

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
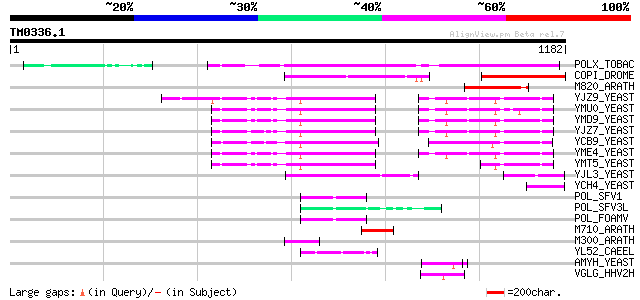
Query= TM0336.1
(1182 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 372 e-102
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 154 2e-36
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 133 2e-30
YJZ9_YEAST (P47100) Transposon Ty1 protein B 86 8e-16
YMU0_YEAST (Q04670) Transposon Ty1 protein B 84 2e-15
YMD9_YEAST (Q03434) Transposon Ty1 protein B 84 2e-15
YJZ7_YEAST (P47098) Transposon Ty1 protein B 83 4e-15
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 82 7e-15
YME4_YEAST (Q04711) Transposon Ty1 protein B 82 9e-15
YMT5_YEAST (Q04214) Transposon Ty1 protein B 80 3e-14
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 79 7e-14
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 60 5e-08
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 59 6e-08
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 58 2e-07
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 54 2e-06
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 53 6e-06
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 52 7e-06
YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III 52 1e-05
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 49 8e-05
VGLG_HHV2H (P13290) Glycoprotein G 47 4e-04
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 372 bits (956), Expect = e-102
Identities = 248/769 (32%), Positives = 375/769 (48%), Gaps = 82/769 (10%)
Query: 421 TSPAQSWYPDSGASHHVTSESQNVHQFTPFAGQ-DQLFVGNGQGLPISGIGSSTLSSG-- 477
+ P W D+ ASHH T ++ AG + +GN I+GIG + +
Sbjct: 288 SGPESEWVVDTAASHHATPVRDLFCRYV--AGDFGTVKMGNTSYSKIAGIGDICIKTNVG 345
Query: 478 --LVLNNLLHVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSVGADGL 535
LVL ++ HVP + NL+S RD G +
Sbjct: 346 CTLVLKDVRHVPDLRMNLISGIALDRD----------------------------GYESY 377
Query: 536 YKFHEFKLSHLPVSLSAPVQHPVSVHTSVS-SSGHAPVEQPSVVSSVCTANLWHFRLGHP 594
+ +++L+ + ++ V T+ G Q + + +LWH R+GH
Sbjct: 378 FANQKWRLTKGSLVIAKGVARGTLYRTNAEICQGELNAAQDEI-----SVDLWHKRMGHM 432
Query: 595 SLNVLRKALQFCKIPVSNKMDVDFCKACVMGKSHRLPSSLSTTEYTSPLELVYSDLWGPS 654
S L+ + I + V C C+ GK HR+ S+ + L+LVYSD+ GP
Sbjct: 433 SEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQTSSERKLNILDLVYSDVCGPM 492
Query: 655 PSPSSSGYLYYITFIDAYSRYTWIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDWG 714
S G Y++TFID SR W+Y+LK K + F +F A E + K+K L++D G
Sbjct: 493 EIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNG 552
Query: 715 GEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDF 772
GE+ R F + GI H P T NGV ER +R IV+ S+L A +P ++W
Sbjct: 553 GEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGE 612
Query: 773 AFSTAVYLINRLPTSSLQFDIPLYKLFKQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQ 832
A TA YLINR P+ L F+IP + Y+ LKVFGC F + KLD +S
Sbjct: 613 AVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSI 672
Query: 833 ECVFLGYSPSHRGYKCL-ASSGRIYISKDVIFDEHTFPYPKLFAQNSNSPSLSPAVVLPS 891
C+F+GY GY+ ++ S+DV+F E ++ + +S V
Sbjct: 673 PCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRE---------SEVRTAADMSEKVKNGI 723
Query: 892 LPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPATSEASAEIPSQ-VTI 950
+P+ F+ P S++ + ++ E E+ Q +
Sbjct: 724 IPN-------------------FVTIPSTSNNPTSAESTTDEVSEQGEQPGEVIEQGEQL 764
Query: 951 PTSSGQVSAPSSSGNVHPMITRSKNGIVQPRVQPT----LLTVQLEPTTVKQALAHP--- 1003
+V P+ H + RS+ V+ R P+ L++ EP ++K+ L+HP
Sbjct: 765 DEGVEEVEHPTQGEEQHQPLRRSERPRVESRRYPSTEYVLISDDREPESLKEVLSHPEKN 824
Query: 1004 QWLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFH 1063
Q ++AMQ E ++LQKN T+ LV LP G+ + CKWVF++K + D L +YKARLV KGF
Sbjct: 825 QLMKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFE 884
Query: 1064 QQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGF 1123
Q+ G D+ E FSPV+K +IR IL+LA + ++QLDV AFL+GDL EE+YM+QP+GF
Sbjct: 885 QKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGF 944
Query: 1124 E--HDKKLVCRLHKALYGLKQAPRAWFEKLTSALVQFGFVASKCDPSLF 1170
E K +VC+L+K+LYGLKQAPR W+ K S + ++ + DP ++
Sbjct: 945 EVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVY 993
Score = 49.3 bits (116), Expect = 6e-05
Identities = 59/279 (21%), Positives = 106/279 (37%), Gaps = 55/279 (19%)
Query: 29 YTISEKLSDRNFLLWKQQVEPIIKAHNLHVLVQDPEIPPRFVTERDRVAGTLNPAYSVWE 88
Y +++ D F W++++ ++ LH ++ P + D W
Sbjct: 6 YEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAED------------WA 53
Query: 89 SKDQYLLSWLQSSLSTPVLSRMIGCAHSFQLWEKIHSFFHNQTRAKSTQLRTELRSETLG 148
D+ S ++ LS V++ +I + +W ++ S + ++T L+ +L + +
Sbjct: 54 DLDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMS 113
Query: 149 NRT-ISEFLLQIKSIIDSLFAIGDPISPREHLDVILQGLPSDYEDLITLI---STRFEEH 204
T L +I L +G I + ++L LPS Y++L T I T E
Sbjct: 114 EGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKTTIELK 173
Query: 205 SIDELEALLLAKEARLGKYRTPSATGEISVNVALSNPQTPSSSAVSTQSASHTTSQGESQ 264
D ALLL ++ R + P G+ + T +G
Sbjct: 174 --DVTSALLLNEKMR----KKPENQGQALI----------------------TEGRGR-- 203
Query: 265 DSDPMDSYDSSGHGRGSSRGRGARGRGRGRSSIQCQLCY 303
SY S + G S GARG+ + RS + + CY
Sbjct: 204 ------SYQRSSNNYGRS---GARGKSKNRSKSRVRNCY 233
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 154 bits (388), Expect = 2e-36
Identities = 79/180 (43%), Positives = 111/180 (60%), Gaps = 2/180 (1%)
Query: 1005 WLQAMQAEFDALQKNKTWSLVPLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQ 1064
W +A+ E +A + N TW++ P + + +WVF VK+N G +YKARLVA+GF Q
Sbjct: 906 WEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQ 965
Query: 1065 QPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFE 1124
+ DY ETF+PV + + R IL+L + + + Q+DV AFLNG L EE+YM+ PQG
Sbjct: 966 KYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGIS 1025
Query: 1125 HDKKLVCRLHKALYGLKQAPRAWFEKLTSALVQFGFVASKCDPSLFVLSKKPL--VIYVL 1182
+ VC+L+KA+YGLKQA R WFE AL + FV S D +++L K + IYVL
Sbjct: 1026 CNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVL 1085
Score = 137 bits (345), Expect = 2e-31
Identities = 101/335 (30%), Positives = 163/335 (48%), Gaps = 31/335 (9%)
Query: 586 LWHFRLGHPS----LNVLRKALQFCKIPVSN-KMDVDFCKACVMGKSHRLPSSL--STTE 638
LWH R GH S L + RK + + ++N ++ + C+ C+ GK RLP T
Sbjct: 417 LWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQARLPFKQLKDKTH 476
Query: 639 YTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSEAISAFHQFKASS 698
PL +V+SD+ GP + Y++ F+D ++ Y YL+K KS+ S F F A S
Sbjct: 477 IKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKS 536
Query: 699 ELQLNAKIKALQTDWGGEFRVFPNFLKDF----GIHHRLICPHTHHQNGVVERKHRHIVD 754
E N K+ L D G E+ N ++ F GI + L PHT NGV ER R I +
Sbjct: 537 EAHFNLKVVYLYIDNGREY--LSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITE 594
Query: 755 LGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSL--QFDIPLYKLFKQIPDYTFLKVFG 812
++++ A + ++W A TA YLINR+P+ +L P + P L+VFG
Sbjct: 595 KARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFG 654
Query: 813 CTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCL-ASSGRIYISKDVIFDE------ 865
T + ++ K D +S + +F+GY P+ G+K A + + +++DV+ DE
Sbjct: 655 ATVYVHIK-NKQGKFDDKSFKSIFVGYEPN--GFKLWDAVNEKFIVARDVVVDETNMVNS 711
Query: 866 HTFPYPKLF------AQNSNSPSLSPAVVLPSLPS 894
+ +F ++N N P+ S ++ P+
Sbjct: 712 RAVKFETVFLKDSKESENKNFPNDSRKIIQTEFPN 746
Score = 37.4 bits (85), Expect = 0.24
Identities = 31/120 (25%), Positives = 53/120 (43%), Gaps = 3/120 (2%)
Query: 87 WESKDQYLLSWLQSSLSTPVLSRMIGCAHSFQLWEKIHSFFHNQTRAKSTQLRTELRSET 146
W+ ++ S + LS L+ + Q+ E + + + ++ A LR L S
Sbjct: 48 WKKAERCAKSTIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLK 107
Query: 147 LGNRT--ISEFLLQIKSIIDSLFAIGDPISPREHLDVILQGLPSDYEDLITLISTRFEEH 204
L + +S F + +I L A G I + + +L LPS Y+ +IT I T EE+
Sbjct: 108 LSSEMSLLSHFHI-FDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEEN 166
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 133 bits (335), Expect = 2e-30
Identities = 71/140 (50%), Positives = 93/140 (65%), Gaps = 10/140 (7%)
Query: 969 MITRSKNGI--VQPRVQPTLLT-VQLEPTTVKQALAHPQWLQAMQAEFDALQKNKTWSLV 1025
M+TRSK GI + P+ T+ T ++ EP +V AL P W QAMQ E DAL +NKTW LV
Sbjct: 1 MLTRSKAGINKLNPKYSLTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILV 60
Query: 1026 PLPPGREAIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRV 1085
P P + +GCKWVF+ K + DGTL++ KARLVAKGFHQ+ G + ET+SPV++ TIR
Sbjct: 61 PPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRT 120
Query: 1086 ILTLALTNHWTIQQLDVNNA 1105
IL +A QQL+V +
Sbjct: 121 ILNVA-------QQLEVGQS 133
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 85.5 bits (210), Expect = 8e-16
Identities = 80/307 (26%), Positives = 132/307 (42%), Gaps = 40/307 (13%)
Query: 871 PKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSS-- 928
P ++ + S+ + LP LP + + P PF P +S Q SS
Sbjct: 1115 PTTVSEQNTEESIIADLPLPDLPP----------ESPTEFPDPFKELPPINSRQTNSSLG 1164
Query: 929 -----------TSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGI 977
S + +E ++ + + P S +H +I K
Sbjct: 1165 GIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIH-LIAAVK--- 1220
Query: 978 VQPRVQPTLLTVQLEP--TTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGRE--- 1032
++P T++ + T K ++++A E + L K KTW +E
Sbjct: 1221 AVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDP 1280
Query: 1033 --AIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLA 1090
I ++F K DGT +KAR VA+G Q P + S + + L+LA
Sbjct: 1281 KRVINSMFIFNKKR--DGT---HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLA 1335
Query: 1091 LTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHDKKLVCRLHKALYGLKQAPRAWFEK 1150
L N++ I QLD+++A+L D+ EE+Y++ P + KL+ RL K+LYGLKQ+ W+E
Sbjct: 1336 LDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLI-RLKKSLYGLKQSGANWYET 1394
Query: 1151 LTSALVQ 1157
+ S L+Q
Sbjct: 1395 IKSYLIQ 1401
Score = 79.3 bits (194), Expect = 6e-14
Identities = 114/488 (23%), Positives = 201/488 (40%), Gaps = 63/488 (12%)
Query: 323 QKPPQNSPQNSQWNSSPQWHSNSQWSSTPQWNSQWSAPVSSPWQHSSSQWPPRYPPAPSG 382
Q+ +NS N + N S + + + +++T + P + +S S+ + + S
Sbjct: 350 QQGSRNSKPNYRRNPSDEKNDSRSYTNTTKPKVIARNPQKT--NNSKSKTARAHNVSTSN 407
Query: 383 YAPMASSPGVPYRGPAPSPMGYAQAHVAFQTST-STPQATSPAQSWYP-----DSGASHH 436
+P + + P + + Q T ST T+ + P DSGAS
Sbjct: 408 NSPSTDNDSISKSTTEPIQLNNKHDLILGQKLTESTVNHTNHSDDELPGHLLLDSGASRT 467
Query: 437 VTSESQNVHQFTPFAGQDQLFVGNGQ--GLPISGIGSSTL---SSGLVLNNLLHVPSITK 491
+ + ++H + + V + Q +PI+ IG + +LH P+I
Sbjct: 468 LIRSAHHIHSAS---SNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVLHTPNIAY 524
Query: 492 NLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEF----KLSHLP 547
+L+S+++ A + C K+ G+V A + K+ +F K LP
Sbjct: 525 DLLSLNELAAVDITA-------CFTKNVLERS---DGTVLAP-IVKYGDFYWVSKKYLLP 573
Query: 548 VSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLRKALQFCK 607
++S P + +VHTS S+ + P + H L H + +R +L+
Sbjct: 574 SNISVPTIN--NVHTSESTRKYP---YPFI----------HRMLAHANAQTIRYSLKNNT 618
Query: 608 IPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSDLWGPSPS 656
I N+ DVD+ C C++GKS H S L P + +++D++GP +
Sbjct: 619 ITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHN 678
Query: 657 PSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIKALQTDWG 714
S Y+I+F D +++ W+Y L + E + F A + Q A + +Q D G
Sbjct: 679 LPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRG 738
Query: 715 GEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDF 772
E+ R FL+ GI +GV ER +R ++D + L + +P W
Sbjct: 739 SEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFS 798
Query: 773 A--FSTAV 778
A FST V
Sbjct: 799 AIEFSTIV 806
Score = 32.7 bits (73), Expect = 6.0
Identities = 31/112 (27%), Positives = 42/112 (36%), Gaps = 10/112 (8%)
Query: 314 HRFNPNFVPQKPPQNSPQNSQWNSSPQWHSNSQWSSTPQWNSQWSAPVSSPWQHSSSQWP 373
H +P PQN P Q + + S WS + S +P+Q S P
Sbjct: 68 HHASPQTAQSHSPQNGPYPQQCMMTQNQANPSGWS----FYGHPSMIPYTPYQMS----P 119
Query: 374 PRYPPAPSGYAPM-ASSPGVPYRGPAP-SPMGYAQAHVAFQTSTSTPQATSP 423
+PP P P SS G P P+P S + + A TST + P
Sbjct: 120 MYFPPGPQSQFPQYPSSVGTPLSTPSPESGNTFTDSSSADSDMTSTKKYVRP 171
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 84.3 bits (207), Expect = 2e-15
Identities = 82/312 (26%), Positives = 136/312 (43%), Gaps = 50/312 (16%)
Query: 871 PKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSS-- 928
P ++ + S+ + LP LP + + P PF P +S Q SS
Sbjct: 688 PTTVSEQNTEESIIADLPLPDLPP----------ESPTEFPDPFKELPPINSRQTNSSLG 737
Query: 929 -----------TSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGI 977
S + +E ++ + + P S +H +I K
Sbjct: 738 GIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIH-LIAAVK--- 793
Query: 978 VQPRVQPTLLTVQLEP--TTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGRE--- 1032
++P T++ + T K +++QA E + L K KTW +E
Sbjct: 794 AVKSIKPIRTTLRYDEAITYNKDIKEKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDP 853
Query: 1033 --AIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIR-----V 1085
I ++F K DGT +KAR VA+G Q P +T+ P ++ T+
Sbjct: 854 KRVINSMFIFNRKR--DGT---HKARFVARGDIQHP-----DTYDPGMQSNTVHHYALMT 903
Query: 1086 ILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHDKKLVCRLHKALYGLKQAPR 1145
L+LAL N++ I QLD+++A+L D+ EE+Y++ P + KL+ RL K+LYGLKQ+
Sbjct: 904 SLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLI-RLKKSLYGLKQSGA 962
Query: 1146 AWFEKLTSALVQ 1157
W+E + S L++
Sbjct: 963 NWYETIKSYLIK 974
Score = 77.8 bits (190), Expect = 2e-13
Identities = 95/375 (25%), Positives = 160/375 (42%), Gaps = 55/375 (14%)
Query: 430 DSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQ--GLPISGIGSSTL---SSGLVLNNLL 484
DSGAS + + ++H + + V + Q +PI+ IG + +L
Sbjct: 34 DSGASRTLIRSAHHIHSAS---SNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVL 90
Query: 485 HVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEF--- 541
H P+I +L+S+++ A + C K+ G+V A + K+ +F
Sbjct: 91 HTPNIAYDLLSLNELAAVDITA-------CFTKNVLERS---DGTVLAP-IVKYGDFYWV 139
Query: 542 -KLSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLR 600
K LP ++S P + +VHTS S+ + P + H L H + +R
Sbjct: 140 SKKYLLPSNISVPTIN--NVHTSESTRKYP---YPFI----------HRMLAHANAQTIR 184
Query: 601 KALQFCKIPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSD 649
+L+ I N+ DVD+ C C++GKS H S L P + +++D
Sbjct: 185 YSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTD 244
Query: 650 LWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIK 707
++GP + S Y+I+F D +++ W+Y L + E + F A + Q A +
Sbjct: 245 IFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVL 304
Query: 708 ALQTDWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASI 765
+Q D G E+ R FL+ GI +GV ER +R ++D + L + +
Sbjct: 305 VIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGL 364
Query: 766 PLTYWDFA--FSTAV 778
P W A FST V
Sbjct: 365 PNHLWFSAIEFSTIV 379
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 84.3 bits (207), Expect = 2e-15
Identities = 79/307 (25%), Positives = 132/307 (42%), Gaps = 40/307 (13%)
Query: 871 PKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSS-- 928
P ++ + S+ + LP LP + + P PF P +S Q SS
Sbjct: 688 PTTVSEQNTEESIIADLPLPDLPP----------ESPTEFPDPFKELPPINSHQTNSSLG 737
Query: 929 -----------TSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGI 977
S + +E ++ + + P S +H +I K
Sbjct: 738 GIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIH-LIAAVK--- 793
Query: 978 VQPRVQPTLLTVQLEP--TTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGRE--- 1032
++P T++ + T K ++++A E + L K KTW +E
Sbjct: 794 AVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDP 853
Query: 1033 --AIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLA 1090
I ++F K DGT +KAR VA+G Q P + S + + L+LA
Sbjct: 854 KRVINSMFIFNKKR--DGT---HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLA 908
Query: 1091 LTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHDKKLVCRLHKALYGLKQAPRAWFEK 1150
L N++ I QLD+++A+L D+ EE+Y++ P + KL+ RL K+LYGLKQ+ W+E
Sbjct: 909 LDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLI-RLKKSLYGLKQSGANWYET 967
Query: 1151 LTSALVQ 1157
+ S L++
Sbjct: 968 IKSYLIK 974
Score = 76.3 bits (186), Expect = 5e-13
Identities = 95/375 (25%), Positives = 159/375 (42%), Gaps = 55/375 (14%)
Query: 430 DSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQ--GLPISGIGSSTL---SSGLVLNNLL 484
DSGAS + + ++H + + V + Q +PI+ IG + +L
Sbjct: 34 DSGASRTLIRSAHHIHSAS---SNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVL 90
Query: 485 HVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEF--- 541
H P+I +L+S+++ A + C K+ G+V A + K+ +F
Sbjct: 91 HTPNIAYDLLSLNELAAVDITA-------CFTKNVLERS---DGTVLAP-IVKYGDFYWV 139
Query: 542 -KLSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLR 600
K LP ++S P + +VHTS S+ + P + H L H + +R
Sbjct: 140 SKKYLLPSNISVPTIN--NVHTSESTRKYP---YPFI----------HRMLAHANAQTIR 184
Query: 601 KALQFCKIPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSD 649
+L+ I N+ DVD C C++GKS H S L P + +++D
Sbjct: 185 YSLKNNTITYFNESDVDRSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTD 244
Query: 650 LWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIK 707
++GP + S Y+I+F D +++ W+Y L + E + F A + Q A +
Sbjct: 245 IFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVL 304
Query: 708 ALQTDWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASI 765
+Q D G E+ R FL+ GI +GV ER +R ++D + L + +
Sbjct: 305 VIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGL 364
Query: 766 PLTYWDFA--FSTAV 778
P W A FST V
Sbjct: 365 PNHLWFSAIEFSTIV 379
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 83.2 bits (204), Expect = 4e-15
Identities = 79/307 (25%), Positives = 131/307 (41%), Gaps = 40/307 (13%)
Query: 871 PKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSS-- 928
P ++ + S+ + LP LP + + P PF P +S Q SS
Sbjct: 1115 PTTVSEQNTEESIIADLPLPDLPP----------ESPTEFPDPFKELPPINSHQTNSSLG 1164
Query: 929 -----------TSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGI 977
S + +E ++ + + P S +H +I K
Sbjct: 1165 GIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIH-LIAAVK--- 1220
Query: 978 VQPRVQPTLLTVQLEP--TTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGRE--- 1032
++P T++ + T K ++++A E + L K KTW +E
Sbjct: 1221 AVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDP 1280
Query: 1033 --AIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLA 1090
I ++F K DGT +KAR VA+G Q P T S + + L+LA
Sbjct: 1281 KRVINSMFIFNKKR--DGT---HKARFVARGDIQHPDTYDTGMQSNTVHHYALMTSLSLA 1335
Query: 1091 LTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHDKKLVCRLHKALYGLKQAPRAWFEK 1150
L N++ I QLD+++A+L D+ EE+Y++ P + KL+ RL K+ YGLKQ+ W+E
Sbjct: 1336 LDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLI-RLKKSHYGLKQSGANWYET 1394
Query: 1151 LTSALVQ 1157
+ S L++
Sbjct: 1395 IKSYLIK 1401
Score = 77.4 bits (189), Expect = 2e-13
Identities = 94/373 (25%), Positives = 158/373 (42%), Gaps = 51/373 (13%)
Query: 430 DSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQ--GLPISGIGSSTL---SSGLVLNNLL 484
DSGAS + + ++H + + V + Q +PI+ IG + +L
Sbjct: 461 DSGASRTLIRSAHHIHSAS---SNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVL 517
Query: 485 HVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKS--QASSKVLLQGSVGADGLYKFHEFK 542
H P+I +L+S+++ A + C K+ + S +L V Y +
Sbjct: 518 HTPNIAYDLLSLNELAAVDITA-------CFTKNVLERSDGTVLAPIVQYGDFYWVSKRY 570
Query: 543 LSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLRKA 602
L LP ++S P + +VHTS S+ + P + H L H + +R +
Sbjct: 571 L--LPSNISVPTIN--NVHTSESTRKYP---YPFI----------HRMLAHANAQTIRYS 613
Query: 603 LQFCKIPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSDLW 651
L+ I N+ DVD+ C C++GKS H S L P + +++D++
Sbjct: 614 LKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIF 673
Query: 652 GPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIKAL 709
GP + S Y+I+F D +++ W+Y L + E + F A + Q A + +
Sbjct: 674 GPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVI 733
Query: 710 QTDWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPL 767
Q D G E+ R FL+ GI +GV ER +R ++D + L + +P
Sbjct: 734 QMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPN 793
Query: 768 TYWDFA--FSTAV 778
W A FST V
Sbjct: 794 HLWFSAIEFSTIV 806
Score = 34.3 bits (77), Expect = 2.1
Identities = 32/112 (28%), Positives = 43/112 (37%), Gaps = 10/112 (8%)
Query: 314 HRFNPNFVPQKPPQNSPQNSQWNSSPQWHSNSQWSSTPQWNSQWSAPVSSPWQHSSSQWP 373
H +P PPQN P Q + + S WS + S +P+Q S P
Sbjct: 68 HHASPQPASVPPPQNGPYPQQCMMTQNQANPSGWS----FYGHPSMIPYTPYQMS----P 119
Query: 374 PRYPPAPSGYAPM-ASSPGVPYRGPAP-SPMGYAQAHVAFQTSTSTPQATSP 423
+PP P P SS G P P+P S + + A TST + P
Sbjct: 120 MYFPPGPQSQFPQYPSSVGTPLSTPSPESGNTFTDSSSADSDMTSTKKYVRP 171
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 82.4 bits (202), Expect = 7e-15
Identities = 89/375 (23%), Positives = 159/375 (41%), Gaps = 45/375 (12%)
Query: 430 DSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQGLPISGIGSS--TLSSGLVLN-NLLHV 486
DSGAS + + +H TP + + + Q +PI+ IG+ +G + LH
Sbjct: 457 DSGASQTLVRSAHYLHHATPNS-EINIVDAQKQDIPINAIGNLHFNFQNGTKTSIKALHT 515
Query: 487 PSITKNLVSVSKFARDNGVFFEFHPYYCCVKS--QASSKVLLQGSVGADGLYKFHEFKLS 544
P+I +L+S+S+ A N C ++ + S +L V Y + L
Sbjct: 516 PNIAYDLLSLSELANQNITA-------CFTRNTLERSDGTVLAPIVKHGDFYWLSKKYL- 567
Query: 545 HLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLRKALQ 604
+P +S + V+ SV+ + L H LGH + ++K+L+
Sbjct: 568 -IPSHISKLTINNVNKSKSVNKYPYP---------------LIHRMLGHANFRSIQKSLK 611
Query: 605 FCKIPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSDLWGP 653
+ + D+++ C C++GKS H S L E P + +++D++GP
Sbjct: 612 KNAVTYLKESDIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGP 671
Query: 654 SPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIKALQT 711
S Y+I+F D +R+ W+Y L + E ++ F A + Q NA++ +Q
Sbjct: 672 VHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQM 731
Query: 712 DWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTY 769
D G E+ + F + GI +GV ER +R +++ +LL + +P
Sbjct: 732 DRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHL 791
Query: 770 WDFAFSTAVYLINRL 784
W A + + N L
Sbjct: 792 WFSAVEFSTIIRNSL 806
Score = 77.0 bits (188), Expect = 3e-13
Identities = 72/277 (25%), Positives = 122/277 (43%), Gaps = 21/277 (7%)
Query: 892 LPSVEHFSQPAIAQPSSSLP---SPFLPSPIASSDQNPSSTSPASAPAT---SEASAEIP 945
LP + H S + S +P S S + D + T+ S + +E E+
Sbjct: 1148 LPDLTHQSPTDTSDVSKDIPHIHSRQTNSSLGGMDDSNVLTTTKSKKRSLEDNETEIEVS 1207
Query: 946 SQVTIPTSSGQVSAPSSSGNVHPMITRSKNGIVQPRVQPTLLTVQLEPTTVKQALAHPQW 1005
+ + P S ++ + + G+ + T L T K ++
Sbjct: 1208 RDTWNNKNMRSLEPPRSKKRIN--LIAAIKGVKSIKPVRTTLRYDEAITYNKDNKEKDRY 1265
Query: 1006 LQAMQAEFDALQKNKTWSLVP------LPPGREAIGCKWVFRVKHNPDGTLNKYKARLVA 1059
++A E L K TW + P ++ I ++F K DGT +KAR VA
Sbjct: 1266 VEAYHKEISQLLKMNTWDTNKYYDRNDIDP-KKVINSMFIFNKKR--DGT---HKARFVA 1319
Query: 1060 KGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQQ 1119
+G Q P ++ S + + L++AL N + I QLD+++A+L D+ EE+Y++
Sbjct: 1320 RGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDISSAYLYADIKEELYIRP 1379
Query: 1120 PQGFEHDKKLVCRLHKALYGLKQAPRAWFEKLTSALV 1156
P + KL+ RL K+LYGLKQ+ W+E + S L+
Sbjct: 1380 PPHLGLNDKLL-RLRKSLYGLKQSGANWYETIKSYLI 1415
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 82.0 bits (201), Expect = 9e-15
Identities = 78/307 (25%), Positives = 131/307 (42%), Gaps = 40/307 (13%)
Query: 871 PKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSS-- 928
P ++ + S+ + LP LP + + P PF P +S Q SS
Sbjct: 688 PTTVSEQNTEESIIADLPLPDLPP----------ESPTEFPDPFKELPPINSHQTNSSLG 737
Query: 929 -----------TSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGI 977
S + +E ++ + + P S +H +I K
Sbjct: 738 GIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNTKNMRSLEPPRSKKRIH-LIAAVK--- 793
Query: 978 VQPRVQPTLLTVQLEP--TTVKQALAHPQWLQAMQAEFDALQKNKTWSLVPLPPGRE--- 1032
++P T++ + T K ++++A E + L K TW +E
Sbjct: 794 AVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDP 853
Query: 1033 --AIGCKWVFRVKHNPDGTLNKYKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLA 1090
I ++F K DGT +KAR VA+G Q P + S + + L+LA
Sbjct: 854 KRVINSMFIFNRKR--DGT---HKARFVARGDIQHPDTYDSGMQSNTVHHYALMTSLSLA 908
Query: 1091 LTNHWTIQQLDVNNAFLNGDLAEEVYMQQPQGFEHDKKLVCRLHKALYGLKQAPRAWFEK 1150
L N++ I QLD+++A+L D+ EE+Y++ P + KL+ RL K+LYGLKQ+ W+E
Sbjct: 909 LDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDKLI-RLKKSLYGLKQSGANWYET 967
Query: 1151 LTSALVQ 1157
+ S L++
Sbjct: 968 IKSYLIK 974
Score = 77.8 bits (190), Expect = 2e-13
Identities = 95/375 (25%), Positives = 160/375 (42%), Gaps = 55/375 (14%)
Query: 430 DSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQ--GLPISGIGSSTL---SSGLVLNNLL 484
DSGAS + + ++H + + V + Q +PI+ IG + +L
Sbjct: 34 DSGASRTLIRSAHHIHSAS---SNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVL 90
Query: 485 HVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEF--- 541
H P+I +L+S+++ A + C K+ G+V A + K+ +F
Sbjct: 91 HTPNIAYDLLSLNELAAVDITA-------CFTKNVLERS---DGTVLAP-IVKYGDFYWV 139
Query: 542 -KLSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLR 600
K LP ++S P + +VHTS S+ + P + H L H + +R
Sbjct: 140 SKKYLLPSNISVPTIN--NVHTSESTRKYP---YPFI----------HRMLAHANAQTIR 184
Query: 601 KALQFCKIPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSD 649
+L+ I N+ DVD+ C C++GKS H S L P + +++D
Sbjct: 185 YSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTD 244
Query: 650 LWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIK 707
++GP + S Y+I+F D +++ W+Y L + E + F A + Q A +
Sbjct: 245 IFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVL 304
Query: 708 ALQTDWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASI 765
+Q D G E+ R FL+ GI +GV ER +R ++D + L + +
Sbjct: 305 VIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGL 364
Query: 766 PLTYWDFA--FSTAV 778
P W A FST V
Sbjct: 365 PNHLWFSAIEFSTIV 379
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 80.5 bits (197), Expect = 3e-14
Identities = 54/159 (33%), Positives = 86/159 (53%), Gaps = 11/159 (6%)
Query: 1004 QWLQAMQAEFDALQKNKTWSLVPLPPGRE-----AIGCKWVFRVKHNPDGTLNKYKARLV 1058
++++A E + L K KTW +E I ++F K DGT +KAR V
Sbjct: 822 KYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRVINSMFIFNRKR--DGT---HKARFV 876
Query: 1059 AKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLAEEVYMQ 1118
A+G Q P + S + + L+LAL N++ I QLD+++A+L D+ EE+Y++
Sbjct: 877 ARGDIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIR 936
Query: 1119 QPQGFEHDKKLVCRLHKALYGLKQAPRAWFEKLTSALVQ 1157
P + KL+ RL K+LYGLKQ+ W+E + S L++
Sbjct: 937 PPPHLGMNDKLI-RLKKSLYGLKQSGANWYETIKSYLIK 974
Score = 77.8 bits (190), Expect = 2e-13
Identities = 95/375 (25%), Positives = 160/375 (42%), Gaps = 55/375 (14%)
Query: 430 DSGASHHVTSESQNVHQFTPFAGQDQLFVGNGQ--GLPISGIGSSTL---SSGLVLNNLL 484
DSGAS + + ++H + + V + Q +PI+ IG + +L
Sbjct: 34 DSGASRTLIRSAHHIHSAS---SNPDINVVDAQKRNIPINAIGDLQFHFQDNTKTSIKVL 90
Query: 485 HVPSITKNLVSVSKFARDNGVFFEFHPYYCCVKSQASSKVLLQGSVGADGLYKFHEF--- 541
H P+I +L+S+++ A + C K+ G+V A + K+ +F
Sbjct: 91 HTPNIAYDLLSLNELAAVDITA-------CFTKNVLERS---DGTVLAP-IVKYGDFYWV 139
Query: 542 -KLSHLPVSLSAPVQHPVSVHTSVSSSGHAPVEQPSVVSSVCTANLWHFRLGHPSLNVLR 600
K LP ++S P + +VHTS S+ + P + H L H + +R
Sbjct: 140 SKKYLLPSNISVPTIN--NVHTSESTRKYP---YPFI----------HRMLAHANAQTIR 184
Query: 601 KALQFCKIPVSNKMDVDF-------CKACVMGKS----HRLPSSLSTTEYTSPLELVYSD 649
+L+ I N+ DVD+ C C++GKS H S L P + +++D
Sbjct: 185 YSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTD 244
Query: 650 LWGPSPSPSSSGYLYYITFIDAYSRYTWIYLLKAKSE--AISAFHQFKASSELQLNAKIK 707
++GP + S Y+I+F D +++ W+Y L + E + F A + Q A +
Sbjct: 245 IFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVL 304
Query: 708 ALQTDWGGEF--RVFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDLGLSLLAHASI 765
+Q D G E+ R FL+ GI +GV ER +R ++D + L + +
Sbjct: 305 VIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGL 364
Query: 766 PLTYWDFA--FSTAV 778
P W A FST V
Sbjct: 365 PNHLWFSAIEFSTIV 379
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 79.0 bits (193), Expect = 7e-14
Identities = 46/129 (35%), Positives = 71/129 (54%), Gaps = 5/129 (3%)
Query: 1053 YKARLVAKGFHQQPGFDYTETFSPVIKPVTIRVILTLALTNHWTIQQLDVNNAFLNGDLA 1112
YKAR+V +G Q P Y+ + + I++ L +A + ++ LD+N+AFL L
Sbjct: 1337 YKARIVCRGDTQSPD-TYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLE 1395
Query: 1113 EEVYMQQPQGFEHDKKLVCRLHKALYGLKQAPRAWFEKLTSALVQFGFVASKCDPSLFVL 1172
EE+Y+ P HD++ V +L+KALYGLKQ+P+ W + L L G + P L+
Sbjct: 1396 EEIYIPHP----HDRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLYQT 1451
Query: 1173 SKKPLVIYV 1181
K L+I V
Sbjct: 1452 EDKNLMIAV 1460
Score = 63.5 bits (153), Expect = 3e-09
Identities = 64/294 (21%), Positives = 120/294 (40%), Gaps = 17/294 (5%)
Query: 588 HFRLGHPSLNVLRKALQFC----KIPVSNKMDVDFCKACVMGKS----HRLPSSLSTTEY 639
H R+GH + + +++ + + + + +C+ C + K+ H S + +
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 640 TSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRY--TWIYLLKAKSEAISAFHQFKAS 697
P D++GP S ++ Y + +D +RY T + K ++ +
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 698 SELQLNAKIKALQTDWGGEFR--VFPNFLKDFGIHHRLICPHTHHQNGVVERKHRHIVDL 755
E Q + K++ + +D G EF + GIHH L H NG ER R I+
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 756 GLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIPLYKLFKQIPDYTFLKVFGCTC 815
+LL +++ + +W++A ++A + N L S +PL + +Q +
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKSTG-KLPLKAISRQPVTVRLMSFLPFGE 800
Query: 816 FPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCLASSGRIYISKDVIFDEHTFP 869
++ +N KL + L P+ GYK S +K V D +T P
Sbjct: 801 KGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPSK----NKIVTSDNYTIP 850
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 59.7 bits (143), Expect = 5e-08
Identities = 31/84 (36%), Positives = 47/84 (55%), Gaps = 2/84 (2%)
Query: 1100 LDVNNAFLNGDLAEEVYMQQPQGF--EHDKKLVCRLHKALYGLKQAPRAWFEKLTSALVQ 1157
+DV+ AFLN + E +Y++QP GF E + V L+ +YGLKQAP W E + + L +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1158 FGFVASKCDPSLFVLSKKPLVIYV 1181
GF + + L+ S IY+
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYI 84
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 59.3 bits (142), Expect = 6e-08
Identities = 45/146 (30%), Positives = 68/146 (45%), Gaps = 10/146 (6%)
Query: 619 CKACVMGKSHRL--PSSLSTTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYT 676
CK C++ + L P L + P + Y D GP P S+GYL+ + +D+ + +
Sbjct: 858 CKQCLVTNATNLTSPPILRPVKPLKPFDKFYIDYIGPLPP--SNGYLHVLVVVDSMTGFV 915
Query: 677 WIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDWGGEF--RVFPNFLKDFGIHHRLI 734
W+Y KA S + + KA + L A K L +D G F F ++ K+ GI
Sbjct: 916 WLYPTKAPSTSATV----KALNMLTSIAIPKVLHSDQGAAFTSSTFADWAKEKGIQLEFS 971
Query: 735 CPHTHHQNGVVERKHRHIVDLGLSLL 760
P+ +G VERK+ I L LL
Sbjct: 972 TPYHPQSSGKVERKNSDIKRLLTKLL 997
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 57.8 bits (138), Expect = 2e-07
Identities = 76/305 (24%), Positives = 116/305 (37%), Gaps = 61/305 (20%)
Query: 619 CKACVMGKSHRL--PSSLSTTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYT 676
CK C++ + L P L P + + D GP P S+GYL+ + +D+ + +
Sbjct: 860 CKQCLVTNAATLAAPPILRPERPVKPFDKFFIDYIGPLPP--SNGYLHVLVVVDSMTGFV 917
Query: 677 WIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDWGGEF--RVFPNFLKDFGIHHRLI 734
W+Y KA S + + KA + L A K + +D G F F ++ K+ GI
Sbjct: 918 WLYPTKAPSTSATV----KALNMLTSIAVPKVIHSDQGAAFTSATFADWAKNKGIQLEFS 973
Query: 735 CPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIP 794
P+ +G VERK+ I L LL P ++D + L N SS
Sbjct: 974 TPYHPQSSGKVERKNSDIKRLLTKLL--VGRPAKWYDLLPVVQLALNNSYSPSS------ 1025
Query: 795 LYKLFKQIPDYTFLKVFGCTCFPLMRPYNSHKLDFRSQECVFLGYSPSHRGYKCLASSGR 854
K P + T F NS LD +E + L + + SS
Sbjct: 1026 -----KYTPHQLLFGIDSNTPFA-----NSDTLDLSREEELSL--------LQEIRSS-- 1065
Query: 855 IYISKDVIFDEHTFPYPKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPF 914
+Y+ PS PA + PSV Q +A+P+S P
Sbjct: 1066 LYL-----------------------PSTPPASIRAWSPSVGQLVQERVARPASLRPRWH 1102
Query: 915 LPSPI 919
P+P+
Sbjct: 1103 KPTPV 1107
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 54.3 bits (129), Expect = 2e-06
Identities = 42/146 (28%), Positives = 63/146 (42%), Gaps = 10/146 (6%)
Query: 619 CKACVMGKSHRLPSS--LSTTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRYT 676
C+ C++ + S L P + + D GP P S GYLY + +D + +T
Sbjct: 650 CQQCLITNASNKASGPILRPDRPQKPFDKFFIDYIGPLPP--SQGYLYVLVVVDGMTGFT 707
Query: 677 WIYLLKAKSEAISAFHQFKASSELQLNAKIKALQTDWGGEF--RVFPNFLKDFGIHHRLI 734
W+Y KA S + + K+ + L A K + +D G F F + K+ GIH
Sbjct: 708 WLYPTKAPSTSATV----KSLNVLTSIAIPKVIHSDQGAAFTSSTFAEWAKERGIHLEFS 763
Query: 735 CPHTHHQNGVVERKHRHIVDLGLSLL 760
P+ VERK+ I L LL
Sbjct: 764 TPYHPQSGSKVERKNSDIKRLLTKLL 789
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 52.8 bits (125), Expect = 6e-06
Identities = 22/68 (32%), Positives = 43/68 (62%)
Query: 749 HRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLINRLPTSSLQFDIPLYKLFKQIPDYTFL 808
+R I++ S+L +P T+ A +TAV++IN+ P++++ F +P F+ +P Y++L
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYL 61
Query: 809 KVFGCTCF 816
+ FGC +
Sbjct: 62 RRFGCVAY 69
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 52.4 bits (124), Expect = 7e-06
Identities = 26/74 (35%), Positives = 38/74 (51%)
Query: 586 LWHFRLGHPSLNVLRKALQFCKIPVSNKMDVDFCKACVMGKSHRLPSSLSTTEYTSPLEL 645
LWH RL H S + ++ + S + FC+ C+ GK+HR+ S +PL+
Sbjct: 71 LWHSRLAHMSQRGMELLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFSTGQHTTKNPLDY 130
Query: 646 VYSDLWGPSPSPSS 659
V+SDLWG P S
Sbjct: 131 VHSDLWGAPSVPLS 144
>YL52_CAEEL (P34431) Hypothetical protein F44E2.2 in chromosome III
Length = 2186
Score = 52.0 bits (123), Expect = 1e-05
Identities = 52/169 (30%), Positives = 77/169 (44%), Gaps = 14/169 (8%)
Query: 619 CKACVMGKSH-RLPSSLSTTEYTSPLELVYSDLWGPSPSPSSSGYLYYITFIDAYSRY-T 676
C C+ H +L SSL+ T PLE+V DL S G Y +T ID +++Y T
Sbjct: 1510 CAKCLCANDHSKLTSSLTPYRMTFPLEIVACDLM--DVGLSVQGNRYILTIIDLFTKYGT 1567
Query: 677 WIYLLKAKSEAI-SAFHQFKASSELQLNAKIKALQTDWGGEF--RVFPNFLKDFGIHHRL 733
+ + K+E + AF + A E ++ K L TD G EF +F F I H
Sbjct: 1568 AVPIPDKKAETVLKAFVERWAIGEGRIPLK---LLTDQGKEFVNGLFAQFTHMLKIEHIT 1624
Query: 734 ICPHTHHQNGVVERKHRHIVDLGLSLLAHASIPLTYWDFAFSTAVYLIN 782
+ NG VER ++ I+ + + ++P+ WD AVY N
Sbjct: 1625 TKGYNSRANGAVERFNKTIMHI---MKKKTAVPME-WDDQVVYAVYAYN 1669
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 48.9 bits (115), Expect = 8e-05
Identities = 32/89 (35%), Positives = 44/89 (48%), Gaps = 2/89 (2%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P SS+ S P+P SS SS++P ++
Sbjct: 571 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSST 630
Query: 937 TSEASAEI--PSQVTIPTSSGQVSAPSSS 963
T +SA + PS T +SS V PSSS
Sbjct: 631 TESSSAPVPTPSSSTTESSSAPVPTPSSS 659
Score = 48.5 bits (114), Expect = 1e-04
Identities = 33/104 (31%), Positives = 50/104 (47%), Gaps = 5/104 (4%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P SS+ S P P SS N +S++P+S P
Sbjct: 769 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPF 828
Query: 937 TSEASAE-----IPSQVTIPTSSGQVSAPSSSGNVHPMITRSKN 975
+S + PS T +SS VS+ ++ +V P+ T S +
Sbjct: 829 SSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSS 872
Score = 47.8 bits (112), Expect = 2e-04
Identities = 32/89 (35%), Positives = 43/89 (47%), Gaps = 2/89 (2%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P SS+ S P P SS SS++P ++
Sbjct: 697 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSST 756
Query: 937 TSEASAEI--PSQVTIPTSSGQVSAPSSS 963
T +SA + PS T +SS V PSSS
Sbjct: 757 TESSSAPVPTPSSSTTESSSAPVPTPSSS 785
Score = 44.7 bits (104), Expect = 0.002
Identities = 29/93 (31%), Positives = 49/93 (52%), Gaps = 4/93 (4%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P + + S +P +P+P +S+ ++ S+ P + +
Sbjct: 727 SSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAP-VPTPSSSTTESSSAPVPTPSSS 785
Query: 937 TSEASAE---IPSQVTIPTSSGQVSAPSSSGNV 966
T+E+S+ PS T +S V PSSS N+
Sbjct: 786 TTESSSAPVPTPSSSTTESSVAPVPTPSSSSNI 818
Score = 43.5 bits (101), Expect = 0.003
Identities = 34/125 (27%), Positives = 53/125 (42%), Gaps = 8/125 (6%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P SS+ S P P SS SS++P ++
Sbjct: 448 SSTTESSSAPVPTPSSSTTESSSAPVT---SSTTESSSAPVPTPSSSTTESSSAPVTSST 504
Query: 937 TSEASAEIPSQVTIPTSSGQVSAPSSSGNV-----HPMITRSKNGIVQPRVQPTLLTVQL 991
T +SA +P+ + T S AP+ S + P+ + + P P+ T +
Sbjct: 505 TESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTES 564
Query: 992 EPTTV 996
T V
Sbjct: 565 SSTPV 569
Score = 43.1 bits (100), Expect = 0.004
Identities = 32/101 (31%), Positives = 43/101 (41%), Gaps = 14/101 (13%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P SS+ S P P SS SS++P ++
Sbjct: 628 SSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSST 687
Query: 937 TSEASAEI--------------PSQVTIPTSSGQVSAPSSS 963
T +SA + PS T +SS V PSSS
Sbjct: 688 TESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSS 728
Score = 42.4 bits (98), Expect = 0.008
Identities = 31/89 (34%), Positives = 42/89 (46%), Gaps = 5/89 (5%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S PS + E S P SS+ S P P SS SS++P ++
Sbjct: 517 SSTTESSSAPAPTPSSSTTESSSAPVT---SSTTESSSAPVPTPSSSTTESSSTPVTSST 573
Query: 937 TSEASAEI--PSQVTIPTSSGQVSAPSSS 963
T +SA + PS T +SS V PSSS
Sbjct: 574 TESSSAPVPTPSSSTTESSSAPVPTPSSS 602
Score = 41.2 bits (95), Expect = 0.017
Identities = 31/101 (30%), Positives = 45/101 (43%), Gaps = 2/101 (1%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQP--SSSLPSPFLPSPIASSDQNPSSTSPASA 934
+S + S S V PS + E S P + SSS P P S S P T +S
Sbjct: 544 SSTTESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSST 603
Query: 935 PATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKN 975
+S A A PS T +SS V++ ++ + P+ T S +
Sbjct: 604 TESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSS 644
Score = 40.0 bits (92), Expect = 0.037
Identities = 30/101 (29%), Positives = 50/101 (48%), Gaps = 4/101 (3%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S++ S A V S + E S P SS+ S P+P SS SS++P ++
Sbjct: 489 SSSTTESSSAPVTSS--TTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSST 546
Query: 937 TSEASAEI--PSQVTIPTSSGQVSAPSSSGNVHPMITRSKN 975
T +SA + PS T +SS V++ ++ + P+ T S +
Sbjct: 547 TESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSS 587
Score = 39.7 bits (91), Expect = 0.049
Identities = 29/87 (33%), Positives = 38/87 (43%), Gaps = 1/87 (1%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S S V PS + E S P + + S +P S SS P T +S
Sbjct: 658 SSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSA-PVPTPSSSTTE 716
Query: 937 TSEASAEIPSQVTIPTSSGQVSAPSSS 963
+S A PS T +SS V PSSS
Sbjct: 717 SSSAPVPTPSSSTTESSSAPVPTPSSS 743
Score = 39.7 bits (91), Expect = 0.049
Identities = 30/94 (31%), Positives = 44/94 (45%), Gaps = 6/94 (6%)
Query: 876 QNSNSPSLSPAVVLPSLP----SVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSP 931
++S++P S S P + E S P SS+ S P P SS SS++P
Sbjct: 677 ESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAP 736
Query: 932 ASAPA--TSEASAEIPSQVTIPTSSGQVSAPSSS 963
P+ T+E+S+ + T +SS V PSSS
Sbjct: 737 VPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSS 770
Score = 38.5 bits (88), Expect = 0.11
Identities = 30/116 (25%), Positives = 47/116 (39%), Gaps = 2/116 (1%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQP--SSSLPSPFLPSPIASSDQNPSSTSPASA 934
+S + S S PS + E S P + SSS P P S S P T +S
Sbjct: 601 SSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSST 660
Query: 935 PATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGIVQPRVQPTLLTVQ 990
+S A PS T +SS V++ ++ + P+ + + P P+ T +
Sbjct: 661 TESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 716
Score = 37.4 bits (85), Expect = 0.24
Identities = 33/115 (28%), Positives = 48/115 (41%), Gaps = 12/115 (10%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S++ S S V PS + E S P SS+ S P P SS N +S++P+S P
Sbjct: 829 SSSTESSSVPVPTPSSSTTESSSAPV---SSSTTESSVAPVPTPSSSSNITSSAPSSIPF 885
Query: 937 TSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKNGIVQPRVQPTLLTVQL 991
+S T S+G PSSS S + + + PT T +
Sbjct: 886 SS---------TTESFSTGTTVTPSSSKYPGSQTETSVSSTTETTIVPTKTTTSV 931
Score = 37.4 bits (85), Expect = 0.24
Identities = 27/111 (24%), Positives = 52/111 (46%), Gaps = 5/111 (4%)
Query: 869 PYPKLFAQNSNSPSLSPAVVLPSLP----SVEHFSQPAIAQPSSSLPSPFLPSPIASSDQ 924
P ++S++P S S P + E S P + + S +P +P+P +S+ +
Sbjct: 409 PVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAP-VPTPSSSTTE 467
Query: 925 NPSSTSPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHPMITRSKN 975
+ S+ +S +S A PS T +SS V++ ++ + P+ T S +
Sbjct: 468 SSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSS 518
Score = 37.0 bits (84), Expect = 0.32
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 2/92 (2%)
Query: 901 PAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPATSEASAEI--PSQVTIPTSSGQVS 958
P SS+ S P P SS SS++P ++ T +SA + PS T +SS V+
Sbjct: 313 PVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVT 372
Query: 959 APSSSGNVHPMITRSKNGIVQPRVQPTLLTVQ 990
+ ++ + P+ + + P P+ T +
Sbjct: 373 SSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 404
Score = 36.2 bits (82), Expect = 0.54
Identities = 31/88 (35%), Positives = 42/88 (47%), Gaps = 6/88 (6%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQNPSSTSPASAPA 936
+S + S V PS S S P+ SSS S +P P SS S+T +SAP
Sbjct: 799 SSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSS----STTESSSAPV 854
Query: 937 TSEASAEIPSQVTIPTSSGQV--SAPSS 962
+S + + V P+SS + SAPSS
Sbjct: 855 SSSTTESSVAPVPTPSSSSNITSSAPSS 882
Score = 35.4 bits (80), Expect = 0.92
Identities = 45/173 (26%), Positives = 72/173 (41%), Gaps = 24/173 (13%)
Query: 326 PQNSPQNSQWNSSPQWHSNSQWSSTPQWNS---QWSAPVSSPWQHSSSQWPPRYPPAPSG 382
P S ++ +S+P S ++ SS P +S SAPV +P SSS P P+
Sbjct: 669 PTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTP---SSSTTESSSAPVPTP 725
Query: 383 YAPMASSPGVPYRGPAPSPMGYAQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQ 442
+ S P P+ S + A V TS++T +++P + P S T+ES
Sbjct: 726 SSSTTESSSAPVPTPSSSTTESSSAPV---TSSTTESSSAPVPT--PSSS-----TTESS 775
Query: 443 NVHQFTPFAGQDQLFVGNGQGLPISGIGSSTLSSGLVLNNLLHVPSITKNLVS 495
+ TP + + P+ SST S + + PS + N+ S
Sbjct: 776 SAPVPTPSSSTTE-----SSSAPVPTPSSSTTESSVA---PVPTPSSSSNITS 820
Score = 33.5 bits (75), Expect = 3.5
Identities = 32/151 (21%), Positives = 54/151 (35%)
Query: 326 PQNSPQNSQWNSSPQWHSNSQWSSTPQWNSQWSAPVSSPWQHSSSQWPPRYPPAPSGYAP 385
P S ++ +S+P S ++ SS P S SS +SS P P+ +
Sbjct: 528 PTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSS 587
Query: 386 MASSPGVPYRGPAPSPMGYAQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQNVH 445
S P P+ S + A +S++T +++P S +S ++ T S
Sbjct: 588 TTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTE 647
Query: 446 QFTPFAGQDQLFVGNGQGLPISGIGSSTLSS 476
+ P+ SST S
Sbjct: 648 SSSAPVPTPSSSTTESSSAPVPTPSSSTTES 678
Score = 33.5 bits (75), Expect = 3.5
Identities = 29/107 (27%), Positives = 43/107 (40%), Gaps = 20/107 (18%)
Query: 877 NSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASSDQ------NPSSTS 930
+S + S S V PS + E S P + + S +P S SS SS++
Sbjct: 385 SSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSA 444
Query: 931 PASAPATSEASAEIPS--------------QVTIPTSSGQVSAPSSS 963
P ++ T +SA +P+ T +SS V PSSS
Sbjct: 445 PVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSS 491
Score = 33.1 bits (74), Expect = 4.6
Identities = 29/119 (24%), Positives = 51/119 (42%), Gaps = 8/119 (6%)
Query: 326 PQNSPQNSQWNSSPQWHSNSQWSSTPQWNSQWSAPVSSPWQHSSSQWPPRYPPAPSGYAP 385
P S ++ +S+P S ++ SS P P S SS P S AP
Sbjct: 330 PTPSSSTTESSSAPVTSSTTESSSAP-------VPTPSSSTTESSSAPVTSSTTESSSAP 382
Query: 386 MASSPGVPYRGPAPSP-MGYAQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQN 443
+ SS P P+P ++ A TS++T +++P S +S ++ +S +++
Sbjct: 383 VTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTES 441
Score = 32.3 bits (72), Expect = 7.8
Identities = 33/118 (27%), Positives = 50/118 (41%), Gaps = 10/118 (8%)
Query: 325 PPQNSPQNSQWNSSPQWHSNSQWSSTPQWNSQWSAPVSSPWQHSSSQWPPRYPPAPSGYA 384
P +S S SS + S+++ SS P S SS SSS P P+
Sbjct: 812 PSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPT--- 868
Query: 385 PMASSPGVPYRGPAPSPMGYAQAHVAFQTSTSTPQATSPAQSWYPDSGASHHVTSESQ 442
P +SS APS + ++ +F T T+ +P+ S YP S V+S ++
Sbjct: 869 PSSSS---NITSSAPSSIPFSSTTESFSTGTT----VTPSSSKYPGSQTETSVSSTTE 919
Score = 32.3 bits (72), Expect = 7.8
Identities = 34/132 (25%), Positives = 56/132 (41%), Gaps = 17/132 (12%)
Query: 867 TFPYPKLFAQNSNSPSLSPAVVLPSLPSVEHFSQPA-----------IAQPSSSLPSPFL 915
T P P +S + S S V PS + E S P + PSSS +
Sbjct: 311 TTPVPT--PSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSS-TTESS 367
Query: 916 PSPIASSDQNPSSTSPASAPATSEASAEI--PSQVTIPTSSGQVSAPSSSGNVHPMITRS 973
+P+ SS SS++P ++ T +SA + PS T +SS V++ ++ + P+ + +
Sbjct: 368 SAPVTSS-TTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSST 426
Query: 974 KNGIVQPRVQPT 985
P T
Sbjct: 427 TESSSAPVTSST 438
>VGLG_HHV2H (P13290) Glycoprotein G
Length = 699
Score = 46.6 bits (109), Expect = 4e-04
Identities = 25/99 (25%), Positives = 42/99 (42%), Gaps = 5/99 (5%)
Query: 875 AQNSNSPSLSPAVVLPSLPSVEHFSQPAIAQPSSSLPSPFLPSPIASS-----DQNPSST 929
A + +P +PA P S +P P ++ P P P P+ +S D +++
Sbjct: 441 ASAAKTPPTTPAPTTPPPTSTHATPRPTTPGPQTTPPGPATPGPVGASAAPTADSPLTAS 500
Query: 930 SPASAPATSEASAEIPSQVTIPTSSGQVSAPSSSGNVHP 968
PA+AP S A+ + + P + G P + HP
Sbjct: 501 PPATAPGPSAANVSVAATTATPGTRGTARTPPTDPKTHP 539
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.132 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 150,373,080
Number of Sequences: 164201
Number of extensions: 6947694
Number of successful extensions: 42137
Number of sequences better than 10.0: 540
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 464
Number of HSP's that attempted gapping in prelim test: 35883
Number of HSP's gapped (non-prelim): 4715
length of query: 1182
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1061
effective length of database: 40,105,733
effective search space: 42552182713
effective search space used: 42552182713
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0336.1


