

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
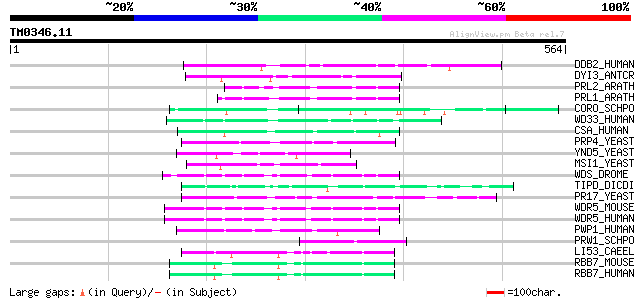
Query= TM0346.11
(564 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DDB2_HUMAN (Q92466) DNA damage binding protein 2 (Damage-specifi... 145 4e-34
DYI3_ANTCR (Q16960) Dynein intermediate chain 3, ciliary 57 1e-07
PRL2_ARATH (Q39190) PP1/PP2A phosphatases pleiotropic regulator ... 55 4e-07
PRL1_ARATH (Q42384) PP1/PP2A phosphatases pleiotropic regulator ... 55 4e-07
CORO_SCHPO (O13923) Coronin-like protein crn1 54 1e-06
WD33_HUMAN (Q9C0J8) WD-repeat protein 33 (WD-repeat protein WDC146) 53 2e-06
CSA_HUMAN (Q13216) Cockayne syndrome WD-repeat protein CSA (DNA ... 52 3e-06
PRP4_YEAST (P20053) U4/U6 small nuclear ribonucleoprotein PRP4 51 7e-06
YND5_YEAST (P53962) Hypothetical 43.8 kDa Trp-Asp repeats contai... 51 9e-06
MSI1_YEAST (P13712) Chromatin assembly factor 1 P50 subunit (CAF... 51 9e-06
WDS_DROME (Q9V3J8) Will die slowly protein 50 2e-05
TIPD_DICDI (O15736) Protein tipD 49 3e-05
PR17_YEAST (P40968) Pre-mRNA splicing factor PRP17 (Cell divisio... 49 3e-05
WDR5_MOUSE (P61965) WD-repeat protein 5 (WD-repeat protein BIG-3) 49 3e-05
WDR5_HUMAN (P61964) WD-repeat protein 5 49 3e-05
PWP1_HUMAN (Q13610) Periodic tryptophan protein 1 homolog (Kerat... 49 3e-05
PRW1_SCHPO (O14021) RbAp48-related WD40-repeat protein prw1 49 3e-05
LI53_CAEEL (P90916) Trp-Asp repeats containing protein lin-53 (A... 49 5e-05
RBB7_MOUSE (Q60973) Histone acetyltransferase type B subunit 2 (... 48 8e-05
RBB7_HUMAN (Q16576) Histone acetyltransferase type B subunit 2 (... 48 8e-05
>DDB2_HUMAN (Q92466) DNA damage binding protein 2 (Damage-specific
DNA binding protein 2) (DDB p48 subunit) (DDBb)
(UV-damaged DNA-binding protein 2) (UV-DDB 2)
Length = 427
Score = 145 bits (365), Expect = 4e-34
Identities = 102/333 (30%), Positives = 165/333 (48%), Gaps = 32/333 (9%)
Query: 177 RRITCLEFHPTKNNIILSGDKKGQLGVWDF-VRVHEKVVYGNIHSCLLNNMKFNPTNDCM 235
RR T L +HPT + + G K G + +W+F ++ + G + +KFNP N
Sbjct: 112 RRATSLAWHPTHPSTVAVGSKGGDIMLWNFGIKDKPTFIKGIGAGGSITGLKFNPLNTNQ 171
Query: 236 VYSASSDGTISFTDLETGI-----SSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVA 290
Y++S +GT D + I SS +N+ W +D+++ +V+
Sbjct: 172 FYASSMEGTTRLQDFKGNILRVFASSDTINI----------W--FCSLDVSASSRMVVTG 219
Query: 291 DNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRM 350
DN G + L++M + K + +HKK KV + NP L T D ++WD+R++
Sbjct: 220 DNVGNVILLNM--DGKELWNLRMHKK--KVTHVALNPCCDWFLATASVDQTVKIWDLRQV 275
Query: 351 ETGSS-IYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSHD 409
+S +Y L H V++A FSP G ++LTT + +RV+ + + D P I H H
Sbjct: 276 RGKASFLYSLPHRHPVNAACFSP-DGARLLTTDQKSEIRVYSA--SQWDCPLGLIPHPHR 332
Query: 410 FNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAA---LHPIDFIDVSTGKLVAEVMDP 466
++LTP KA W P+ +L VVGRY N+ L ID D ++GK++ ++ DP
Sbjct: 333 HFQHLTPIKAAWHPR---YNLIVVGRYPDPNFKSCTPYELRTIDVFDGNSGKMMCQLYDP 389
Query: 467 NITTISPVNKLHPNDDILASGSSSSLFIWKPKE 499
+ IS +N+ +P D LAS + IW +E
Sbjct: 390 ESSGISSLNEFNPMGDTLASAMGYHILIWSQEE 422
>DYI3_ANTCR (Q16960) Dynein intermediate chain 3, ciliary
Length = 597
Score = 57.0 bits (136), Expect = 1e-07
Identities = 57/234 (24%), Positives = 102/234 (43%), Gaps = 23/234 (9%)
Query: 179 ITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKV----VYGNIHSCLLNNMKFNPTNDC 234
+ CLE++P ++++ G GQ+ WD + + V V + H + +
Sbjct: 218 LVCLEYNPKDVHVLIGGCYNGQVAFWDTRKGSQAVEMSPVEHSHHDPVYKTIWLQSKTGT 277
Query: 235 MVYSASSDGTISFTDL-ETGISSSLMNLNP------EGWQGPSTWKMLYGMDINSEKGLV 287
+SAS+DG + + D+ + G + + ++P E QG + Y I ++
Sbjct: 278 ECFSASTDGQVLWWDMRKLGEPTEKLIMDPSKKGKMENAQG--VISLEYEPTIPTK---F 332
Query: 288 LVADNFGFLHLVDMRSNNKTGNAILIHKKNT-KVVGIHCNPIEPDILLTCGNDHYARVW- 345
+V G + + ++ + I+K++ V + NP P LT G D AR+W
Sbjct: 333 MVGTEQGTIISCNRKAKTPPEKIVAIYKEHIGPVYSLQRNPFFPKNFLTVG-DWTARIWS 391
Query: 346 -DIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMD 398
DIR S ++ H ++ +SPV TT +D L VWD +F + D
Sbjct: 392 EDIR---DSSIMWTKYHMSYLTDGCWSPVRPAVFFTTKMDGSLDVWDYLFKQKD 442
>PRL2_ARATH (Q39190) PP1/PP2A phosphatases pleiotropic regulator
PRL2
Length = 479
Score = 55.5 bits (132), Expect = 4e-07
Identities = 47/180 (26%), Positives = 85/180 (47%), Gaps = 22/180 (12%)
Query: 219 HSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGM 278
H + ++ F+P+N+ + S+D TI D+ TG+ + L G G + G+
Sbjct: 169 HLGWVRSVAFDPSNEWFC-TGSADRTIKIWDVATGV----LKLTLTGHIG-----QVRGL 218
Query: 279 DINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEP--DILLTC 336
+++ + A + + D+ N +I + + G++C + P D++LT
Sbjct: 219 AVSNRHTYMFSAGDDKQVKCWDLEQNK------VIRSYHGHLHGVYCLALHPTLDVVLTG 272
Query: 337 GNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGK 396
G D RVWDIR T I+ L H+ V S P + +++T S D+ ++ WD +GK
Sbjct: 273 GRDSVCRVWDIR---TKMQIFVLPHDSDVFSVLARP-TDPQVITGSHDSTIKFWDLRYGK 328
Score = 35.8 bits (81), Expect = 0.31
Identities = 28/88 (31%), Positives = 39/88 (43%), Gaps = 18/88 (20%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWD-------FVRVHEKVVYGNIHSCLLNNMK 227
H + CL HPT +++L+G + VWD FV H+ V+ ++
Sbjct: 253 HLHGVYCLALHPTL-DVVLTGGRDSVCRVWDIRTKMQIFVLPHDSDVF---------SVL 302
Query: 228 FNPTNDCMVYSASSDGTISFTDLETGIS 255
PT D V + S D TI F DL G S
Sbjct: 303 ARPT-DPQVITGSHDSTIKFWDLRYGKS 329
>PRL1_ARATH (Q42384) PP1/PP2A phosphatases pleiotropic regulator
PRL1
Length = 486
Score = 55.5 bits (132), Expect = 4e-07
Identities = 48/188 (25%), Positives = 88/188 (46%), Gaps = 25/188 (13%)
Query: 212 KVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPST 271
+V+ G H + ++ F+P+N+ + S+D TI D+ TG+ + + E
Sbjct: 170 RVIQG--HLGWVRSVAFDPSNEWFC-TGSADRTIKIWDVATGVLKLTLTGHIE------- 219
Query: 272 WKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEP- 330
+ G+ +++ + A + + D+ N +I + + G++C + P
Sbjct: 220 --QVRGLAVSNRHTYMFSAGDDKQVKCWDLEQNK------VIRSYHGHLSGVYCLALHPT 271
Query: 331 -DILLTCGNDHYARVWDIRRMETGSSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLR 388
D+LLT G D RVWDIR T I+ L H+ V S + P + +++T S D ++
Sbjct: 272 LDVLLTGGRDSVCRVWDIR---TKMQIFALSGHDNTVCSVFTRP-TDPQVVTGSHDTTIK 327
Query: 389 VWDSIFGK 396
WD +GK
Sbjct: 328 FWDLRYGK 335
>CORO_SCHPO (O13923) Coronin-like protein crn1
Length = 601
Score = 53.9 bits (128), Expect = 1e-06
Identities = 83/420 (19%), Positives = 162/420 (37%), Gaps = 60/420 (14%)
Query: 163 IPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVV-YGNIH-- 219
+PDQVN + R H+ + +++P + ++ SG ++ +W + + Y ++H
Sbjct: 70 LPDQVN--LFRGHTAAVLDTDWNPFHDQVLASGGDDSKIMIWKVPEDYTVMEPYEDVHPI 127
Query: 220 ------SCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWK 273
S + ++++PT ++ S+S+D TI D E G++ + ++
Sbjct: 128 AELKGHSRKVGLVQYHPTAANVLASSSADNTIKLWDCEKGVAHVSLKMD----------V 177
Query: 274 MLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDIL 333
M M N++ ++ + + D R++ K V H P ++
Sbjct: 178 MCQSMSFNADGTRLVTTSRDKKVRVWDPRTD-----------KPVSVGNGHAGAKNPRVV 226
Query: 334 LTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDS- 392
D +A + + +++D + P+ G L T + WD
Sbjct: 227 WLGSLDRFATTGFSKMSDRQIALWD-------PTNLSEPIGGFTTLDTGSGILMPFWDDG 279
Query: 393 -----IFGKMDSPSREIVHSHDFNRYLTPFKAEWDPKDPSESLAVVGRYISEN------- 440
+ GK D R + +D YL+ FK+ DP+ L G +SEN
Sbjct: 280 TKVIYLAGKGDGNIRYYEYENDVFHYLSEFKSV-DPQRGIAFLPKRGVNVSENEVMRAYK 338
Query: 441 -YNGAALHPIDFIDVSTGKLVAEVMDPNITTISPVNKLHPNDDILASGSSS--SLFIWKP 497
N + + PI FI +E +I +P K + ASG + L
Sbjct: 339 SVNDSIIEPISFIVPRR----SESFQSDIYPPAPSGKPSLTAEEWASGKDAQPDLLDMST 394
Query: 498 KEKSEPVHEKDESKIVVCGRAEKKRGKKRGADTDESDNEGFNSKLKKSKFQKTSKSQKTK 557
+S+ EK S V A+ ++ + +T + + + + + ++ QK SK + K
Sbjct: 395 LYESKGTVEKAVSATVPSAGAQVQKHNEEKVETPKPEAQPVSKPKESAEEQKPSKEPEVK 454
Score = 46.2 bits (108), Expect = 2e-04
Identities = 45/230 (19%), Positives = 87/230 (37%), Gaps = 46/230 (20%)
Query: 294 GFLHLVDMRSNNKTGNAILIHKKNTK-VVGIHCNPIEPDILLTCGNDHYARVW----DIR 348
G L ++ + K + + + + +T V+ NP +L + G+D +W D
Sbjct: 57 GALAVIPLNERGKLPDQVNLFRGHTAAVLDTDWNPFHDQVLASGGDDSKIMIWKVPEDYT 116
Query: 349 RMETGSSIYDLK----HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFG------KMD 398
ME ++ + H+R V + P + N + ++S DN +++WD G KMD
Sbjct: 117 VMEPYEDVHPIAELKGHSRKVGLVQYHPTAANVLASSSADNTIKLWDCEKGVAHVSLKMD 176
Query: 399 SPSREIVHSHDFNRYLTPFKAE----WDPKDPSESLAVVGRYISENYNGAALHPIDFIDV 454
+ + + D R +T + + WDP+ V
Sbjct: 177 VMCQSMSFNADGTRLVTTSRDKKVRVWDPRTDK-------------------------PV 211
Query: 455 STGKLVAEVMDPNITTISPVNKLHPNDDILASGSSSSLFIWKPKEKSEPV 504
S G A +P + + +++ + S + +W P SEP+
Sbjct: 212 SVGNGHAGAKNPRVVWLGSLDRFATTG--FSKMSDRQIALWDPTNLSEPI 259
>WD33_HUMAN (Q9C0J8) WD-repeat protein 33 (WD-repeat protein WDC146)
Length = 1336
Score = 52.8 bits (125), Expect = 2e-06
Identities = 60/283 (21%), Positives = 113/283 (39%), Gaps = 23/283 (8%)
Query: 160 KYVIPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIH 219
KY + N + + H I F PT N D G + +WDF+R HE+ + H
Sbjct: 186 KYWQSNMNNVKMFQAHKEAIREASFSPTDNKFATCSDD-GTVRIWDFLRCHEERILRG-H 243
Query: 220 SCLLNNMKFNPTNDCMVY-SASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGM 278
+ + ++PT +V S S I F D +TG S + ++ + + +
Sbjct: 244 GADVKCVDWHPTKGLVVSGSKDSQQPIKFWDPKTGQSLATLHAHK---------NTVMEV 294
Query: 279 DINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGN 338
+N +L A L D+R+ + HKK V H P+ + + G+
Sbjct: 295 KLNLNGNWLLTASRDHLCKLFDIRNLKEELQVFRGHKKEATAVAWH--PVHEGLFASGGS 352
Query: 339 DHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMD 398
D W + +E ++ H ++ S + P+ G+ + + S D+ + W +
Sbjct: 353 DGSLLFWHV-GVEKEVGGMEMAHEGMIWSLAWHPL-GHILCSGSNDHTSKFW-----TRN 405
Query: 399 SPSREIVHSHDFNRY--LTPFKAEWDPKDPSESLAVVGRYISE 439
P ++ ++ N ++ E+D +P+ + G I E
Sbjct: 406 RPGDKMRDRYNLNLLPGMSEDGVEYDDLEPNSLAVIPGMGIPE 448
>CSA_HUMAN (Q13216) Cockayne syndrome WD-repeat protein CSA (DNA
excision repair protein ERCC-8)
Length = 396
Score = 52.4 bits (124), Expect = 3e-06
Identities = 55/252 (21%), Positives = 102/252 (39%), Gaps = 37/252 (14%)
Query: 171 VIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYG------------NI 218
V R H I L+ P + +LSG G + ++D + Y ++
Sbjct: 38 VERIHGGGINTLDIEPVEGRYMLSGGSDGVIVLYDLENSSRQSYYTCKAVCSIGRDHPDV 97
Query: 219 HSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGM 278
H + +++ P + M S+S D T+ D T ++ + N T +
Sbjct: 98 HRYSVETVQWYPHDTGMFTSSSFDKTLKVWDTNTLQTADVFNFE-------ETVYSHHMS 150
Query: 279 DINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGN 338
++++ LV V + L D++S + + ++ +++ + +P IL T
Sbjct: 151 PVSTKHCLVAVGTRGPKVQLCDLKSGSCSH---ILQGHRQEILAVSWSPRYDYILATASA 207
Query: 339 DHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVS--------------GNKILTTSLD 384
D ++WD+RR +G I +HN S A S + G +LT D
Sbjct: 208 DSRVKLWDVRRA-SGCLITLDQHNGKKSQAVESANTAHNGKVNGLCFTSDGLHLLTVGTD 266
Query: 385 NRLRVWDSIFGK 396
NR+R+W+S G+
Sbjct: 267 NRMRLWNSSNGE 278
>PRP4_YEAST (P20053) U4/U6 small nuclear ribonucleoprotein PRP4
Length = 465
Score = 51.2 bits (121), Expect = 7e-06
Identities = 52/222 (23%), Positives = 94/222 (41%), Gaps = 18/222 (8%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKV-VYGNI--HSCLLNNMKFNPT 231
H +I +++HP NN ++S + G + + + + + G++ H ++++K++P+
Sbjct: 217 HVGKIGAIDWHPDSNNQMISCAEDGLIKNFQYSNEEGGLRLLGDLVGHERRISDVKYHPS 276
Query: 232 NDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVAD 291
+ SAS D T D T L + +G ++ + + LV
Sbjct: 277 GK-FIGSASHDMTWRLWDASTHQELLLQEGHDKG---------VFSLSFQCDGSLVCSGG 326
Query: 292 NFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRME 351
L D+RS +K + H K V N + + T G D VWDIR+ +
Sbjct: 327 MDSLSMLWDIRSGSKV-MTLAGHSKPIYTVAWSPNGYQ---VATGGGDGIINVWDIRKRD 382
Query: 352 TGSSIYDLKHNRVVSSAYFSPV-SGNKILTTSLDNRLRVWDS 392
G L H +V+ FS G K+++ DN + V+ S
Sbjct: 383 EGQLNQILAHRNIVTQVRFSKEDGGKKLVSCGYDNLINVYSS 424
>YND5_YEAST (P53962) Hypothetical 43.8 kDa Trp-Asp repeats
containing protein in NCE3-HHT2 intergenic region
Length = 389
Score = 50.8 bits (120), Expect = 9e-06
Identities = 50/190 (26%), Positives = 84/190 (43%), Gaps = 23/190 (12%)
Query: 170 AVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVR------VHEKVVYGNIHSCLL 223
++I H +TC++FHP+ NI+LSG G ++D + +H+ + Y +IHSC
Sbjct: 142 SLIDSHHDDVTCIKFHPSDVNILLSGSTDGYTNIYDLKQDEEEDALHQVINYASIHSCGW 201
Query: 224 NNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNP-EGWQGPSTWKMLYGMDINS 282
+ K +++ S T + +L S L P + W Y +DI
Sbjct: 202 LSPK-------RIFTLSHMETFAIHELNDK-SDELKEPQPLDFGDVREIWNCDYVVDI-- 251
Query: 283 EKGLVLVA---DNFGFLHLVDMRSNN-KTGNAILI-HKKNTKVV-GIHCNPIEPDILLTC 336
GL+ +N G L L+ + T N I+I H +VV I ++L +C
Sbjct: 252 YPGLIATGKTQENCGELCLLPFKDEKVDTENGIVIPHAHGDEVVRDIFIPAQHSNMLYSC 311
Query: 337 GNDHYARVWD 346
G D ++W+
Sbjct: 312 GEDGCVKIWE 321
>MSI1_YEAST (P13712) Chromatin assembly factor 1 P50 subunit (CAF-1
P50 subunit) (MSI1 protein) (IRA1 multicopy suppressor)
Length = 422
Score = 50.8 bits (120), Expect = 9e-06
Identities = 49/182 (26%), Positives = 81/182 (43%), Gaps = 18/182 (9%)
Query: 180 TCLEFHPTKNNIILSGDKKGQLGVWDFVRV-HEK------VVYGNIHSCLLNNMKFNPTN 232
T L ++ + ++LS GQ+ VWD + HE +V N +N++ + PT+
Sbjct: 204 TSLAWNLQQEALLLSSHSNGQVQVWDIKQYSHENPIIDLPLVSINSDGTAVNDVTWMPTH 263
Query: 233 DCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLV-AD 291
D + + + +S DL T + N E G + N + L+L AD
Sbjct: 264 DSLFAACTEGNAVSLLDLRT--KKEKLQSNREKHDGG-----VNSCRFNYKNSLILASAD 316
Query: 292 NFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCG-NDHYARVWDIRRM 350
+ G L+L D+R+ NK+ A + H T V + +P +L T G D ++WD
Sbjct: 317 SNGRLNLWDIRNMNKSPIATMEH--GTSVSTLEWSPNFDTVLATAGQEDGLVKLWDTSCE 374
Query: 351 ET 352
ET
Sbjct: 375 ET 376
Score = 36.2 bits (82), Expect = 0.23
Identities = 24/114 (21%), Positives = 48/114 (42%), Gaps = 6/114 (5%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H + F+ + I+ S D G+L +WD +++ + H ++ ++++P D
Sbjct: 295 HDGGVNSCRFNYKNSLILASADSNGRLNLWDIRNMNKSPIATMEHGTSVSTLEWSPNFDT 354
Query: 235 MVYSA-SSDGTISF--TDLETGI---SSSLMNLNPEGWQGPSTWKMLYGMDINS 282
++ +A DG + T E I ++ +N W W M + NS
Sbjct: 355 VLATAGQEDGLVKLWDTSCEETIFTHGGHMLGVNDISWDAHDPWLMCSVANDNS 408
>WDS_DROME (Q9V3J8) Will die slowly protein
Length = 361
Score = 49.7 bits (117), Expect = 2e-05
Identities = 56/243 (23%), Positives = 109/243 (44%), Gaps = 27/243 (11%)
Query: 156 SIKPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWD-FVRVHEKVV 214
S+KP Y + + H++ ++ ++F P + S K + +W + EK +
Sbjct: 57 SVKPNYTLK-----FTLAGHTKAVSAVKFSPNGEWLASSSADK-LIKIWGAYDGKFEKTI 110
Query: 215 YGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKM 274
G H ++++ ++ + +V S S D T+ +L TG S + +G S +
Sbjct: 111 SG--HKLGISDVAWSSDSRLLV-SGSDDKTLKVWELSTGKSLKTL-------KGHSNY-- 158
Query: 275 LYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAI-LIHKKNTKVVGIHCNPIEPDIL 333
++ + N + L++ + + D+R TG + + + V +H N + ++
Sbjct: 159 VFCCNFNPQSNLIVSGSFDESVRIWDVR----TGKCLKTLPAHSDPVSAVHFNR-DGSLI 213
Query: 334 LTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSI 393
++ D R+WD + ++ D N VS FSP +G IL +LDN L++WD
Sbjct: 214 VSSSYDGLCRIWDTASGQCLKTLID-DDNPPVSFVKFSP-NGKYILAATLDNTLKLWDYS 271
Query: 394 FGK 396
GK
Sbjct: 272 KGK 274
Score = 36.6 bits (83), Expect = 0.18
Identities = 46/206 (22%), Positives = 84/206 (40%), Gaps = 20/206 (9%)
Query: 189 NNIILSGDKKGQLGVWDFVRVHE-KVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISF 247
+ +++SG L VW+ K + G HS + FNP ++ +V S S D ++
Sbjct: 126 SRLLVSGSDDKTLKVWELSTGKSLKTLKG--HSNYVFCCNFNPQSNLIV-SGSFDESVRI 182
Query: 248 TDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNN-- 305
D+ TG + P+ + + N + L++ + G + D S
Sbjct: 183 WDVRTGKCLKTL---------PAHSDPVSAVHFNRDGSLIVSSSYDGLCRIWDTASGQCL 233
Query: 306 KTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVV 365
KT LI N V + +P +L D+ ++WD + + + K+ +
Sbjct: 234 KT----LIDDDNPPVSFVKFSP-NGKYILAATLDNTLKLWDYSKGKCLKTYTGHKNEKYC 288
Query: 366 SSAYFSPVSGNKILTTSLDNRLRVWD 391
A FS G I++ S DN + +W+
Sbjct: 289 IFANFSVTGGKWIVSGSEDNMVYIWN 314
>TIPD_DICDI (O15736) Protein tipD
Length = 612
Score = 49.3 bits (116), Expect = 3e-05
Identities = 81/343 (23%), Positives = 129/343 (36%), Gaps = 61/343 (17%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H+ I C+ F+ N + G K + VWD + +K +++ + F+P ND
Sbjct: 323 HNSEIYCMAFNSIGNLLATGGGDKC-VKVWDVISGQQKSTLLGASQSIVS-VSFSP-NDE 379
Query: 235 MVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNFG 294
+ S+D + + E G S + G G K+ G INS + V+ +
Sbjct: 380 SILGTSNDNSARLWNTELGRSRHTLT----GHIG----KVYTGKFINSNR--VVTGSHDR 429
Query: 295 FLHLVDMRSNNKTGNAILIHKKNTKVV----GIHCNPIEPDILLTCGN-DHYARVWDIRR 349
+ L D++ T N V+ G H L G+ DH R WD
Sbjct: 430 TIKLWDLQKGYCTRTIFCFSSCNDLVILGGSGTH---------LASGHVDHSVRFWDSNA 480
Query: 350 METGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSHD 409
E + + H ++S SP + N+ILT S D+ L++ D + +
Sbjct: 481 GEPTQVLSSI-HEGQITSITNSPTNTNQILTNSRDHTLKIIDI----RTFDTIRTFKDPE 535
Query: 410 FNRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVAEVMDPNIT 469
+ L KA W P GRYI+ +G+ I D + GK
Sbjct: 536 YRNGLNWTKASWSPD---------GRYIA---SGSIDGSICIWDATNGK----------- 572
Query: 470 TISPVNKLHPNDDILASGSSSSLFIWKPKEKSEPVHEKDESKI 512
T+ + K+H N GSS W P +KD++ I
Sbjct: 573 TVKVLTKVHNN------GSSVCCCSWSPLANIFISADKDKNII 609
>PR17_YEAST (P40968) Pre-mRNA splicing factor PRP17 (Cell division
control protein 40)
Length = 455
Score = 49.3 bits (116), Expect = 3e-05
Identities = 74/326 (22%), Positives = 133/326 (40%), Gaps = 39/326 (11%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H T L+F P ++ILSG + +WDF +E + H+ + ++F T DC
Sbjct: 161 HPEGTTALKFLPKTGHLILSGGNDHTIKIWDFYHDYECLRDFQGHNKPIKALRF--TEDC 218
Query: 235 MVY-SASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSEKGLVLVADNF 293
+ S+S D ++ D ETG + ++LN ST + N + +V ++ N
Sbjct: 219 QSFLSSSFDRSVKIWDTETGKVKTRLHLN-------STPADVESRPTNPHEFIVGLS-NS 270
Query: 294 GFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPD--ILLTCGNDHYARVWDIRRME 351
LH D S N+ L+ + + I PD ++ D R+W+ +
Sbjct: 271 KILHYDDRVSENQG----LVQTYDHHLSSILALKYFPDGSKFISSSEDKTVRIWENQINV 326
Query: 352 TGSSIYD-LKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSREIVHSHDF 410
I D +H+ + + S N S+DNR+ + S+ K ++I H
Sbjct: 327 PIKQISDTAQHSMPFLNVH---PSQNYFCAQSMDNRIYSF-SLKPKYKRHPKKIFKGHSS 382
Query: 411 NRYLTPFKAEWDPKDPSESLAVVGRYISENYNGAALHPIDFIDVSTGKLVAEVMDPNITT 470
Y D GRYI + + L D+ +T +L+ + P
Sbjct: 383 AGYGISLAFSGD-----------GRYICSGDSKSRLFTWDW---NTSRLLNNIKIPGNKP 428
Query: 471 ISPVNKLHPND--DILASGSSSSLFI 494
I+ V+ HP + ++ SG++ +++
Sbjct: 429 ITQVD-WHPQETSKVICSGAAGKIYV 453
>WDR5_MOUSE (P61965) WD-repeat protein 5 (WD-repeat protein BIG-3)
Length = 334
Score = 48.9 bits (115), Expect = 3e-05
Identities = 55/241 (22%), Positives = 109/241 (44%), Gaps = 22/241 (9%)
Query: 158 KPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWD-FVRVHEKVVYG 216
KP V P+ + H++ ++ ++F P + S K + +W + EK + G
Sbjct: 27 KPTPVKPNYALKFTLAGHTKAVSSVKFSPNGEWLASSSADK-LIKIWGAYDGKFEKTISG 85
Query: 217 NIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLY 276
H ++++ ++ ++ +V SAS D T+ D+ +G + +G S + ++
Sbjct: 86 --HKLGISDVAWSSDSNLLV-SASDDKTLKIWDVSSGKCLKTL-------KGHSNY--VF 133
Query: 277 GMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAI-LIHKKNTKVVGIHCNPIEPDILLT 335
+ N + L++ + + D+ KTG + + + V +H N + ++++
Sbjct: 134 CCNFNPQSNLIVSGSFDESVRIWDV----KTGKCLKTLPAHSDPVSAVHFNR-DGSLIVS 188
Query: 336 CGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFG 395
D R+WD + ++ D N VS FSP +G IL +LDN L++WD G
Sbjct: 189 SSYDGLCRIWDTASGQCLKTLID-DDNPPVSFVKFSP-NGKYILAATLDNTLKLWDYSKG 246
Query: 396 K 396
K
Sbjct: 247 K 247
>WDR5_HUMAN (P61964) WD-repeat protein 5
Length = 334
Score = 48.9 bits (115), Expect = 3e-05
Identities = 55/241 (22%), Positives = 109/241 (44%), Gaps = 22/241 (9%)
Query: 158 KPKYVIPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWD-FVRVHEKVVYG 216
KP V P+ + H++ ++ ++F P + S K + +W + EK + G
Sbjct: 27 KPTPVKPNYALKFTLAGHTKAVSSVKFSPNGEWLASSSADK-LIKIWGAYDGKFEKTISG 85
Query: 217 NIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLY 276
H ++++ ++ ++ +V SAS D T+ D+ +G + +G S + ++
Sbjct: 86 --HKLGISDVAWSSDSNLLV-SASDDKTLKIWDVSSGKCLKTL-------KGHSNY--VF 133
Query: 277 GMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAI-LIHKKNTKVVGIHCNPIEPDILLT 335
+ N + L++ + + D+ KTG + + + V +H N + ++++
Sbjct: 134 CCNFNPQSNLIVSGSFDESVRIWDV----KTGKCLKTLPAHSDPVSAVHFNR-DGSLIVS 188
Query: 336 CGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFG 395
D R+WD + ++ D N VS FSP +G IL +LDN L++WD G
Sbjct: 189 SSYDGLCRIWDTASGQCLKTLID-DDNPPVSFVKFSP-NGKYILAATLDNTLKLWDYSKG 246
Query: 396 K 396
K
Sbjct: 247 K 247
>PWP1_HUMAN (Q13610) Periodic tryptophan protein 1 homolog
(Keratinocyte protein IEF SSP 9502)
Length = 501
Score = 48.9 bits (115), Expect = 3e-05
Identities = 44/210 (20%), Positives = 93/210 (43%), Gaps = 18/210 (8%)
Query: 170 AVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFN 229
A + H+ ++ L+FHP + ++SG + ++D E S + + +N
Sbjct: 293 ASLAVHTDKVQTLQFHPFEAQTLISGSYDKSVALYDCRSPDESHRMWRF-SGQIERVTWN 351
Query: 230 PTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPSTWKMLYGMDINSE-KGLVL 288
+ C +++ DG + +L+ + LN + + G+D++S+ KG ++
Sbjct: 352 HFSPCHFLASTDDGFVY--NLDARSDKPIFTLNAHNDE-------ISGLDLSSQIKGCLV 402
Query: 289 VADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPD---ILLTCGNDHYARVW 345
A ++ + D+ + + L+H ++ K+ + C+ PD I G RVW
Sbjct: 403 TASADKYVKIWDILGDRPS----LVHSRDMKMGVLFCSSCCPDLPFIYAFGGQKEGLRVW 458
Query: 346 DIRRMETGSSIYDLKHNRVVSSAYFSPVSG 375
DI + + + + + V+ SA S +SG
Sbjct: 459 DISTVSSVNEAFGRRERLVLGSARNSSISG 488
>PRW1_SCHPO (O14021) RbAp48-related WD40-repeat protein prw1
Length = 431
Score = 48.9 bits (115), Expect = 3e-05
Identities = 33/111 (29%), Positives = 52/111 (46%), Gaps = 4/111 (3%)
Query: 295 FLHLVDMRSNN-KTGNAILIHKKNTKVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETG 353
+LH+ D+R + T A +H + + + NP IL TC D +WD+R +
Sbjct: 258 YLHVHDIRRPDASTKPARSVHAHSGPIHSVAFNPHNDFILATCSTDKTIALWDLRNL--N 315
Query: 354 SSIYDLK-HNRVVSSAYFSPVSGNKILTTSLDNRLRVWDSIFGKMDSPSRE 403
++ L+ H +V+ FSP + +TS D R VWD D P+ E
Sbjct: 316 QRLHTLEGHEDIVTKISFSPHEEPILASTSADRRTLVWDLSRIGEDQPAEE 366
Score = 36.2 bits (82), Expect = 0.23
Identities = 29/118 (24%), Positives = 52/118 (43%), Gaps = 14/118 (11%)
Query: 284 KGLVLVADNFGFLHLVDMRSNNKTGNAILI------HKKNTKVVGIHCNPIEPDILLTCG 337
KG ++ L D+ + N++ +A ++ H+K V H D+L +
Sbjct: 197 KGTLVSGSQDATLSCWDLNAYNESDSASVLKVHISSHEKQVSDVRFHYK--HQDLLASVS 254
Query: 338 NDHYARVWDIRRMETGS----SIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWD 391
D Y V DIRR + + S++ H+ + S F+P + + T S D + +WD
Sbjct: 255 YDQYLHVHDIRRPDASTKPARSVH--AHSGPIHSVAFNPHNDFILATCSTDKTIALWD 310
Score = 33.9 bits (76), Expect = 1.2
Identities = 22/82 (26%), Positives = 40/82 (47%), Gaps = 3/82 (3%)
Query: 172 IRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYG---NIHSCLLNNMKF 228
I H ++++ + FH +++ S L V D R + HS ++++ F
Sbjct: 230 ISSHEKQVSDVRFHYKHQDLLASVSYDQYLHVHDIRRPDASTKPARSVHAHSGPIHSVAF 289
Query: 229 NPTNDCMVYSASSDGTISFTDL 250
NP ND ++ + S+D TI+ DL
Sbjct: 290 NPHNDFILATCSTDKTIALWDL 311
>LI53_CAEEL (P90916) Trp-Asp repeats containing protein lin-53
(Abnormal cell lineage protein 53)
Length = 385
Score = 48.5 bits (114), Expect = 5e-05
Identities = 49/232 (21%), Positives = 94/232 (40%), Gaps = 27/232 (11%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLN---------N 225
H + + P K+NII + + ++D+++ H V N + L+
Sbjct: 87 HEGEVNRARYMPQKSNIIATKSPHADVYIFDYLK-HSAVPRDNTFNPLIRLKGHTKEGYG 145
Query: 226 MKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPEGWQGPST------WKMLYGMD 279
+ +NP + ++ SAS D T+ D+ + + + ++G + W +L+
Sbjct: 146 LSWNPNKEGLILSASDDQTVCHWDINANQNVAGELQAKDVFKGHESVVEDVAWHVLHDGV 205
Query: 280 INSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNTKVVGIHCNPIEPDILLTCGND 339
S V D+ L + D+R++ G+ I H + NP IL T D
Sbjct: 206 FGS------VGDDKKLL-IWDVRTST-PGHCIDAHSAEVNCLAF--NPYSEFILATGSAD 255
Query: 340 HYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKILTTSLDNRLRVWD 391
+WD+R + ++ H + +SP + + ++ D RL VWD
Sbjct: 256 KTVALWDLRNLRMKLHSFE-SHRDEIFQVQWSPHNETILASSGTDKRLHVWD 306
Score = 37.7 bits (86), Expect = 0.081
Identities = 18/76 (23%), Positives = 37/76 (48%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
HS + CL F+P I+ +G + +WD + K+ H + ++++P N+
Sbjct: 232 HSAEVNCLAFNPYSEFILATGSADKTVALWDLRNLRMKLHSFESHRDEIFQVQWSPHNET 291
Query: 235 MVYSASSDGTISFTDL 250
++ S+ +D + DL
Sbjct: 292 ILASSGTDKRLHVWDL 307
>RBB7_MOUSE (Q60973) Histone acetyltransferase type B subunit 2
(Retinoblastoma binding protein p46)
(Retinoblastoma-binding protein 7) (RBBP-7)
Length = 425
Score = 47.8 bits (112), Expect = 8e-05
Identities = 52/253 (20%), Positives = 95/253 (36%), Gaps = 41/253 (16%)
Query: 163 IPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDF---------------- 206
+ ++ C + H + + P +II + + V+D+
Sbjct: 110 VTGKIECEIKINHEGEVNRARYMPQNPHIIATKTPSSDVLVFDYTKHPAKPDPSGECNPD 169
Query: 207 --VRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPE 264
+R H+K YG + +N + SAS D T+ D+ G +
Sbjct: 170 LRLRGHQKEGYG---------LSWNSNLSGHLLSASDDHTVCLWDINAGPKEGKIVDAKA 220
Query: 265 GWQGPST------WKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNT 318
+ G S W +L+ S VAD+ + + D RSN + + L+
Sbjct: 221 IFTGHSAVVEDVAWHLLHESLFGS------VADDQKLM-IWDTRSNTTSKPSHLVDAHTA 273
Query: 319 KVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKI 378
+V + NP IL T D +WD+R ++ ++ H + ++SP + +
Sbjct: 274 EVNCLSFNPYSEFILATGSADKTVALWDLRNLKLKLHTFE-SHKDEIFQVHWSPHNETIL 332
Query: 379 LTTSLDNRLRVWD 391
++ D RL VWD
Sbjct: 333 ASSGTDRRLNVWD 345
Score = 36.6 bits (83), Expect = 0.18
Identities = 17/76 (22%), Positives = 37/76 (48%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H+ + CL F+P I+ +G + +WD + K+ H + + ++P N+
Sbjct: 271 HTAEVNCLSFNPYSEFILATGSADKTVALWDLRNLKLKLHTFESHKDEIFQVHWSPHNET 330
Query: 235 MVYSASSDGTISFTDL 250
++ S+ +D ++ DL
Sbjct: 331 ILASSGTDRRLNVWDL 346
Score = 31.6 bits (70), Expect = 5.8
Identities = 19/84 (22%), Positives = 35/84 (41%), Gaps = 12/84 (14%)
Query: 319 KVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK------------HNRVVS 366
++ +H +P IL + G D VWD+ ++ S D + H +S
Sbjct: 318 EIFQVHWSPHNETILASSGTDRRLNVWDLSKIGEEQSAEDAEDGPPELLFIHGGHTAKIS 377
Query: 367 SAYFSPVSGNKILTTSLDNRLRVW 390
++P I + S DN +++W
Sbjct: 378 DFSWNPNEPWVICSVSEDNIMQIW 401
>RBB7_HUMAN (Q16576) Histone acetyltransferase type B subunit 2
(Retinoblastoma binding protein P46)
(Retinoblastoma-binding protein 7) (RBBP-7)
Length = 425
Score = 47.8 bits (112), Expect = 8e-05
Identities = 52/253 (20%), Positives = 95/253 (36%), Gaps = 41/253 (16%)
Query: 163 IPDQVNCAVIRYHSRRITCLEFHPTKNNIILSGDKKGQLGVWDF---------------- 206
+ ++ C + H + + P +II + + V+D+
Sbjct: 110 VTGKIECEIKINHEGEVNRARYMPQNPHIIATKTPSSDVLVFDYTKHPAKPDPSGECNPD 169
Query: 207 --VRVHEKVVYGNIHSCLLNNMKFNPTNDCMVYSASSDGTISFTDLETGISSSLMNLNPE 264
+R H+K YG + +N + SAS D T+ D+ G +
Sbjct: 170 LRLRGHQKEGYG---------LSWNSNLSGHLLSASDDHTVCLWDINAGPKEGKIVDAKA 220
Query: 265 GWQGPST------WKMLYGMDINSEKGLVLVADNFGFLHLVDMRSNNKTGNAILIHKKNT 318
+ G S W +L+ S VAD+ + + D RSN + + L+
Sbjct: 221 IFTGHSAVVEDVAWHLLHESLFGS------VADDQKLM-IWDTRSNTTSKPSHLVDAHTA 273
Query: 319 KVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLKHNRVVSSAYFSPVSGNKI 378
+V + NP IL T D +WD+R ++ ++ H + ++SP + +
Sbjct: 274 EVNCLSFNPYSEFILATGSADKTVALWDLRNLKLKLHTFE-SHKDEIFQVHWSPHNETIL 332
Query: 379 LTTSLDNRLRVWD 391
++ D RL VWD
Sbjct: 333 ASSGTDRRLNVWD 345
Score = 36.6 bits (83), Expect = 0.18
Identities = 17/76 (22%), Positives = 37/76 (48%)
Query: 175 HSRRITCLEFHPTKNNIILSGDKKGQLGVWDFVRVHEKVVYGNIHSCLLNNMKFNPTNDC 234
H+ + CL F+P I+ +G + +WD + K+ H + + ++P N+
Sbjct: 271 HTAEVNCLSFNPYSEFILATGSADKTVALWDLRNLKLKLHTFESHKDEIFQVHWSPHNET 330
Query: 235 MVYSASSDGTISFTDL 250
++ S+ +D ++ DL
Sbjct: 331 ILASSGTDRRLNVWDL 346
Score = 31.6 bits (70), Expect = 5.8
Identities = 19/84 (22%), Positives = 35/84 (41%), Gaps = 12/84 (14%)
Query: 319 KVVGIHCNPIEPDILLTCGNDHYARVWDIRRMETGSSIYDLK------------HNRVVS 366
++ +H +P IL + G D VWD+ ++ S D + H +S
Sbjct: 318 EIFQVHWSPHNETILASSGTDRRLNVWDLSKIGEEQSAEDAEDGPPELLFIHGGHTAKIS 377
Query: 367 SAYFSPVSGNKILTTSLDNRLRVW 390
++P I + S DN +++W
Sbjct: 378 DFSWNPNEPWVICSVSEDNIMQIW 401
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.134 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 73,366,718
Number of Sequences: 164201
Number of extensions: 3453662
Number of successful extensions: 22924
Number of sequences better than 10.0: 567
Number of HSP's better than 10.0 without gapping: 259
Number of HSP's successfully gapped in prelim test: 320
Number of HSP's that attempted gapping in prelim test: 16197
Number of HSP's gapped (non-prelim): 3278
length of query: 564
length of database: 59,974,054
effective HSP length: 115
effective length of query: 449
effective length of database: 41,090,939
effective search space: 18449831611
effective search space used: 18449831611
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0346.11


