

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
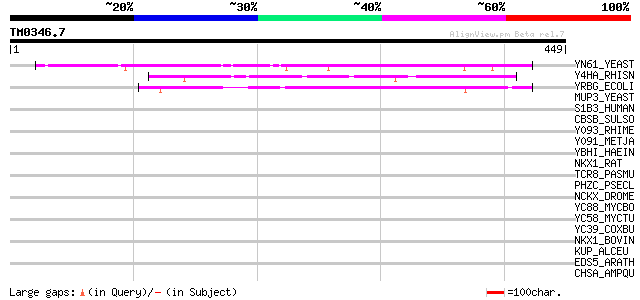
Query= TM0346.7
(449 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YN61_YEAST (P42839) Hypothetical 102.5 kDa protein in KRE1-HXT14... 108 4e-23
Y4HA_RHISN (P55471) Putative ionic transporter y4hA 54 6e-07
YRBG_ECOLI (P45394) Hypothetical protein yrbG 47 1e-04
MUP3_YEAST (P38734) Low-affinity methionine permease 34 0.88
S1B3_HUMAN (Q9NPD5) Solute carrier organic anion transporter fam... 33 1.1
CBSB_SULSO (P58030) Cytochrome b558/566 subunit B 33 1.5
Y093_RHIME (O87394) Hypothetical transport protein R00093 33 2.0
Y091_METJA (Q57556) Hypothetical protein MJ0091 33 2.0
YBHI_HAEIN (Q57048) Hypothetical protein HI0020 32 2.6
NKX1_RAT (Q9QZM6) Sodium/potassium/calcium exchanger 1 precursor... 32 4.4
TCR8_PASMU (P51564) Tetracycline resistance protein, class H (TE... 31 5.7
PHZC_PSECL (Q51517) Probable phospho-2-dehydro-3-deoxyheptonate ... 31 5.7
NCKX_DROME (Q9U6A0) Sodium/potassium/calcium exchanger (Na(+)/K(... 31 5.7
YC88_MYCBO (P64784) Hypothetical protein Mb1288c 31 7.4
YC58_MYCTU (P64783) Hypothetical protein Rv1258c/MT1297 31 7.4
YC39_COXBU (Q83C89) Hypothetical UPF0078 protein CBU1239 30 9.7
NKX1_BOVIN (Q28139) Sodium/potassium/calcium exchanger 1 precurs... 30 9.7
KUP_ALCEU (Q7WXK8) Probable potassium transport system protein kup 30 9.7
EDS5_ARATH (Q945F0) Enhanced disease susceptibility 5 (Eds5) (Sa... 30 9.7
CHSA_AMPQU (Q12564) Chitin synthase A (EC 2.4.1.16) (Chitin-UDP ... 30 9.7
>YN61_YEAST (P42839) Hypothetical 102.5 kDa protein in KRE1-HXT14
intergenic region
Length = 908
Score = 108 bits (269), Expect = 4e-23
Identities = 104/452 (23%), Positives = 202/452 (44%), Gaps = 56/452 (12%)
Query: 22 NEETSHENLSRSMVKRSDSVLVTNNINARFQMLRIFMTNLRVVLLGTKLVVLFPAVPLAV 81
N SHEN + + + ++L+ A + + + V+++ +V F V
Sbjct: 456 NSSQSHEN-EITQQQLNKNILLCTFRAAGWHYYKYTVDGTNVIVVNLISIVFFTIFDFYV 514
Query: 82 AADFYKFGRPW------IFAFSLLGLAPLAERVSFLTEQIAYFTGPTVGGLLNATCGNAT 135
+F + + W IF L PLA + I+ T VG ++NA
Sbjct: 515 LKNFLHW-KTWFTYESSIFILCLTSTIPLAFYIGQAVASISAQTSMGVGAVINAFFSTIV 573
Query: 136 EMIIAILALRKNKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADVNS 195
E+ + +AL++ K +V+ S++GS+L +LL+ G S +CGG N ++ QRY+ A V+S
Sbjct: 574 EIFLYCVALQQKKGLLVEGSMIGSILGAVLLLPGLS-MCGGALN-RKTQRYNPASAGVSS 631
Query: 196 LLLLLGLLCHLLPLLFKYSLAGGDYSI-----------------------ATSTLQ-LSR 231
LL+ ++ +P + Y + GG YS+ T +Q +S
Sbjct: 632 ALLIFSMIVMFVPTVL-YEIYGG-YSVNCADGANDRDCTFSHPPLKFNRLFTHVIQPMSI 689
Query: 232 ASSIVMLLAYVAYIFFQLKTHRRLF------DAQEVIIDIEEDDEEKAVIGFWSAFTWLV 285
+ +IV+ AY+ ++F L+TH ++ D + +E + A S T ++
Sbjct: 690 SCAIVLFCAYIIGLWFTLRTHAKMIWQLPIADPTSTAPEQQEQNSHDAPNWSRSKSTCIL 749
Query: 286 GM-TLVISLLSEYVVGTIEAA-SDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLD 343
M TL+ ++++E +V ++A D ++ F+ + + ++ N E +I FA +
Sbjct: 750 LMSTLLYAIIAEILVSCVDAVLEDIPSLNPKFLGLTIFALIPNTTEFLNAISFAIHGNVA 809
Query: 344 ISLGVAMGSATQISMFVVPLSVI--VAWIMGIKMDLDFNLLETGCLAF----------AI 391
+S+ + A Q+ + +P VI + + +K + + L F ++
Sbjct: 810 LSMEIGSAYALQVCLLQIPSLVIYSIFYTWNVKKSMINIRTQMFPLVFPRWDIFGAMTSV 869
Query: 392 IVTAFTLQDGTSHYMKGVVLTLCYLVIAACFF 423
+ + +G S+Y KG +L L Y++I F+
Sbjct: 870 FMFTYLYAEGKSNYFKGSMLILLYIIIVVGFY 901
>Y4HA_RHISN (P55471) Putative ionic transporter y4hA
Length = 367
Score = 54.3 bits (129), Expect = 6e-07
Identities = 71/307 (23%), Positives = 130/307 (42%), Gaps = 25/307 (8%)
Query: 113 EQIAYFTGPTVGGLLNATCGNATEMIIA---ILALRKNKVNVVKFSLLGSVLSNLLLVLG 169
E +A G G +L A C E+ I +L+ + V + ++ +V+ L V+G
Sbjct: 62 EILAARVGQPSGAILLAVCVTIIEVAIIGSLMLSGAEGNEEVARDTVFAAVMIVLNGVIG 121
Query: 170 SSLLCGGLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYSIATSTLQL 229
L+ GG + REQ + A ++ L +LG L L +L + AG A L +
Sbjct: 122 LCLVLGGRRH--REQSFQLNAA--SAALAVLGTLATLSLVLPNFVTAGKPQQFAAIQLVV 177
Query: 230 SRASSIVMLLAYVAYIFFQLKTHRRLF-DAQEVIIDIEEDDEEKAVIGFWSAFTWLVGMT 288
S+V+ Y ++F Q HR F D ++ + + + +A LV
Sbjct: 178 IGLVSVVL---YGVFLFVQTVRHRDYFIDDEDAASPPATHETPRNPL---AAGALLVLAL 231
Query: 289 LVISLLSEYVVGTIEAASDSWG-----ISVSFISIILLPIVGNAAEHAGSIIFAFKNKLD 343
+ + LL+ + +++A ++ G + V+ ++LLP E S+ A N+L
Sbjct: 232 IAVILLAMLLSYPLDSAVEALGLPQAVVGVTIAGVVLLP------EGITSVKAALMNRLQ 285
Query: 344 ISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQDGTS 403
S+ + +GSA +P+ ++ +G + L L + V TL G +
Sbjct: 286 NSINLVLGSALASIGVTIPVVAAISVALGRDLALGLAPQNLIMLILTLFVGTITLGTGRT 345
Query: 404 HYMKGVV 410
++G V
Sbjct: 346 TVLQGAV 352
>YRBG_ECOLI (P45394) Hypothetical protein yrbG
Length = 325
Score = 46.6 bits (109), Expect = 1e-04
Identities = 70/324 (21%), Positives = 136/324 (41%), Gaps = 30/324 (9%)
Query: 105 AERVSFLTEQIAYFTG--PTVGGLLNATCGNAT-EMIIAILALRKNKVNVVKFSLLGSVL 161
A+R+ F + G P + G+ + G + E+I+++ A + ++ + LGS +
Sbjct: 19 ADRLVFAASILCRTFGIPPLIIGMTVVSIGTSLPEVIVSLAASLHEQRDLAVGTALGSNI 78
Query: 162 SNLLLVLGSSLLCGGLANLKREQRYDRKQADVNSLLLLLGLLCHLLPLLFKYSLAGGDYS 221
N+LL+LG + L V + +L LPL+ S+ G
Sbjct: 79 INILLILGLAAL-------------------VRPFTVHSDVLRRELPLMLLVSVVAGS-- 117
Query: 222 IATSTLQLSRASSIVMLLAYVAYIFFQLKTHRRLFDAQEVIIDIEEDDEEKAVIGFWSAF 281
QLSR+ I +L V ++ F +K R+ + E+ E G AF
Sbjct: 118 -VLYDGQLSRSDGIFLLFLAVLWLLFIVKLARQAERQGTDSLTREQLAELPRDGGLPVAF 176
Query: 282 TWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNK 341
WL +++ + + VV ++ + IS + + + I + E A +I K +
Sbjct: 177 LWLGIALIIMPVATRMVVDNATVLANYFAISELTMGLTAIAIGTSLPELATAIAGVRKGE 236
Query: 342 LDISLGVAMGSATQISMFVVPLSVIV--AWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQ 399
DI++G +G+ + V+ L ++ I + D++++ + FA++ + Q
Sbjct: 237 NDIAVGNIIGANIFNIVIVLGLPALITPGEIDPLAYSRDYSVMLLVSIIFALLCWRRSPQ 296
Query: 400 DGTSHYMKGVVLTLCYLVIAACFF 423
G GV+LT ++V A +
Sbjct: 297 PGRG---VGVLLTGGFIVWLAMLY 317
>MUP3_YEAST (P38734) Low-affinity methionine permease
Length = 546
Score = 33.9 bits (76), Expect = 0.88
Identities = 30/120 (25%), Positives = 50/120 (41%), Gaps = 1/120 (0%)
Query: 278 WSAFTWLVGMTLVISLL-SEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIF 336
+S ++ L G L S++ +YV+ DSW VS II ++ + G I
Sbjct: 151 FSCYSVLTGYALTGSIVFGKYVLSAFGVTDDSWSKYVSISFIIFAVLIHGVSVRHGVFIQ 210
Query: 337 AFKNKLDISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAF 396
L + + V M A ++F + VAW + + +LL +A A I + F
Sbjct: 211 NALGGLKLIMIVLMCFAGLYTLFFYKSTGQVAWDLPVTQVEKDSLLSVSSIATAFISSFF 270
>S1B3_HUMAN (Q9NPD5) Solute carrier organic anion transporter family
member 1B3 (Solute carrier family 21 member 8) (Organic
anion transporter 8) (Organic anion transporting
polypeptide 8) (OATP8) (Liver-specific organic anion
transporter 2) (LST-2)
Length = 702
Score = 33.5 bits (75), Expect = 1.1
Identities = 37/135 (27%), Positives = 58/135 (42%), Gaps = 25/135 (18%)
Query: 43 VTNNINARFQMLRIFMTNLRVVLLGTKLVVLFPAVPLAVAADFYKFGRPWIFAFSLLGLA 102
VT N+ FQ L+ +TN L V+F + L + F ++F +
Sbjct: 318 VTKNVTGFFQSLKSILTN--------PLYVIFLLLTLLQVSSFIG-SFTYVFKYMEQQYG 368
Query: 103 PLAERVSFLTEQIAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVV---KFSLLGS 159
A +FL + T PTV AT M + ++K K+++V KFS L S
Sbjct: 369 QSASHANFL---LGIITIPTV----------ATGMFLGGFIIKKFKLSLVGIAKFSFLTS 415
Query: 160 VLSNLLLVLGSSLLC 174
++S L +L L+C
Sbjct: 416 MISFLFQLLYFPLIC 430
>CBSB_SULSO (P58030) Cytochrome b558/566 subunit B
Length = 314
Score = 33.1 bits (74), Expect = 1.5
Identities = 35/137 (25%), Positives = 59/137 (42%), Gaps = 5/137 (3%)
Query: 297 YVV--GTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSAT 354
YV+ G I++ + ISV +S+I N H + L +LG+ + +
Sbjct: 173 YVISWGKIKSLRNYIAISVGLLSMIPFIFFENIISHNRYLEILMNMILPSTLGITLYNPY 232
Query: 355 QISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQDGTSHYMKGVVLTLC 414
I++ V+ L + MGI L + +G F II T F DG S + + +
Sbjct: 233 HITLLVMALGLST---MGIVTSLIKGNVSSGVGYFLIITTVFLGIDGFSLLIYMLTPIIG 289
Query: 415 YLVIAACFFVNKTAQIN 431
+LVI + +K I+
Sbjct: 290 FLVITSSEIESKRRLID 306
>Y093_RHIME (O87394) Hypothetical transport protein R00093
Length = 465
Score = 32.7 bits (73), Expect = 2.0
Identities = 31/131 (23%), Positives = 55/131 (41%), Gaps = 12/131 (9%)
Query: 283 WLVGMTLV-ISLLSEYVVGTIEAASDSW-------GISVSFISIILLPIVGNAAEHAGSI 334
W+ G+ ++++ V T+ AA + G ++F + L + E A +I
Sbjct: 58 WVAGLLYTCVAMIDSEVTSTVAAAGGQYAQAKHIVGPLMAFNVGLFLVMAYTMLEAANAI 117
Query: 335 IFAFKNKLDISLGVAMGSATQISMFVVPLSVIVAWI--MGIKMDLDFNLLETGCLAFAII 392
F LD G+ + F+V + +AW+ G+ L FNL+ T AI+
Sbjct: 118 TVGFL--LDTVAGMQGQTGLNQQPFIVLAIMFLAWLNYRGVLATLTFNLVITAIAFLAIV 175
Query: 393 VTAFTLQDGTS 403
++Q G S
Sbjct: 176 ALFVSVQFGAS 186
>Y091_METJA (Q57556) Hypothetical protein MJ0091
Length = 302
Score = 32.7 bits (73), Expect = 2.0
Identities = 38/151 (25%), Positives = 74/151 (48%), Gaps = 6/151 (3%)
Query: 283 WLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKL 342
+L+G+ L+ +V+G+ E + + +S I ++ I + E S ++ +
Sbjct: 9 FLLGLILLYYGSDWFVLGS-ERIARHFNVSNFVIGATVMAIGTSLPEILTSAYASYMHAP 67
Query: 343 DISLGVAMGSATQISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQDGT 402
IS+G A+GS V+ LS I++ I+ + +L N+L L F I + DG
Sbjct: 68 GISIGNAIGSCICNIGLVLGLSAIISPII-VDKNLQKNILV--YLLFVIFAAVIGI-DGF 123
Query: 403 SHYMKGVVLTLCYLVIAACFFVNKTAQINQH 433
S ++ GVVL + +++ N +A+I ++
Sbjct: 124 S-WIDGVVLLILFIIYLRWTVKNGSAEIEEN 153
>YBHI_HAEIN (Q57048) Hypothetical protein HI0020
Length = 479
Score = 32.3 bits (72), Expect = 2.6
Identities = 39/171 (22%), Positives = 66/171 (37%), Gaps = 33/171 (19%)
Query: 196 LLLLLGLLCHLLPLLFKYSLAG----GDYSIATSTLQLSRASSIVMLLAYVAYIFFQLKT 251
+L+ + L+ LLP SL G Y L L V+LLA +A
Sbjct: 11 ILMAIPLITFLLPAPDGLSLIAWRLLGVYIATIVGLVLKPYGEPVILLAAIA-------- 62
Query: 252 HRRLFDAQEVIIDIEEDDEEKAVIG-----FWSAFTWLVGMTLVISL----------LSE 296
VII E +E +G + S TWL+ +S ++
Sbjct: 63 ------VSGVIIGNTEGAKELVKVGNMLDGYKSGTTWLIFTAFTLSSAFVITGLGKRIAY 116
Query: 297 YVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLG 347
+++G + + + G F+ ++L P + +G IIF N + ++LG
Sbjct: 117 HMIGAMGSTTLRLGYVTMFLDLLLSPATPSNTARSGGIIFPIINSVVVALG 167
>NKX1_RAT (Q9QZM6) Sodium/potassium/calcium exchanger 1 precursor
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger)
Length = 1181
Score = 31.6 bits (70), Expect = 4.4
Identities = 23/82 (28%), Positives = 37/82 (45%), Gaps = 4/82 (4%)
Query: 284 LVGMTLVISLLS----EYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFK 339
+ GMT V L+ EY V + +D IS + G+A E S+I F
Sbjct: 426 IFGMTYVFVALAIVCDEYFVPALGVITDKLQISEDVAGATFMAAGGSAPELFTSLIGVFI 485
Query: 340 NKLDISLGVAMGSATQISMFVV 361
+ ++ +G +GSA +FV+
Sbjct: 486 SHSNVGIGTIVGSAVFNILFVI 507
>TCR8_PASMU (P51564) Tetracycline resistance protein, class H
(TETA(H))
Length = 400
Score = 31.2 bits (69), Expect = 5.7
Identities = 36/117 (30%), Positives = 52/117 (43%), Gaps = 6/117 (5%)
Query: 87 KFGRPWIFAFSLLGLAPLAERVSFLTEQIAYFTGPTVGGLLNATCGNATEMIIAILALRK 146
K+GR I FSLLG A ++F T + G + G+ AT G ++ + K
Sbjct: 67 KYGRKPILLFSLLGAALDYLLMAFSTTLWMLYIGRIIAGITGAT-GAVCASAMSDVTPAK 125
Query: 147 NKVNVVKFSLLGSVLSNLLLVLGSSLLCGGLANLKREQRYDRKQADVNSLLLLLGLL 203
N+ F LG V + L++G L GGL A +S+LL+L LL
Sbjct: 126 NRTRY--FGFLGGVF-GVGLIIGPML--GGLLGDISAHMPFIFAAISHSILLILSLL 177
>PHZC_PSECL (Q51517) Probable phospho-2-dehydro-3-deoxyheptonate
aldolase (EC 2.5.1.54) (Phospho-2-keto-3-deoxyheptonate
aldolase) (DAHP synthetase)
(3-deoxy-D-arabino-heptulosonate 7-phosphate synthase)
Length = 400
Score = 31.2 bits (69), Expect = 5.7
Identities = 16/41 (39%), Positives = 21/41 (51%)
Query: 101 LAPLAERVSFLTEQIAYFTGPTVGGLLNATCGNATEMIIAI 141
L PL E V +I + + P G + A CGN T M+ AI
Sbjct: 282 LPPLVEAVRQAGHKIIWLSDPMHGNTIVAPCGNKTRMVQAI 322
>NCKX_DROME (Q9U6A0) Sodium/potassium/calcium exchanger
(Na(+)/K(+)/Ca(2+)-exchange protein)
Length = 856
Score = 31.2 bits (69), Expect = 5.7
Identities = 25/123 (20%), Positives = 56/123 (45%), Gaps = 5/123 (4%)
Query: 296 EYVVGTIEAASDSWGISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQ 355
E+ V +++ + GI+ + G+A E S+I F + D+ +G +GSA
Sbjct: 353 EFFVPSLDVIIEKLGITDDVAGATFMAAGGSAPELFTSVIGVFVSFDDVGIGTIVGSAVF 412
Query: 356 ISMFVVPLSVIVAWIMGIKMDLDFNLLETGCLAFAI--IVTAFTLQDGTSHYMKGVVLTL 413
+FV+ + + + + + L + L C ++I +V + +D + + ++L
Sbjct: 413 NILFVIGMCALFSKTV---LSLTWWPLFRDCSFYSISLLVLIYFFRDNRIFWWEALILFT 469
Query: 414 CYL 416
Y+
Sbjct: 470 IYI 472
>YC88_MYCBO (P64784) Hypothetical protein Mb1288c
Length = 419
Score = 30.8 bits (68), Expect = 7.4
Identities = 15/45 (33%), Positives = 23/45 (50%)
Query: 115 IAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGS 159
+A+ GP +GGL+ AT G T M I A + + + L G+
Sbjct: 152 LAFIVGPAIGGLMIATVGGITTMWITATAFGLSILAIAALQLEGA 196
>YC58_MYCTU (P64783) Hypothetical protein Rv1258c/MT1297
Length = 419
Score = 30.8 bits (68), Expect = 7.4
Identities = 15/45 (33%), Positives = 23/45 (50%)
Query: 115 IAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGS 159
+A+ GP +GGL+ AT G T M I A + + + L G+
Sbjct: 152 LAFIVGPAIGGLMIATVGGITTMWITATAFGLSILAIAALQLEGA 196
>YC39_COXBU (Q83C89) Hypothetical UPF0078 protein CBU1239
Length = 193
Score = 30.4 bits (67), Expect = 9.7
Identities = 24/110 (21%), Positives = 50/110 (44%), Gaps = 16/110 (14%)
Query: 310 GISVSFISIILLPIVGNAAEHAGSIIFAFKNKLDIS--LGVAMGSATQISMFVVPLSVIV 367
G+S++F+ ++ ++G H + F F+ ++ +GV +G + I +FV+ VIV
Sbjct: 78 GVSLAFVGLVA--VLG----HLFPVYFKFRGGKGVATMMGVLLGLSFWIGLFVIATWVIV 131
Query: 368 AWIMGIKMDLDFNLLETGCLAFAIIVTAFTLQDGTSHYMKGVVLTLCYLV 417
I F L A+ +T+ G + Y+ V++ ++
Sbjct: 132 VSI--------FRYSSVAALVSAVAAPIYTIIAGRTDYLFPVLIIAILII 173
>NKX1_BOVIN (Q28139) Sodium/potassium/calcium exchanger 1 precursor
(Na(+)/K(+)/Ca(2+)-exchange protein 1) (Retinal rod
Na-Ca+K exchanger)
Length = 1216
Score = 30.4 bits (67), Expect = 9.7
Identities = 23/100 (23%), Positives = 42/100 (42%), Gaps = 3/100 (3%)
Query: 262 IIDIEEDDEEKAVIGFWSAFTWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIILL 321
+ +EE + V+ + V + +V EY V + +D IS +
Sbjct: 438 LFSVEERRQGWVVLHIFGMMYVFVALAIVCD---EYFVPALGVITDKLQISEDVAGATFM 494
Query: 322 PIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQISMFVV 361
G+A E S+I F + ++ +G +GSA +FV+
Sbjct: 495 AAGGSAPELFTSLIGVFISHSNVGIGTIVGSAVFNILFVI 534
>KUP_ALCEU (Q7WXK8) Probable potassium transport system protein kup
Length = 632
Score = 30.4 bits (67), Expect = 9.7
Identities = 27/110 (24%), Positives = 54/110 (48%), Gaps = 20/110 (18%)
Query: 273 AVIGFWSAFTWLVGMTLVISLLSEYVVGTIEAASDSWGISVSFISIIL------------ 320
AV+G S W M +V+S+ +YVV + A +D G ++ ++++L
Sbjct: 55 AVLGVISILFW--AMIIVVSI--KYVVFVMRADNDGEGGVLALMALVLRTVAQRSRKAKI 110
Query: 321 ---LPIVGNAAEHAGSIIFAFKNKLDISLGVAMGSATQISMFVVPLSVIV 367
L I G + ++I + L G+ + +A Q+S FV+P+++++
Sbjct: 111 LMMLGIFGACMFYGDAVITPAISVLSAVEGLEI-AAPQLSQFVIPITLVI 159
>EDS5_ARATH (Q945F0) Enhanced disease susceptibility 5 (Eds5)
(Salicylic acid induction deficient 1) (Sid1)
Length = 543
Score = 30.4 bits (67), Expect = 9.7
Identities = 29/106 (27%), Positives = 52/106 (48%), Gaps = 18/106 (16%)
Query: 113 EQIAYFTGPTVGGLLNATCGNATEMIIAILALRKNKVNVVKFSLLGSVLSN-------LL 165
++I FTGP +G + CG +I ++ + + + + G+VL + L
Sbjct: 100 KEIVKFTGPAMGMWI---CGPLMSLIDTVVIGQGSSIELAALGP-GTVLCDHMSYVFMFL 155
Query: 166 LVLGSSLLCGGLANLKREQRYDRKQADVN-SLLLLLGLLCHLLPLL 210
V S+++ LA + D+K+A S+LL +GL+C L+ LL
Sbjct: 156 SVATSNMVATSLA------KQDKKEAQHQISVLLFIGLVCGLMMLL 195
>CHSA_AMPQU (Q12564) Chitin synthase A (EC 2.4.1.16) (Chitin-UDP
acetyl-glucosaminyl transferase A) (Class-I chitin
synthase A)
Length = 910
Score = 30.4 bits (67), Expect = 9.7
Identities = 23/63 (36%), Positives = 33/63 (51%), Gaps = 8/63 (12%)
Query: 288 TLVISLLSEYVVGTIEAA--SDSWGISVSFISIILL-PIVGNAAEHAGSIIFAFKNKLDI 344
TL+ISLLS YV+ + + D W + SFI +L+ P N ++AF N DI
Sbjct: 701 TLIISLLSTYVMWLVVSIIFLDPWHMFTSFIQYLLMTPTYINILN-----VYAFCNTHDI 755
Query: 345 SLG 347
+ G
Sbjct: 756 TWG 758
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.139 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,085,802
Number of Sequences: 164201
Number of extensions: 1751009
Number of successful extensions: 6640
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 6617
Number of HSP's gapped (non-prelim): 35
length of query: 449
length of database: 59,974,054
effective HSP length: 113
effective length of query: 336
effective length of database: 41,419,341
effective search space: 13916898576
effective search space used: 13916898576
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0346.7


