571) Hypothetical protein MPN205 (GT9_orf438V)
Length = 438
Score = 32.0 bits (71), Expect = 2.9
Identities = 13/49 (26%), Positives = 25/49 (50%), Gaps = 3/49 (6%)
Query: 360 YGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTKAIY 408
+G +G +G P+ W + +++P+ V DP P+ F + P +Y
Sbjct: 72 FGGFGLSGSAPQQWNEVKNKVPVEVAQ---DPTDPYRFAVLLVPRSVVY 117
>AROP_CORGL (Q46065) Aromatic amino acid transport protein aroP
(General aromatic amino acid permease)
Length = 463
Score = 31.2 bits (69), Expect = 5.0
Identities = 16/35 (45%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Query: 331 VIEPYLALIDALVAGWLPGSEGQGVADVLYGDYGF 365
VI +L + AL+ GWLPGS G ++ + GD+GF
Sbjct: 159 VIIAFLIIGIALIFGWLPGSTFVGTSNFI-GDHGF 192
>NUM1_YEAST (Q00402) Nuclear migration protein NUM1
Length = 2748
Score = 30.8 bits (68), Expect = 6.5
Identities = 21/67 (31%), Positives = 28/67 (41%), Gaps = 5/67 (7%)
Query: 260 ETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAIT 319
E V KENPD EF++ VV Y+E+E + P E + E I
Sbjct: 1334 EDLVKCKENPDMEFLKEKSAKLGHTVVSNKEYSELE-----KKLEQPSLEYLVKHAEQIQ 1388
Query: 320 CVVIIIS 326
+I IS
Sbjct: 1389 SKIISIS 1395
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,753,114
Number of Sequences: 164201
Number of extensions: 2357190
Number of successful extensions: 5175
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 5119
Number of HSP's gapped (non-prelim): 46
length of query: 409
length of database: 59,974,054
effective HSP length: 113
effective length of query: 296
effective length of database: 41,419,341
effective search space: 12260124936
effective search space used: 12260124936
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0371a.1


