

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0280b.3
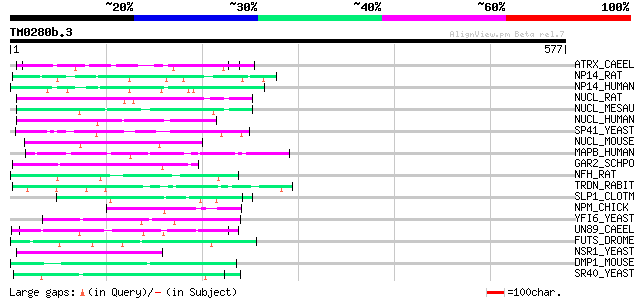
(577 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 57 1e-07
NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 k... 54 1e-06
NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130... 54 1e-06
NUCL_RAT (P13383) Nucleolin (Protein C23) 53 2e-06
NUCL_MESAU (P08199) Nucleolin (Protein C23) 53 2e-06
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 52 3e-06
SP41_YEAST (P38904) SPP41 protein 51 9e-06
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 51 9e-06
MAPB_HUMAN (P46821) Microtubule-associated protein 1B (MAP 1B) [... 50 2e-05
GAR2_SCHPO (P41891) Protein gar2 50 2e-05
NFH_RAT (P16884) Neurofilament triplet H protein (200 kDa neurof... 49 3e-05
TRDN_RABIT (Q28820) Triadin 49 5e-05
SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer... 49 5e-05
NPM_CHICK (P16039) Nucleophosmin (NPM) (Nucleolar phosphoprotein... 49 5e-05
YFI6_YEAST (P43597) Hypothetical 137.7 kDa protein in UGS1-FAB1 ... 48 6e-05
UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89 (Uncoo... 48 6e-05
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 48 8e-05
NSR1_YEAST (P27476) Nuclear localization sequence binding protei... 47 1e-04
DMP1_MOUSE (O55188) Dentin matrix acidic phosphoprotein 1 precur... 47 1e-04
SR40_YEAST (P32583) Suppressor protein SRP40 47 2e-04
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 57.0 bits (136), Expect = 1e-07
Identities = 59/249 (23%), Positives = 105/249 (41%), Gaps = 31/249 (12%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
S+S S E+ +S+ K + +K KSK +SS ED SDEE E +
Sbjct: 90 SESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSS---EDEDSDEEREQKSKKKSKK 146
Query: 68 NEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS-SKKG 126
+++T S + E + + ++E + + + +EE+ E PS SKKG
Sbjct: 147 TKKQTSSESSEE---SEEERKVKKSKKNKEKSVKKRAET----SEESDEDEKPSKKSKKG 199
Query: 127 DKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAP 186
K S+ S+ + D+ +V + ++ +K + EAP
Sbjct: 200 LKKKAKSE---------------SESESEDEKEV----KKSKKKSKKVVKKESESEDEAP 240
Query: 187 SVPQ-KKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIV 245
+ +K K S T+ + + K + ++S + KKK P+ V + SD E++ D+
Sbjct: 241 EKKKTEKRKRSKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDVE 300
Query: 246 LAEMKKFAG 254
+ KK G
Sbjct: 301 VLPQKKKRG 309
Score = 50.8 bits (120), Expect = 9e-06
Identities = 58/239 (24%), Positives = 98/239 (40%), Gaps = 27/239 (11%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLE-DVTSDEEEEPFHDAPAN 66
SK S KT+K S + RK KSK + S E SDE+E+P +
Sbjct: 140 SKKKSKKTKKQTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKG 199
Query: 67 I-----NEEETLMGSDRDVVPDAEASAPQDTNVSQ-ENPIPENPDVEDVIAEETVSTETP 120
+ +E E+ +++V + S S+ E+ PE E +T S E+
Sbjct: 200 LKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEESS 259
Query: 121 SSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPES--------VGA 172
S K ++ + + V+ S ++S++ DV E LP+ +
Sbjct: 260 ESEKSDEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDV---EVLPQKKKRGAVTLISD 316
Query: 173 RLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMS----EKKKKAP 227
K + + SEA V +K +K K+KK++ G ++E S++ KKK+ P
Sbjct: 317 SEDEKDQKSESEASDVEEKVSK-----KKAKKQESSESGSDSSEGSITVNRKSKKKEKP 370
Score = 44.7 bits (104), Expect = 7e-04
Identities = 50/229 (21%), Positives = 93/229 (39%), Gaps = 26/229 (11%)
Query: 14 KTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEETL 73
K + L + R++ P K +K SSS E+ DEEE P + + ++
Sbjct: 34 KRAQKLKEKREREGKPPPKKRPAKKRKASSS----EEDDDDEEESPRKSSKKSRKRAKSE 89
Query: 74 MGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVST--ETPSSSKKGDKGSV 131
SD E+ +D S+ + E + T S+ + S ++ K
Sbjct: 90 SESD-------ESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDEDSDEEREQKSKK 142
Query: 132 SSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQK 191
SK+T Q +S+ + +++ + ++ + KR + S+ P K
Sbjct: 143 KSKKTKK--------QTSSESSEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSK 194
Query: 192 KTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVS-ESDSDVEA 239
K+K KK + + ++ EK + + KKK+ +V ES+S+ EA
Sbjct: 195 KSKKG----LKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEA 239
Score = 40.8 bits (94), Expect = 0.010
Identities = 35/153 (22%), Positives = 61/153 (38%), Gaps = 22/153 (14%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKAR------------TPARKYYKSKSHGVSSSHVLLEDV 51
K+ S + S+ EK + +S+KK++ P +K + + +SS E
Sbjct: 203 KAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEESSESE 262
Query: 52 TSDEEEEPFHDAP----------ANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIP 101
SDEEEE +P ++ +E SD +V+P + E+
Sbjct: 263 KSDEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDVEVLPQKKKRGAVTLISDSEDEKD 322
Query: 102 ENPDVEDVIAEETVSTETPSSSKKGDKGSVSSK 134
+ + E EE VS + + + GS SS+
Sbjct: 323 QKSESEASDVEEKVSKKKAKKQESSESGSDSSE 355
Score = 32.3 bits (72), Expect = 3.5
Identities = 30/140 (21%), Positives = 56/140 (39%), Gaps = 25/140 (17%)
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDE--------PIQVLVQNNSDGDDSDDDDV 160
VI +E + +++ +K + KE E P + ++S+ DD D+++
Sbjct: 15 VIEDEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDDDDEEES 74
Query: 161 PLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMS 220
P ++ RKR + SE+ +++ + K K K K +K
Sbjct: 75 P-------RKSSKKSRKRAKSESESDESDEEEDR----------KKSKSKKKVDQKKKEK 117
Query: 221 EKKKKAPVIVSESDSDVEAD 240
KKK+ + DSD E +
Sbjct: 118 SKKKRTTSSSEDEDSDEERE 137
Score = 31.2 bits (69), Expect = 7.7
Identities = 28/114 (24%), Positives = 49/114 (42%), Gaps = 12/114 (10%)
Query: 142 IQVLVQNNSDGDDSDDDDVPLFETL----PESVGARLKRKRRTTASEAPSVPQKKTKPSP 197
+ V +SDG +D+D+ + + E +LK KR E P K +P+
Sbjct: 3 VGVSESEDSDGHVIEDEDLEMARQIENERKEKRAQKLKEKR-----EREGKPPPKKRPAK 57
Query: 198 TTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKK 251
S ++ + + KS + +K+A SES+SD + D ++ KK
Sbjct: 58 KRKASSSEEDDDDEEESPRKSSKKSRKRAK---SESESDESDEEEDRKKSKSKK 108
>NP14_RAT (P41777) Nucleolar phosphoprotein p130 (Nucleolar 130 kDa
protein) (140 kDa nucleolar phosphoprotein) (Nopp140)
(Nucleolar and coiled-body phosphoprotein 1)
Length = 704
Score = 53.9 bits (128), Expect = 1e-06
Identities = 83/330 (25%), Positives = 130/330 (39%), Gaps = 76/330 (23%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLL-----------EDVT 52
K+GSS S+SS + S+ KKA P +K K V+ + V + ED +
Sbjct: 223 KAGSSSSSSSSSSSDDSEEEKKAAAPLKK-TAPKKQVVAKAPVKVTAAPTQKSSSSEDSS 281
Query: 53 SDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAE 112
S+EEEE + VP S + +V ++P + +
Sbjct: 282 SEEEEEQ--------KKPMKKKAGPYSSVPPPSVSLSK-KSVGAQSPKKAAAQTQPADSS 332
Query: 113 ETVSTETPSSS---KKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSD--DDDVP------ 161
S E+ SSS KK +V SK P++ +++SD DSD +D+ P
Sbjct: 333 ADSSEESDSSSEEEKKTPAKTVVSKTPAKPAPVKKKAESSSDSSDSDSSEDEAPAKPVSA 392
Query: 162 ------------------LFETLPESVGARLKRKRR-----TTASEAPSVPQKKTKPSPT 198
T + G+ K + R ++ E+ S ++ TK S T
Sbjct: 393 TKSPLSKPAVTPKPPAAKAVATPKQPAGSGQKPQSRKADSSSSEEESSSSEEEATKKSVT 452
Query: 199 TPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEAD-------VPDIVLAEMKK 251
TPK+ + TA+ + S K+AP +S SD E+ P A+ KK
Sbjct: 453 TPKA---------RVTAKAAPSLPAKQAPRAGGDSSSDSESSSSEEEKKTPPKPPAK-KK 502
Query: 252 FAGRRIPKNVP----AAPLDNVSFHSEENA 277
AG +PK P AA + S SE+++
Sbjct: 503 AAGAAVPKPTPVKKAAAESSSSSSSSEDSS 532
Score = 40.8 bits (94), Expect = 0.010
Identities = 51/231 (22%), Positives = 92/231 (39%), Gaps = 41/231 (17%)
Query: 2 GLKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFH 61
G K S K+ SS +E+ S S ++A + VT+ +
Sbjct: 422 GQKPQSRKADSSSSEEESSSSEEEATK--------------------KSVTTPKARVTAK 461
Query: 62 DAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENP----------IPENPDVEDVIA 111
AP+ ++ G D D+E+S+ ++ + P +P+ V+ A
Sbjct: 462 AAPSLPAKQAPRAGGDSS--SDSESSSSEEEKKTPPKPPAKKKAAGAAVPKPTPVKKAAA 519
Query: 112 EETVSTETPSSSKKGDKGSVSSKETHSDEPIQV----LVQNNSDGDDSDDDDVPLFETLP 167
E + S+ + S + +K SK T + + QN G +S++++ +
Sbjct: 520 ESSSSSSSSEDSSEEEKKKPKSKATPKPQAGKANGVPASQNGKAGKESEEEEEDTEQNKK 579
Query: 168 ES---VGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA 215
+ G+ KRK TA EA + KK K TP + K KG+ +A++
Sbjct: 580 AAGTKPGSGKKRKHNETADEAATPQSKKVKLQ--TPNTFPKRKKGEKRASS 628
Score = 40.4 bits (93), Expect = 0.013
Identities = 44/214 (20%), Positives = 76/214 (34%), Gaps = 13/214 (6%)
Query: 35 KSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNV 94
K +S+G + E +SD E+ + E++ + + + P AS PQ
Sbjct: 67 KLQSNGPVAKKAKKETSSSDSSEDSSEE------EDKAQVPTQKAAAPAKRASLPQHAGK 120
Query: 95 SQENPIPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDD 154
+ + E EE E K + +V + P + + S+ D
Sbjct: 121 AAAKASESSSSEESSEEEE----EKDKKKKPVQQKAVKPQAKAVRPPPKKAESSESESDS 176
Query: 155 SDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKK-KDLKGKGKA 213
S +D+ P +T A + ++AP+ P K P K +
Sbjct: 177 SSEDEAP--QTQKPKAAATAAKAPTKAQTKAPAKPGPPAKAQPKAANGKAGSSSSSSSSS 234
Query: 214 TAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLA 247
+++ S EKK AP+ + V A P V A
Sbjct: 235 SSDDSEEEKKAAAPLKKTAPKKQVVAKAPVKVTA 268
Score = 36.6 bits (83), Expect = 0.18
Identities = 53/264 (20%), Positives = 91/264 (34%), Gaps = 28/264 (10%)
Query: 16 EKSLSQSRKKARTPARKYYKSKSHGVSSSH-------------VLLEDVTSDEEEEPFHD 62
+K++ K R P +K S+S SSS + T + + P
Sbjct: 150 QKAVKPQAKAVRPPPKKAESSESESDSSSEDEAPQTQKPKAAATAAKAPTKAQTKAPAKP 209
Query: 63 APANINEEETLMGSDRDVVPDAEASAPQDTNVSQE--NPIPENPDVEDVIAEETVS-TET 119
P + + G + +S+ D+ ++ P+ + + V+A+ V T
Sbjct: 210 GPPAKAQPKAANGKAGSSSSSSSSSSSDDSEEEKKAAAPLKKTAPKKQVVAKAPVKVTAA 269
Query: 120 PSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRR 179
P+ + S S +E +P++ P +SVGA+ +K
Sbjct: 270 PTQKSSSSEDSSSEEEEEQKKPMK------KKAGPYSSVPPPSVSLSKKSVGAQSPKKAA 323
Query: 180 TTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEA 239
A S + ++ + KK K T K + KKKA SDSD
Sbjct: 324 AQTQPADSSADSSEESDSSSEEEKKTPAKTVVSKTPAKP-APVKKKAESSSDSSDSDSSE 382
Query: 240 DVPDIVLAEMKKFAGRRIPKNVPA 263
D A K + + P + PA
Sbjct: 383 DE-----APAKPVSATKSPLSKPA 401
Score = 32.7 bits (73), Expect = 2.7
Identities = 49/218 (22%), Positives = 77/218 (34%), Gaps = 21/218 (9%)
Query: 13 SKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLE---DVTSDEEEEPFHDAPANINE 69
S T+ LS+ + PA K + S D +S EEE ++ +E
Sbjct: 391 SATKSPLSKPAVTPKPPAAKAVATPKQPAGSGQKPQSRKADSSSSEEE-------SSSSE 443
Query: 70 EETLMGSDRDVVPDAEASAPQDTNVSQENPI--PENPDVEDVIAEETVSTETPSSSKKGD 127
EE S A A Q + D E +EE T +KK
Sbjct: 444 EEATKKSVTTPKARVTAKAAPSLPAKQAPRAGGDSSSDSESSSSEEEKKTPPKPPAKKKA 503
Query: 128 KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPS 187
G+ K T P++ +S S +D + P+S K + A +A
Sbjct: 504 AGAAVPKPT----PVKKAAAESSSSSSSSEDSSEEEKKKPKS-----KATPKPQAGKANG 554
Query: 188 VPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKK 225
VP + + + +++D + KA K S KK+K
Sbjct: 555 VPASQNGKAGKESEEEEEDTEQNKKAAGTKPGSGKKRK 592
Score = 32.7 bits (73), Expect = 2.7
Identities = 41/185 (22%), Positives = 66/185 (35%), Gaps = 18/185 (9%)
Query: 84 AEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSS--KKGDKGSVSSKETHSDEP 141
A + QD N S + D+ + ST+ P G + KET S +
Sbjct: 37 ATGATQQDANASS---------LLDIYSFWLKSTKAPKVKLQSNGPVAKKAKKETSSSD- 86
Query: 142 IQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAP-SVPQKKTKPSPTTP 200
+ ++ D VP + + A L + A++A S +++
Sbjct: 87 -----SSEDSSEEEDKAQVPTQKAAAPAKRASLPQHAGKAAAKASESSSSEESSEEEEEK 141
Query: 201 KSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRRIPKN 260
KKK ++ K K++ KKA SESDS E + P + A + K
Sbjct: 142 DKKKKPVQQKAVKPQAKAVRPPPKKAESSESESDSSSEDEAPQTQKPKAAATAAKAPTKA 201
Query: 261 VPAAP 265
AP
Sbjct: 202 QTKAP 206
Score = 31.6 bits (70), Expect = 5.9
Identities = 37/141 (26%), Positives = 53/141 (37%), Gaps = 24/141 (17%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
SS S+SS + S + +K K K++GV +S S+EEEE D N
Sbjct: 521 SSSSSSSSEDSSEEEKKKPKSKATPKPQAGKANGVPASQNGKAGKESEEEEE---DTEQN 577
Query: 67 INEEETLMGSDRDVVPDA---EASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSS 123
T GS + + EA+ PQ V + P T
Sbjct: 578 KKAAGTKPGSGKKRKHNETADEAATPQSKKVKLQTP------------------NTFPKR 619
Query: 124 KKGDKGSVSSKETHSDEPIQV 144
KKG+K + S +E I+V
Sbjct: 620 KKGEKRASSPFRRVREEEIEV 640
>NP14_HUMAN (Q14978) Nucleolar phosphoprotein p130 (Nucleolar 130
kDa protein) (140 kDa nucleolar phosphoprotein)
(Nopp140) (Nucleolar and coiled-body phosphoprotein 1)
Length = 699
Score = 53.5 bits (127), Expect = 1e-06
Identities = 80/311 (25%), Positives = 119/311 (37%), Gaps = 66/311 (21%)
Query: 2 GLKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSK-------------SHGVSSSHVLL 48
G + SS S+SS + S+ K A TP + K + + SSS
Sbjct: 212 GKAASSSSSSSSSSSSDDSEEEKAAATPKKTVPKKQVVAKAPVKAATTPTRKSSSS---- 267
Query: 49 EDVTSDEEEE---PFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPD 105
ED +SDEEEE P + P + P A P+ + + + P
Sbjct: 268 EDSSSDEEEEQKKPMKNKPGPYS-----------YAPPPSAPPPKKS-LGTQPPKKAVEK 315
Query: 106 VEDVIAEETVSTETPSSS---KKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSD---DDD 159
+ V + E S E+ SSS KK +V SK T P + +++SD DSD DD+
Sbjct: 316 QQPVESSEDSSDESDSSSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDE 375
Query: 160 VPLFETLPESVGARLKRKRRTTASE-------APSVP----------------QKKTKPS 196
P + P K T AP P ++ S
Sbjct: 376 AP---SKPAGTTKNSSNKPAVTTKSPAVKPAAAPKQPVGGGQKLLTRKADSSSSEEESSS 432
Query: 197 PTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRR 256
K+KK K KATA+ ++S K+AP +S SD ++ + + K A ++
Sbjct: 433 SEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRDSSSDSDSSSSEEEEEKTSKSAVKK 492
Query: 257 IPKNVP--AAP 265
P+ V AAP
Sbjct: 493 KPQKVAGGAAP 503
Score = 38.1 bits (87), Expect = 0.063
Identities = 75/320 (23%), Positives = 108/320 (33%), Gaps = 88/320 (27%)
Query: 4 KSGSSKSTSSKTEK-------------------SLSQSRKKARTPARKYYKSKSHGVSSS 44
KS SS+ +SS E+ S +K T K K V SS
Sbjct: 263 KSSSSEDSSSDEEEEQKKPMKNKPGPYSYAPPPSAPPPKKSLGTQPPKKAVEKQQPVESS 322
Query: 45 HVLLE--DVTSDEEEEPFHDA----------PANINEEETLMGSDRDVVPDAEA-SAPQD 91
+ D +S+EE++P A PA E + SD D D EA S P
Sbjct: 323 EDSSDESDSSSEEEKKPPTKAVVSKATTKPPPAKKAAESSSDSSDSDSSEDDEAPSKPAG 382
Query: 92 TNVSQENP---IPENPDVEDVIA-EETVSTETPSSSKKGDKGSVSSKETHSDE------- 140
T + N ++P V+ A ++ V ++K D S + + S+E
Sbjct: 383 TTKNSSNKPAVTTKSPAVKPAAAPKQPVGGGQKLLTRKADSSSSEEESSSSEEEKTKKMV 442
Query: 141 ---------------PIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEA 185
P + Q + D D E S A K+ ++ A
Sbjct: 443 ATTKPKATAKAALSLPAKQAPQGSRDSSSDSDSSSSEEEEEKTSKSAVKKKPQKVAGGAA 502
Query: 186 PSVPQKKTKPSPTTPKSKKKD---------LKGKG-------KATAEKSMS--------- 220
PS P K + S D LKGKG KA +++
Sbjct: 503 PSKPASAKKGKAESSNSSSSDDSSEEEEEKLKGKGSPRPQAPKANGTSALTAQNGKAAKN 562
Query: 221 -----EKKKKAPVIVSESDS 235
E+KKKA V+VS+S S
Sbjct: 563 SEEEEEEKKKAAVVVSKSGS 582
Score = 37.7 bits (86), Expect = 0.082
Identities = 47/213 (22%), Positives = 84/213 (39%), Gaps = 23/213 (10%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKS--HGVSSSHVLLEDVTSDEEEEPFHDAP 64
SS S KT+K ++ ++ KA A +K G S + +S+EEEE +
Sbjct: 430 SSSSEEEKTKKMVATTKPKATAKAALSLPAKQAPQGSRDSSSDSDSSSSEEEEEKTSKSA 489
Query: 65 ANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSK 124
++ G+ A+ + +N S + S+E
Sbjct: 490 VKKKPQKVAGGAAPSKPASAKKGKAESSNSSSSDD----------------SSEEEEEKL 533
Query: 125 KGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFET--LPESVGARLKRKRRTTA 182
KG KGS + ++ + QN +S++++ + + G+ KRK+ A
Sbjct: 534 KG-KGSPRPQAPKANGTSALTAQNGKAAKNSEEEEEEKKKAAVVVSKSGSLKKRKQNEAA 592
Query: 183 SEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA 215
EA + KK K TP + K KG+ +A++
Sbjct: 593 KEAETPQAKKIKLQ--TPNTFPKRKKGEKRASS 623
>NUCL_RAT (P13383) Nucleolin (Protein C23)
Length = 712
Score = 53.1 bits (126), Expect = 2e-06
Identities = 66/262 (25%), Positives = 111/262 (42%), Gaps = 26/262 (9%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSS-HVLLEDVTSDEEEEPFHDAPAN 66
+K + +K +Q++ TP +K + + G + + ED DE+EE D+ +
Sbjct: 100 AKVVPTPGKKGAAQAKALVPTPGKKGAVTPAKGAKNGKNAKKEDSDEDEDEEDEDDSDED 159
Query: 67 INEEETLMGSDRDVVPDAEAS--APQDTNVSQENPIPENPDVEDVIAEETVSTE------ 118
+EE+ V A+A+ AP + +E+ E+ D +D EE +E
Sbjct: 160 EDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDEDDDDDDEEEEEEDDSEEEVMEI 219
Query: 119 TPSSSKKGD------KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGA 172
TP+ KK K ++E DE + ++ + D+ DD+D E A
Sbjct: 220 TPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDEEDDEDEDEEEEEEPVKAA 279
Query: 173 RLKRKRRTT-ASEAPSVPQKKTKPS-PTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIV 230
KRK+ T EAP ++K + S PTTP + G KS++E K V +
Sbjct: 280 PGKRKKEMTKQKEAPEAKKQKIEGSEPTTPFN-----LFIGNLNPNKSVAELK----VAI 330
Query: 231 SESDSDVEADVPDIVLAEMKKF 252
SE + + D+ +KF
Sbjct: 331 SELFAKNDLAAVDVRTGTNRKF 352
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 53.1 bits (126), Expect = 2e-06
Identities = 58/264 (21%), Positives = 101/264 (37%), Gaps = 32/264 (12%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSS-HVLLEDVTSDEEEEPFHDAPAN 66
+K+ ++ +K +Q++ TP +K + + G + + ED DE+++ D
Sbjct: 99 AKAVATPGKKGATQAKALVATPGKKGAVTPAKGAKNGKNAKKEDSDEDEDDDDDEDDSDE 158
Query: 67 INEEE--------TLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTE 118
E+E + G V A AS +D +E E+ + ED EE
Sbjct: 159 DEEDEEEDEFEPPVVKGKQGKVAAAAPASEDEDEEEDEEEE-EEDEEEEDDSEEEEAMEI 217
Query: 119 TPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKR 178
TP+ KK V K + V D D+ +D+D E E + +
Sbjct: 218 TPAKGKKAPAKVVPVKAKN--------VAEEDDDDEEEDEDEEEDEEEEEDEEEEEEEEE 269
Query: 179 RTTASEAPSVPQKKTKPSPTTPKSKKKDLKGK----------GKATAEKSMSEKKKKAPV 228
AP +K+ P++KK+ ++G G KS++E K V
Sbjct: 270 EEPVKPAPGKRKKEMTKQKEVPEAKKQKVEGSESTTPFNLFIGNLNPNKSVAELK----V 325
Query: 229 IVSESDSDVEADVPDIVLAEMKKF 252
+SE + + V D+ +KF
Sbjct: 326 AISEPFAKNDLAVVDVRTGTNRKF 349
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 52.4 bits (124), Expect = 3e-06
Identities = 46/212 (21%), Positives = 88/212 (40%), Gaps = 10/212 (4%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
+K+ ++ +K + + TP +K + G + ++ + +EE++ + +
Sbjct: 100 AKAVTTPGKKGATPGKALVATPGKKGAAIPAKGAKNGKNAKKEDSDEEEDDDSEEDEEDD 159
Query: 68 NEEETLMGSDRDVVPDAEASAP---QDTNVSQENPIPENPDVEDVIAEETVSTETPSSSK 124
+E+ A A+AP + + E+ ++ D ED EE + T TP+ K
Sbjct: 160 EDEDEDEDEIEPAAMKAAAAAPASEDEDDEDDEDDEDDDDDEEDDSEEEAMET-TPAKGK 218
Query: 125 KGDKG-SVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTAS 183
K K V +K DE + ++ D DD DD+D E + +
Sbjct: 219 KAAKVVPVKAKNVAEDEDEEEDDEDEDDDDDEDDEDDD-----DEDDEEEEEEEEEEPVK 273
Query: 184 EAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA 215
EAP +K+ P++KK+ ++G TA
Sbjct: 274 EAPGKRKKEMAKQKAAPEAKKQKVEGTEPTTA 305
>SP41_YEAST (P38904) SPP41 protein
Length = 1395
Score = 50.8 bits (120), Expect = 9e-06
Identities = 69/257 (26%), Positives = 105/257 (40%), Gaps = 49/257 (19%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
SSK +S K +K S++R+ ++ + K KS SH S H +D +++ +P
Sbjct: 210 SSKKSSKKKKKDKSKNRESSKDKSSKKSKSSSH--SKKHA--KDRNKEKQSKP------- 258
Query: 67 INEEETLMGSD--RDVVPDAEASA---PQDTNVSQENPIPENPDVEDVIAEETVSTETPS 121
N E TL S+ +++ + + +A + T Q+N +N + EDV A+ V
Sbjct: 259 TNNENTLDLSNILENLIHENDNAAIDTAKQTVDIQDNSHTDNTNNEDVEAQALVEATL-- 316
Query: 122 SSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
E S P + Q S G P S ++ R+ T
Sbjct: 317 --------KAFENELLSSAPTEEPSQEQSIG--------------PVSSRKAVEPPRKPT 354
Query: 182 ASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKA--TAEKS----MSEKKKKAPVIVSESDS 235
A + P + KP P+ KKK KA TA KS + KKKK V ES+
Sbjct: 355 ADDIPLAMLQAFKPKKRPPQEKKKTKSKTSKAASTANKSPASESTSKKKKKKKTVKESNK 414
Query: 236 DVEA---DVPDIVLAEM 249
EA D +LA+M
Sbjct: 415 SQEAYEDDEFSRILADM 431
Score = 33.1 bits (74), Expect = 2.0
Identities = 51/231 (22%), Positives = 90/231 (38%), Gaps = 15/231 (6%)
Query: 9 KSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAP---A 65
K T SKT K+ S + K + + K K V S+ E DE D
Sbjct: 377 KKTKSKTSKAASTANKSPASESTSKKKKKKKTVKESNKSQEAYEDDEFSRILADMVNQVV 436
Query: 66 NINEEETLMGS---DRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSS 122
N + +ET + D + +++ ++P + + E+ N D D+ +
Sbjct: 437 NTSLKETSTHTATQDNKLESESDFTSPVQSQYTTEDASTANDDSLDLNQIMQNAMAMVFQ 496
Query: 123 SKKGDKGSVSSKETHSDEPIQVLVQNNSDGDD---SDDDDVPLFETLPESVGARLKRKRR 179
++ D+ + E + + V + D+ + VP + E A +R +
Sbjct: 497 NQNDDEFDENIVEDFNRGLGDLSVSDLLPHDNLSRMEKKSVPKSSSKSEKKTAISRRASK 556
Query: 180 TTASEAPSV-----PQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKK 225
+ +A SV P K KPS T +KK L+ K + A ++ S +KK
Sbjct: 557 KASRDASSVELTEVPSKPKKPSKTEVSLEKK-LRKKYVSIANEAASVARKK 606
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 50.8 bits (120), Expect = 9e-06
Identities = 48/200 (24%), Positives = 86/200 (43%), Gaps = 15/200 (7%)
Query: 16 EKSLSQSRKKARTPARKYYKSKSHGVSSS-HVLLEDVTSDEEEEPFHDAPANINEEET-- 72
+K +Q++ TP +K + + G + + ED DE+EE D+ + ++EE
Sbjct: 108 KKGAAQAKALVPTPGKKGAATPAKGAKNGKNAKKEDSDEDEDEEDEDDSDEDEDDEEEDE 167
Query: 73 -----LMG-SDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKG 126
+ G P A AS ++ + +++ ++ + ED EE + T K
Sbjct: 168 FEPPIVKGVKPAKAAPAAPASEDEEDDEDEDDEEDDDEEEEDDSEEEVMEITTAKGKKTP 227
Query: 127 DKG-SVSSKETHSDEPIQVLVQNNSDGDD----SDDDDVPLFETLPESVGARLKRKRRTT 181
K + +K +E + +++ D DD +DDD E P ++K T
Sbjct: 228 AKVVPMKAKSVAEEEDDEEEDEDDEDEDDEEEDDEDDDEEEEEEEPVKAAPGKRKKEMTK 287
Query: 182 ASEAPSVPQKKTKPS-PTTP 200
EAP ++K + S PTTP
Sbjct: 288 QKEAPEAKKQKVEGSEPTTP 307
Score = 32.0 bits (71), Expect = 4.5
Identities = 40/176 (22%), Positives = 65/176 (36%), Gaps = 18/176 (10%)
Query: 80 VVPDAEASA-PQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHS 138
V P +A A P N++ IP A+ V T + KG+ + K
Sbjct: 82 VTPGKKAVATPAKKNITPAKVIPTPGKKGAAQAKALVPTPGKKGAATPAKGAKNGKNAKK 141
Query: 139 DEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPT 198
++ + + + D D D+DD E P V K A AP+ P + +
Sbjct: 142 EDSDEDEDEEDEDDSDEDEDDEEEDEFEPPIV------KGVKPAKAAPAAPASEDEEDDE 195
Query: 199 TPKSKKKDLKGKGKATAEKSM---SEKKKKAPV--------IVSESDSDVEADVPD 243
++ D + + + E+ M + K KK P V+E + D E D D
Sbjct: 196 DEDDEEDDDEEEEDDSEEEVMEITTAKGKKTPAKVVPMKAKSVAEEEDDEEEDEDD 251
>MAPB_HUMAN (P46821) Microtubule-associated protein 1B (MAP 1B)
[Contains: MAP1 light chain LC1]
Length = 2468
Score = 49.7 bits (117), Expect = 2e-05
Identities = 65/282 (23%), Positives = 119/282 (42%), Gaps = 31/282 (10%)
Query: 17 KSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEETLMGS 76
K + SR+ + PA K SKS S E + E+P P ++E+ ++
Sbjct: 536 KQRADSRESLK-PAAKPLPSKSVRKESKEETPEVTKVNHVEKP----PKVESKEKVMVKK 590
Query: 77 DRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSK-----KGDKGSV 131
D+ V + + S + S+E P P V+ +AE+ + P ++K K K
Sbjct: 591 DKPVKTETKPSVTEKEVPSKEEPSP----VKAEVAEKQATDVKPKAAKEKTVKKETKVKP 646
Query: 132 SSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQK 191
K+ ++P + V D ++ P E + + V +K++ + P+K
Sbjct: 647 EDKKEEKEKPKKE-VAKKEDKTPIKKEEKPKKEEVKKEVKKEIKKEEKKE-------PKK 698
Query: 192 KTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKK 251
+ K T PK KK++K + K +K E KK+ + ++ ++ P L+E KK
Sbjct: 699 EVKKE-TPPKEVKKEVKKEEKKEVKKEEKEPKKEIKKLPKDAK---KSSTP---LSEAKK 751
Query: 252 FAGRR--IPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATE 291
A + +PK + D+V+ + K K + + A E
Sbjct: 752 PAALKPKVPKKEESVKKDSVAAGKPKEKGKIKVIKKEGKAAE 793
Score = 43.9 bits (102), Expect = 0.001
Identities = 62/237 (26%), Positives = 90/237 (37%), Gaps = 27/237 (11%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSK-------SHGVSSSHVLLEDVTSDEEEEP 59
+S+ + ++EK L+QS P K + S S+ DV + EE P
Sbjct: 2111 ASEEPTEESEKPLTQSGGAPPPPGGKQQGRQCDETPPTSVSESAPSQTDSDVPPETEECP 2170
Query: 60 FHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDV-----EDVIAEET 114
A ANI+ E+ D + P V +P P +PDV E + E+
Sbjct: 2171 SITADANIDSEDESETIPTDKTVTYKHMDPPPAPVQDRSPSPRHPDVSMVDPEALAIEQN 2230
Query: 115 VSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARL 174
+ K+ K +T S P++ SDG P L ES
Sbjct: 2231 LGKALKKDLKEKTKTKKPGTKTKSSSPVK-----KSDGKSKPLAASPKPAGLKESSD--- 2282
Query: 175 KRKRRTTASEAPSVPQKKTKPSPTTPKSK-----KKDLKGKGKATAEKSMSEKKKKA 226
K R + + SV +K KP+ TTP+ K +KD + K A A S S K A
Sbjct: 2283 KVSRVASPKKKESV-EKAAKPT-TTPEVKAARGEEKDKETKNAANASASKSAKTATA 2337
Score = 32.7 bits (73), Expect = 2.7
Identities = 47/222 (21%), Positives = 80/222 (35%), Gaps = 20/222 (9%)
Query: 10 STSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEP-----FHDAP 64
S KT + +S S+ + Y SSH+ E + E+ P F +
Sbjct: 1312 SPEDKTLEVVSPSQSVTGSAGHTPYYQSPTDEKSSHLPTEVI----EKPPAVPVSFEFSD 1367
Query: 65 ANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPI--PENPDVEDVIAEETVS---TET 119
A E + + VPD+E+ + + + P+ E+ + A++ S E+
Sbjct: 1368 AKDENERASVSPMDEPVPDSESPIEKVLSPLRSPPLIGSESAYESFLSADDKASGRGAES 1427
Query: 120 PSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRR 179
P K G +GS S+ L Q+ +G +D F + E G K
Sbjct: 1428 PFEEKSGKQGSPDQVSPVSEMTSTSLYQDKQEGKSTD------FAPIKEDFGQEKKTDDV 1481
Query: 180 TTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSE 221
S P++ + K +P G K + S+SE
Sbjct: 1482 EAMSSQPALALDERKLGDVSPTQIDVSQFGSFKEDTKMSISE 1523
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 49.7 bits (117), Expect = 2e-05
Identities = 48/201 (23%), Positives = 88/201 (42%), Gaps = 11/201 (5%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDA 63
K SS S K+ K +S+KK + + +S S SS E +S E E ++
Sbjct: 59 KRASSPEPSKKSVKKQKKSKKKEESSSESESESSSSESESSSSESES-SSSESESSSSES 117
Query: 64 PANINEEETLMGSDRDVVPDAEASAPQDTNVSQEN--PIPENPDVEDVIAEETVSTETPS 121
++ +EEE ++ ++ +E+S+ ++ +E I E + + E+ S+E+ S
Sbjct: 118 SSSESEEEVIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESES 177
Query: 122 SSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDD------DDVPLFETLPESVGARLK 175
S + ++E + +++SD + S D D ++ ES K
Sbjct: 178 ESSSSESEEEEEVVEKTEEKKEGSSESSSDSESSSDSSSESGDSDSSSDSESESSSEDEK 237
Query: 176 RKRRTTASEAPSVPQKKTKPS 196
+++ ASE P K TKPS
Sbjct: 238 KRKAEPASEER--PAKITKPS 256
Score = 36.6 bits (83), Expect = 0.18
Identities = 32/172 (18%), Positives = 70/172 (40%), Gaps = 12/172 (6%)
Query: 82 PDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEP 141
P ++ Q + +E E+ E+ S+E+ SSS + + S S + S+E
Sbjct: 66 PSKKSVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSESESSSSESSSSESEE- 124
Query: 142 IQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTA----------SEAPSVPQK 191
+V+V+ + S + E +++ K+ +++ SE+ S +
Sbjct: 125 -EVIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESESESSSSE 183
Query: 192 KTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEADVPD 243
+ K+++K ++ +S S+ ++ S SDS+ E+ D
Sbjct: 184 SEEEEEVVEKTEEKKEGSSESSSDSESSSDSSSESGDSDSSSDSESESSSED 235
Score = 32.3 bits (72), Expect = 3.5
Identities = 27/135 (20%), Positives = 60/135 (44%), Gaps = 4/135 (2%)
Query: 176 RKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDS 235
+ ++ T A + ++ +K + KSKK+ + ++KS+ ++KK S S+S
Sbjct: 28 KSKKITKEAAKEIAKQSSKTDVSPKKSKKEAKRASSPEPSKKSVKKQKKSKKKEESSSES 87
Query: 236 DVEADVPDIVLAEMKKFAGRRIPKNVPAAPLDNVSFHSEENALKWKYVYQRRLATEREVG 295
+ E+ + +E ++ ++ S SEE + K ++ ++E
Sbjct: 88 ESESSSSE---SESSSSESESSSSESESSSSESSSSESEEEVIV-KTEEKKESSSESSSS 143
Query: 296 SDVLECKEVMALIEK 310
S+ E +E + IE+
Sbjct: 144 SESEEEEEAVVKIEE 158
>NFH_RAT (P16884) Neurofilament triplet H protein (200 kDa
neurofilament protein) (Neurofilament heavy polypeptide)
(NF-H) (Fragment)
Length = 831
Score = 49.3 bits (116), Expect = 3e-05
Identities = 49/249 (19%), Positives = 101/249 (39%), Gaps = 30/249 (12%)
Query: 1 MGLKSGSSKSTSSKTEKSLSQSR--KKARTPARKYYKSKSHGVSSSHVLL---EDVTSDE 55
+ +KS +K + + K + R ++ ++PA++ KS + + + E+V S
Sbjct: 582 LDVKSPEAKPPAKEEAKRPADIRSPEQVKSPAKEEAKSPEKEETRTEKVAPKKEEVKSPV 641
Query: 56 EEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTN---VSQENPIPENPDVEDVIAE 112
EE + P + EE+T +V + AP++ ++ P+ E P
Sbjct: 642 EEVKAKEPPKKVEEEKTPATPKTEVKESKKDEAPKEAQKPKAEEKEPLTEKPK------- 694
Query: 113 ETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGA 172
++P +KK + + + P ++ V+ + + +D + P
Sbjct: 695 -----DSPGEAKKEEAKEKKAAAPEEETPAKLGVKEEAKPKEKAEDAKAKEPSKPSE--- 746
Query: 173 RLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA---EKSMSEKKKKAPVI 229
K + E P+ P+KK T +SKK++ K K +A A +K + ++ K
Sbjct: 747 ----KEKPKKEEVPAAPEKKDTKEEKTTESKKREEKPKMEAKAKEEDKGLPQEPSKPKTE 802
Query: 230 VSESDSDVE 238
+E S +
Sbjct: 803 KAEKSSSTD 811
Score = 33.5 bits (75), Expect = 1.6
Identities = 35/178 (19%), Positives = 64/178 (35%), Gaps = 20/178 (11%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDA 63
K+ +S + K + + + + P + K + + EEE P A
Sbjct: 663 KTEVKESKKDEAPKEAQKPKAEEKEPLTEKPKDSPGEAKKEEAKEKKAAAPEEETP---A 719
Query: 64 PANINEEETLMGSDRDVVPDAEA---SAPQDTNVSQENPIPENPDVEDVIAEETVSTETP 120
+ EE ++ DA+A S P + ++ +P P+ +D E+T
Sbjct: 720 KLGVKEE----AKPKEKAEDAKAKEPSKPSEKEKPKKEEVPAAPEKKDTKEEKTT----- 770
Query: 121 SSSKKGDKGSVSSKETHSD-----EPIQVLVQNNSDGDDSDDDDVPLFETLPESVGAR 173
S K+ +K + +K D EP + + +D D E PE A+
Sbjct: 771 ESKKREEKPKMEAKAKEEDKGLPQEPSKPKTEKAEKSSSTDQKDSQPSEKAPEDKAAK 828
>TRDN_RABIT (Q28820) Triadin
Length = 705
Score = 48.5 bits (114), Expect = 5e-05
Identities = 77/326 (23%), Positives = 125/326 (37%), Gaps = 63/326 (19%)
Query: 4 KSGSSKSTSSKTEK----SLSQSRKKARTPARKYYKSKSH--GVSSSHVL--------LE 49
K + K K EK ++++ KKART + K+K GV V ++
Sbjct: 182 KKATHKEKLEKKEKPETKTVTKEEKKARTKEKIEEKTKKEVKGVKQEKVKQTVAKAKEVQ 241
Query: 50 DVTSDEEEEPFHDAPANINEEETLMGSDR---------DVVPDAEASAPQDTNVSQEN-- 98
+E+E A + E++ R D+ P + P + Q +
Sbjct: 242 KTPKPKEKESKETAAVSKQEQKDQYAFCRYMIDIFVHGDLKPGQSPAIPPPSPTEQASRP 301
Query: 99 ----PIPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDD 154
P PE + E AE+ V+TET ++K D S KET D ++ G
Sbjct: 302 TPALPTPEEKEGEKKKAEKKVTTETKKKAEKEDAKKKSEKETDID------MKKKEPGKS 355
Query: 155 SDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKAT 214
D T P +V ++ + T E +K KP+ PK KK++ K K +
Sbjct: 356 PD--------TKPGTV--KVTTQAATKKDEKKEDSKKAKKPAEEQPKGKKQEKKEKHEEP 405
Query: 215 AEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRRIPKNVPAAPLDNVSFHSE 274
A+ ++K+ AP SE + + + + V A K K VPA + + E
Sbjct: 406 AKS--TKKEHAAP---SEKQAKAKIERKEEVSAASTK-------KAVPAKKEEKTTKTVE 453
Query: 275 ENALKWK------YVYQRRLATEREV 294
+ K K + + L E+EV
Sbjct: 454 QETRKEKPGKISSVLKDKELTKEKEV 479
Score = 39.7 bits (91), Expect = 0.022
Identities = 49/227 (21%), Positives = 94/227 (40%), Gaps = 45/227 (19%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
++++ + K EK + KKA+ PA + K K +E++ H+ PA
Sbjct: 366 TTQAATKKDEKK--EDSKKAKKPAEEQPKGKK----------------QEKKEKHEEPAK 407
Query: 67 INEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKG 126
++E S++ A+A + VS + P ++ +TV ET +K
Sbjct: 408 STKKEHAAPSEKQ----AKAKIERKEEVSAASTKKAVPAKKEEKTTKTVEQET----RKE 459
Query: 127 DKGSVSS----KETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTA 182
G +SS KE ++ ++V G ++ D+ + PE ++K++ +
Sbjct: 460 KPGKISSVLKDKELTKEKEVKVPASLKEKGSETKKDEKT---SKPEP---QIKKEEK--- 510
Query: 183 SEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVI 229
P K+ KP P P+ KK++ + EK+ K + V+
Sbjct: 511 ------PGKEVKPKPPQPQIKKEEKPEQDIMKPEKTALHGKPEEKVL 551
>SLP1_CLOTM (Q06852) Cell surface glycoprotein 1 precursor (Outer
layer protein B) (S-layer protein 1)
Length = 1664
Score = 48.5 bits (114), Expect = 5e-05
Identities = 52/219 (23%), Positives = 73/219 (32%), Gaps = 26/219 (11%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPEN---PD 105
E SDE P+ EE + D ++ P D + P P + P
Sbjct: 1247 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1306
Query: 106 VEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFET 165
E ++E +ETP D + S + T SDEP SD D+ P E
Sbjct: 1307 DEPTPSDEPTPSETPEEPIPTD--TPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEP 1364
Query: 166 LPESVGARLKRKRRTTASEAPSVPQKKTKPSP---TTPKSKKKDLKGKGKA--------- 213
P T SE P P T P+P TTP S G G
Sbjct: 1365 TPSD---------EPTPSETPEEPTPTTTPTPTPSTTPTSGSGGSGGSGGGGGGGGGTVP 1415
Query: 214 TAEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMKKF 252
T+ K + +E + +DVP + E + +
Sbjct: 1416 TSPTPTPTSKPTSTPAPTEIEEPTPSDVPGAIGGEHRAY 1454
Score = 45.8 bits (107), Expect = 3e-04
Identities = 45/195 (23%), Positives = 68/195 (34%), Gaps = 4/195 (2%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 879 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 938
Query: 109 VIAEETVSTETPSSS-KKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLP 167
EE + T+TPS D+ + S + T SDEP SD D+ P ET
Sbjct: 939 ETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTP-SETPE 997
Query: 168 ESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAP 227
E + T + E P + PS S + + + E + SE ++
Sbjct: 998 EPIPTDTPSDEPTPSDE--PTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPI 1055
Query: 228 VIVSESDSDVEADVP 242
+ SD +D P
Sbjct: 1056 PTDTPSDEPTPSDEP 1070
Score = 44.3 bits (103), Expect = 9e-04
Identities = 44/194 (22%), Positives = 68/194 (34%), Gaps = 14/194 (7%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 836 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 895
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPE 168
EE + T+TPS D+ + S + T SDEP + D+ D P PE
Sbjct: 896 ETPEEPIPTDTPS-----DEPTPSDEPTPSDEP--------TPSDEPTPSDEPTPSETPE 942
Query: 169 SVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPV 228
T S+ P+ P + PS S + + + E + SE ++
Sbjct: 943 EPIPTDTPSDEPTPSDEPT-PSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIP 1001
Query: 229 IVSESDSDVEADVP 242
+ SD +D P
Sbjct: 1002 TDTPSDEPTPSDEP 1015
Score = 44.3 bits (103), Expect = 9e-04
Identities = 44/194 (22%), Positives = 68/194 (34%), Gaps = 14/194 (7%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 1161 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1220
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPE 168
EE + T+TPS D+ + S + T SDEP + D+ D P PE
Sbjct: 1221 ETPEEPIPTDTPS-----DEPTPSDEPTPSDEP--------TPSDEPTPSDEPTPSETPE 1267
Query: 169 SVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPV 228
T S+ P+ P + PS S + + + E + SE ++
Sbjct: 1268 EPIPTDTPSDEPTPSDEPT-PSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIP 1326
Query: 229 IVSESDSDVEADVP 242
+ SD +D P
Sbjct: 1327 TDTPSDEPTPSDEP 1340
Score = 43.9 bits (102), Expect = 0.001
Identities = 37/155 (23%), Positives = 54/155 (33%), Gaps = 7/155 (4%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 1204 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1263
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPE 168
EE + T+TPS D+ + S + T SDEP SD D+ P E P
Sbjct: 1264 ETPEEPIPTDTPS-----DEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1318
Query: 169 SVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSK 203
+ T S+ P+ + T TP +
Sbjct: 1319 ETPE--EPIPTDTPSDEPTPSDEPTPSDEPTPSDE 1351
Score = 42.7 bits (99), Expect = 0.003
Identities = 36/155 (23%), Positives = 53/155 (33%), Gaps = 13/155 (8%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 1118 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1177
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPE 168
EE + T+TPS D+ + S + T SDEP + D+ D P PE
Sbjct: 1178 ETPEEPIPTDTPS-----DEPTPSDEPTPSDEP--------TPSDEPTPSDEPTPSETPE 1224
Query: 169 SVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSK 203
T S+ P+ + T TP +
Sbjct: 1225 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE 1259
Score = 42.7 bits (99), Expect = 0.003
Identities = 36/155 (23%), Positives = 53/155 (33%), Gaps = 13/155 (8%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 793 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 852
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPE 168
EE + T+TPS D+ + S + T SDEP + D+ D P PE
Sbjct: 853 ETPEEPIPTDTPS-----DEPTPSDEPTPSDEP--------TPSDEPTPSDEPTPSETPE 899
Query: 169 SVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSK 203
T S+ P+ + T TP +
Sbjct: 900 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE 934
Score = 42.7 bits (99), Expect = 0.003
Identities = 36/155 (23%), Positives = 53/155 (33%), Gaps = 13/155 (8%)
Query: 49 EDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVED 108
E SDE P+ EE + D ++ P D + P P +
Sbjct: 1032 EPTPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDEPTPS 1091
Query: 109 VIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPE 168
EE + T+TPS D+ + S + T SDEP + D+ D P PE
Sbjct: 1092 ETPEEPIPTDTPS-----DEPTPSDEPTPSDEP--------TPSDEPTPSDEPTPSETPE 1138
Query: 169 SVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSK 203
T S+ P+ + T TP +
Sbjct: 1139 EPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE 1173
Score = 36.2 bits (82), Expect = 0.24
Identities = 28/120 (23%), Positives = 43/120 (35%), Gaps = 13/120 (10%)
Query: 84 AEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQ 143
++ P D + P P + EE + T+TPS D+ + S + T SDEP
Sbjct: 785 SDEPTPSDEPTPSDEPTPSDEPTPSETPEEPIPTDTPS-----DEPTPSDEPTPSDEP-- 837
Query: 144 VLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSK 203
+ D+ D P PE T S+ P+ + T TP +
Sbjct: 838 ------TPSDEPTPSDEPTPSETPEEPIPTDTPSDEPTPSDEPTPSDEPTPSDEPTPSDE 891
>NPM_CHICK (P16039) Nucleophosmin (NPM) (Nucleolar phosphoprotein
B23) (Numatrin) (Nucleolar protein NO38)
Length = 294
Score = 48.5 bits (114), Expect = 5e-05
Identities = 38/146 (26%), Positives = 61/146 (41%), Gaps = 20/146 (13%)
Query: 101 PENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDD- 159
PE+ D E+ ST+ P+S K + DE ++ D +D DDD
Sbjct: 125 PESEDEEEDTKIGNASTKRPASGGGAKTPQKKPKLSEDDEDDDEDEDDDEDDEDDLDDDE 184
Query: 160 ----VPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA 215
P+ + E G +++ ++ PS P KTK TP SKK
Sbjct: 185 EEIKTPMKKPAREPAGKNMQKAKQNGKDSKPSTPASKTK----TPDSKK----------- 229
Query: 216 EKSMSEKKKKAPVIVSESDSDVEADV 241
+KS++ K K P+ + E + ++A V
Sbjct: 230 DKSLTPKTPKVPLSLEEIKAKMQASV 255
>YFI6_YEAST (P43597) Hypothetical 137.7 kDa protein in UGS1-FAB1
intergenic region
Length = 1233
Score = 48.1 bits (113), Expect = 6e-05
Identities = 47/219 (21%), Positives = 96/219 (43%), Gaps = 17/219 (7%)
Query: 35 KSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNV 94
K+ HG + V +E +EEEE + + + +E +++ V + E S ++ +
Sbjct: 452 KNLQHGTNDISVEVEKEEEEEEEEEENSTFSKVKKENVT--GEQEAVRNNEVSGTEEEST 509
Query: 95 SQENPIPENPDVEDVIAEET--VSTETPSSSKKGDKGSVSSKE-----THSDEPIQVLVQ 147
S+ I + + E++ + E ++S K K ++ ++ T ++P +V+
Sbjct: 510 SKGEEIMGGDEKQSEAGEKSSIIEIEGSANSAKISKDNLVLEDEAEAPTQENKPTEVV-- 567
Query: 148 NNSDGDDSDDDDVPLFETLPESV------GARLKRKRRTTASEAPSVPQKKTKPSPTTPK 201
D D+ DDV + E + +++ A+ ++ + P K K + +
Sbjct: 568 GEIDIPDAPRDDVEIVEAVEKNIIPEDLEVAKEDQEGEQVKLDEPVKAMKDDKIAMRGAE 627
Query: 202 SKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEAD 240
S +D+K K + TAE S + KK+ ES VE +
Sbjct: 628 SISEDMKKKQEGTAELSNEKAKKEVDETARESAEGVEVE 666
>UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89
(Uncoordinated protein 89)
Length = 6632
Score = 48.1 bits (113), Expect = 6e-05
Identities = 50/238 (21%), Positives = 99/238 (41%), Gaps = 18/238 (7%)
Query: 3 LKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
+KS KS +S T+K S + ++ ++P +K KS S +E
Sbjct: 1404 VKSPKEKSPASPTKKEKSPAAEEVKSPTKKE-KSPSSPTKKEKSPSSPTKKTGDEVKEKS 1462
Query: 63 APANINEEETLMGSDRDVV-PDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPS 121
P + ++E DV P + +P TN+ + + + E+T +T T
Sbjct: 1463 PPKSPTKKEKSPEKPEDVKSPVKKEKSPDATNIVEVS--------SETTIEKTETTMTTE 1514
Query: 122 SSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
+ + ++ S K+ + E + ++ + D S + +++ E + + +K+++
Sbjct: 1515 MTHESEESRTSVKKEKTPEKVDEKPKSPTKKDKSPE------KSITEEIKSPVKKEKSPE 1568
Query: 182 -ASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVE 238
E P+ P KK K SP P S K + + K+ +K S +K + S + E
Sbjct: 1569 KVEEKPASPTKKEK-SPEKPASPTKKSENEVKSPTKKEKSPEKSVVEELKSPKEKSPE 1625
Score = 46.6 bits (109), Expect = 2e-04
Identities = 54/242 (22%), Positives = 99/242 (40%), Gaps = 28/242 (11%)
Query: 11 TSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEE 70
TS K EK+ + +K ++P +K KS S + + V ++ E + PA+ ++
Sbjct: 1524 TSVKKEKTPEKVDEKPKSPTKK---DKSPEKSITEEIKSPVKKEKSPEKVEEKPASPTKK 1580
Query: 71 E---------TLMGSDRDVVPDAEASAPQDTNVSQ-ENPIPENPD-VEDVIAEETVSTET 119
E T + P + +P+ + V + ++P ++P+ +D T ++
Sbjct: 1581 EKSPEKPASPTKKSENEVKSPTKKEKSPEKSVVEELKSPKEKSPEKADDKPKSPTKKEKS 1640
Query: 120 PSSSKKGDKGSVSSKETH----SDEPIQVLVQNNSDGDDSDDD---------DVPLFETL 166
P S D S + KE ++P + +S +DD+ E
Sbjct: 1641 PEKSATEDVKSPTKKEKSPEKVEEKPTSPTKKESSPTKKTDDEVKSPTKKEKSPQTVEEK 1700
Query: 167 PESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMS-EKKKK 225
P S + K ++ E S +K + + PKS K K K+ AE+ S KK+K
Sbjct: 1701 PASPTKKEKSPEKSVVEEVKSPKEKSPEKAEEKPKSPTKKEKSPEKSAAEEVKSPTKKEK 1760
Query: 226 AP 227
+P
Sbjct: 1761 SP 1762
Score = 43.9 bits (102), Expect = 0.001
Identities = 43/220 (19%), Positives = 88/220 (39%), Gaps = 12/220 (5%)
Query: 13 SKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEET 72
+K EKS + +K +P +K S + + ++ + + PA+ ++E
Sbjct: 1653 TKKEKSPEKVEEKPTSPTKK---ESSPTKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEK 1709
Query: 73 LMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGDKGSVS 132
++ VV E +P++ + + P++P ++ E++ + E S +KK S
Sbjct: 1710 --SPEKSVVE--EVKSPKEKSPEKAEEKPKSPTKKEKSPEKSAAEEVKSPTKKEKSPEKS 1765
Query: 133 SKE-----THSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPS 187
++E T + + + + E P S + K ++ A E S
Sbjct: 1766 AEEKPKSPTKKESSPVKMADDEVKSPTKKEKSPEKVEEKPASPTKKEKTPEKSAAEELKS 1825
Query: 188 VPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAP 227
+K+ PS T K+ + + + EK S KK+P
Sbjct: 1826 PTKKEKSPSSPTKKTGDESKEKSPEKPEEKPKSPTPKKSP 1865
Score = 38.9 bits (89), Expect = 0.037
Identities = 55/241 (22%), Positives = 92/241 (37%), Gaps = 18/241 (7%)
Query: 11 TSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLE--DVTSDEEEEPFHDAPANIN 68
+SS+ +K ++Q K+A A ++ S+ + +VTS +
Sbjct: 1252 SSSEVQKEIAQQVKEASPEATTTITMETSLTSTKTTTMSTTEVTSTVGGVTVETKESESE 1311
Query: 69 EEETLMGSDRDVVPDAEASAPQDTNVSQENPIPE----NPDVEDVIAEETVSTETPSSSK 124
T++G V + S + VS+ + + P AEE + E S +
Sbjct: 1312 SATTVIGGGSGGVTEGSISVSKIEVVSKTDSQTDVREGTPKRRVSFAEEELPKEVIDSDR 1371
Query: 125 KGDKGSVSSKETHSDEPIQVL----VQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRT 180
K K K+ S E + + + S + P T E A + K T
Sbjct: 1372 KKKKSPSPDKKEKSPEKTEEKPASPTKKTGEEVKSPKEKSPASPTKKEKSPAAEEVKSPT 1431
Query: 181 TASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVEAD 240
++PS P KK K SP++P K D + K + KK+K+P E DV++
Sbjct: 1432 KKEKSPSSPTKKEK-SPSSPTKKTGD---EVKEKSPPKSPTKKEKSP----EKPEDVKSP 1483
Query: 241 V 241
V
Sbjct: 1484 V 1484
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 47.8 bits (112), Expect = 8e-05
Identities = 69/288 (23%), Positives = 117/288 (39%), Gaps = 36/288 (12%)
Query: 2 GLKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLED----------- 50
G KS S K S+K EK +K + + V SHVLL++
Sbjct: 1239 GAKSISHKEESAKEEKETDD--EKENKVGEIELGDEPNKVDISHVLLKESVQEVAEKVVV 1296
Query: 51 --VTSDEEEEPFHDAPANINEE--ETLMGSDRDVVPDA---EASAPQDTNVSQENPIPEN 103
T ++++E +A I +E E LM +D E+ + + PE
Sbjct: 1297 IETTVEKKQEEIVEATTVITQENQEDLMEQVKDKEEHEQKIESGIITEKEAKKSASTPEE 1356
Query: 104 PDVEDVIAEETV-------STETPSSSK-KGDKGSVSSKETHSDEPIQVLVQNNSDGDDS 155
+ D+ +++ + +T P S+K + D GS+ S T +E I+V VQ +
Sbjct: 1357 KETSDITSDDELPAQLADPTTVPPKSAKDREDTGSIESPPT-IEEAIEVEVQAKQEAQKP 1415
Query: 156 DDDDVPLFETLPESVGAR-LKRKRRTTASEAPSVPQKKTK--PSPTTPKSKKKDLK---G 209
+T + ++ R T S Q K+K P P+ P+S+ KD K
Sbjct: 1416 VPAPEEAIKTEKSPLASKETSRPESATGSVKEDTEQTKSKKSPVPSRPESEAKDKKSPFA 1475
Query: 210 KGKATAEKSMSEK-KKKAPVIVSESDSDVEADVPDIVLAEMKKFAGRR 256
G+A+ +S++E K +A S +S + + L + K+ RR
Sbjct: 1476 SGEASRPESVAESVKDEAGKAESRRESIAKTHKDESSLDKAKEQESRR 1523
Score = 43.9 bits (102), Expect = 0.001
Identities = 42/199 (21%), Positives = 81/199 (40%), Gaps = 22/199 (11%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN- 66
++S +TEKS +SR+++ T ++ +S ++D +EE ++ A
Sbjct: 3880 AESVKDETEKSKEESRRESVTEKSPLPSKEASRPTSVAESVKDEAEKSKEESRRESVAEK 3939
Query: 67 ---INEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSS 123
++E + S + + D Q+ S+ +PE+ E +
Sbjct: 3940 SPLASKESSRPASVAESIKDEAEGTKQE---SRRESMPESGKAESI-------------- 3982
Query: 124 KKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTAS 183
KGD+ S++SKET + + V++ ++ + D + PESV K ++ S
Sbjct: 3983 -KGDQSSLASKETSRPDSVVESVKDETEKPEGSAIDKSQVASRPESVAVSAKDEKSPLHS 4041
Query: 184 EAPSVPQKKTKPSPTTPKS 202
SV K S +S
Sbjct: 4042 RPESVADKSPDASKEASRS 4060
Score = 41.6 bits (96), Expect = 0.006
Identities = 62/329 (18%), Positives = 125/329 (37%), Gaps = 45/329 (13%)
Query: 8 SKSTSSKTEKSLSQSRKKA---RTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAP 64
++S + EKS +SR+++ ++P S+ V+ S DE E+ ++
Sbjct: 3695 AESVKDEAEKSKEESRRESVAEKSPLPSKEASRPTSVAES-------VKDEAEKSKEESR 3747
Query: 65 ANINEEETLMGSDRDVVPDAEASAPQDT-----------NVSQENPIPENPDVEDVIAEE 113
E++ + S + P + A + +D +V++++P+ E
Sbjct: 3748 RESVAEKSSLASKKASRPASVAESVKDEAEKSKEESRRESVAEKSPLASKEASRPASVAE 3807
Query: 114 TVSTETPSSSKKGDKGSVSSKET----HSDEPIQVLVQNNSDGDDSDDDD---------- 159
+V E S ++ + SV+ K + P V + D S ++
Sbjct: 3808 SVKDEAEKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEADKSKEESRRESGAEKSP 3867
Query: 160 -VPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKS 218
+ + P SV +K + + E+ + P P+ S+ + K AEKS
Sbjct: 3868 LASMEASRPTSVAESVKDETEKSKEESRRESVTEKSPLPSKEASRPTSVAESVKDEAEKS 3927
Query: 219 MSEKKK-----KAPVIVSESD--SDVEADVPDIVLAEMKKFAGRRIPKNVPAAPL--DNV 269
E ++ K+P+ ES + V + D ++ +P++ A + D
Sbjct: 3928 KEESRRESVAEKSPLASKESSRPASVAESIKDEAEGTKQESRRESMPESGKAESIKGDQS 3987
Query: 270 SFHSEENALKWKYVYQRRLATEREVGSDV 298
S S+E + V + TE+ GS +
Sbjct: 3988 SLASKETSRPDSVVESVKDETEKPEGSAI 4016
Score = 39.7 bits (91), Expect = 0.022
Identities = 53/276 (19%), Positives = 109/276 (39%), Gaps = 37/276 (13%)
Query: 8 SKSTSSKTEKSLSQSRKKA---RTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAP 64
++S + EKS +SR+++ ++P S+ V+ S ++D +EE ++
Sbjct: 2067 AESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAES---IKDEAEKSKEESRRESV 2123
Query: 65 ANIN----EEETLMGSDRDVVPDAEASAPQDT---NVSQENPIPENPDVEDVIAEETVST 117
A + +E + S + + D + +++ +V++++P+P E++
Sbjct: 2124 AEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKD 2183
Query: 118 ETPSSSKKGDKGSVSSKET----HSDEPIQVLVQNNSDGDDSDDDD----------VPLF 163
E S ++ + SV+ K + P V + + S ++ +P
Sbjct: 2184 EAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEETRRESVAEKSPLPSK 2243
Query: 164 E-TLPESVGARLK--------RKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKAT 214
E + P SV +K RR +A+E +P K+ P + KD K K
Sbjct: 2244 EASRPASVAESIKDEAEKSKEESRRESAAEKSPLPSKEAS-RPASVAESVKDEADKSKEE 2302
Query: 215 AEKSMSEKKKKAPVIVSESDSDVEADVPDIVLAEMK 250
+ + + KA I + E P+ V +K
Sbjct: 2303 SRRESMAESGKAQSIKGDQSPLKEVSRPESVAESVK 2338
Score = 38.9 bits (89), Expect = 0.037
Identities = 55/273 (20%), Positives = 106/273 (38%), Gaps = 30/273 (10%)
Query: 6 GSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVL--LEDVTSDEEEEPFHDA 63
GS K S E S +S K +R+ + S S+ L++++ E +A
Sbjct: 1875 GSMKDESMSKEPSRRESVKDGAAQSRETSRPASVAESAKDGADDLKELSRPESTTQSKEA 1934
Query: 64 PANINEEETLMGSDRDVVPDAEASAPQDT-----------NVSQENPIPENPDVEDVIAE 112
+I +E++ + S+ P + A + +D +V++++P+P
Sbjct: 1935 -GSIKDEKSPLASEEASRPASVAESVKDEAEKSKEESRRESVAEKSPLPSKEASRPASVA 1993
Query: 113 ETVSTETPSSSKKGDKGSVSSKET----HSDEPIQVLVQNNSDGDDSDDDD--------- 159
E++ E S ++ + SV+ K + P V + + S ++
Sbjct: 1994 ESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKS 2053
Query: 160 -VPLFE-TLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEK 217
+P E + P SV +K + + E+ + P P+ S+ + K AEK
Sbjct: 2054 PLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEK 2113
Query: 218 SMSEKKKKAPVIVSESDSDVEADVPDIVLAEMK 250
S E ++++ S S EA P V +K
Sbjct: 2114 SKEESRRESVAEKSPLPSK-EASRPASVAESIK 2145
Score = 38.9 bits (89), Expect = 0.037
Identities = 53/268 (19%), Positives = 108/268 (39%), Gaps = 42/268 (15%)
Query: 8 SKSTSSKTEKSLSQSRKKA---RTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAP 64
++S + EKS +SR+++ ++P S+ V+ S ++D +EE ++
Sbjct: 3510 AESVQDEAEKSKEESRRESVAEKSPLASKEASRPASVAES---IKDEAEKSKEESRRESV 3566
Query: 65 AN----INEEETLMGSDRDVVPDAEASAPQDTN---VSQENPIPENPDVEDVIAEETVST 117
A ++E + S + V D + ++++ V++++P+ E+V
Sbjct: 3567 AEKSPLASKEASRPTSVAESVKDEAEKSKEESSRDSVAEKSPLASKEASRPASVAESVQD 3626
Query: 118 ETPSSSKKGDKGSVSSKET----HSDEPIQVLVQNNSDGDDSDDDD--------VPLFE- 164
E S ++ + SV+ K + P V D + S ++ PL
Sbjct: 3627 EAEKSKEESRRESVAEKSPLASKEASRPASVAESVKDDAEKSKEESRRESVAEKSPLASK 3686
Query: 165 --TLPESVGARLK--------RKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKAT 214
+ P SV +K RR + +E +P K+ + +S K + + + +
Sbjct: 3687 EASRPASVAESVKDEAEKSKEESRRESVAEKSPLPSKEASRPTSVAESVKDEAEKSKEES 3746
Query: 215 AEKSMSE------KKKKAPVIVSESDSD 236
+S++E KK P V+ES D
Sbjct: 3747 RRESVAEKSSLASKKASRPASVAESVKD 3774
Score = 38.1 bits (87), Expect = 0.063
Identities = 58/272 (21%), Positives = 100/272 (36%), Gaps = 32/272 (11%)
Query: 3 LKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHD 62
LK + + + ++++KS SQ K+ P K +S E V +
Sbjct: 1845 LKGSTRRESVAESDKS-SQPFKETSRPESAVGSMKDESMSKEPSRRESVKDGAAQSRETS 1903
Query: 63 APANINEEETLMGSDRDVVPDA-EASAPQDTNVSQE--------NPIPENPDVEDVIAEE 113
PA++ E S +D D E S P+ T S+E +P+ E
Sbjct: 1904 RPASVAE------SAKDGADDLKELSRPESTTQSKEAGSIKDEKSPLASEEASRPASVAE 1957
Query: 114 TVSTETPSSSKKGDKGSVSSKET----HSDEPIQVLVQNNSDGDDSDDDD---------- 159
+V E S ++ + SV+ K + P V + + S ++
Sbjct: 1958 SVKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSP 2017
Query: 160 VPLFE-TLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKS 218
+P E + P SV +K + + E+ + P P+ S+ + K AEKS
Sbjct: 2018 LPSKEASRPASVAESIKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESIKDEAEKS 2077
Query: 219 MSEKKKKAPVIVSESDSDVEADVPDIVLAEMK 250
E ++++ S S EA P V +K
Sbjct: 2078 KEESRRESVAEKSPLPSK-EASRPASVAESIK 2108
Score = 37.0 bits (84), Expect = 0.14
Identities = 60/267 (22%), Positives = 103/267 (38%), Gaps = 53/267 (19%)
Query: 4 KSGSSKSTSS------KTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEE 57
K+GS K S + EKS +SR+++ + + S VS + E V DE E
Sbjct: 3287 KAGSIKDEKSPLASKDEAEKSKEESRRES---VAEQFPLVSKEVSRPASVAESV-KDEAE 3342
Query: 58 EPFHDAPANINEEETLMGSDRDVVPDAEASAPQDT--NVSQENPIPENPDVEDVIAEETV 115
+ ++P E V +AE S + +V++++P+P E+V
Sbjct: 3343 KSKEESPLMSKEASRPASVAGSVKDEAEKSKEESRRESVAEKSPLPSKEASRPASVAESV 3402
Query: 116 STETPSSSKKG------DKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPES 169
E S ++ +K ++SKE + P V + + S ++
Sbjct: 3403 KDEADKSKEESRRESGAEKSPLASKE--ASRPASVAESIKDEAEKSKEES---------- 3450
Query: 170 VGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKK----- 224
RR + +E +P K+ PT+ KD AEKS E ++
Sbjct: 3451 --------RRESVAEKSPLPSKEAS-RPTSVAESVKD-------EAEKSKEESRRDSVAE 3494
Query: 225 KAPVIVSESDSDVEADVPDIVLAEMKK 251
K+P+ E+ A V + V E +K
Sbjct: 3495 KSPLASKEASR--PASVAESVQDEAEK 3519
Score = 33.9 bits (76), Expect = 1.2
Identities = 38/219 (17%), Positives = 81/219 (36%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
S + S K EKSL S++ +R + + S E VT ++ A
Sbjct: 3199 SVADSIKDEKSLLVSQEASRPESEAESLKDAAAPSQETSRPESVTESVKDGKSPVASKEA 3258
Query: 68 NEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGD 127
+ ++ + +D +++ P+ S+ I + E S E +
Sbjct: 3259 SRPASVAENAKDSADESKEQRPESLPQSKAGSIKDEKSPLASKDEAEKSKEESRRESVAE 3318
Query: 128 KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPS 187
+ + SKE + V++ ++ + + + P SV +K + + E+
Sbjct: 3319 QFPLVSKEVSRPASVAESVKDEAEKSKEESPLMSKEASRPASVAGSVKDEAEKSKEESRR 3378
Query: 188 VPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKA 226
+ P P+ S+ + K A+KS E ++++
Sbjct: 3379 ESVAEKSPLPSKEASRPASVAESVKDEADKSKEESRRES 3417
Score = 31.2 bits (69), Expect = 7.7
Identities = 42/236 (17%), Positives = 100/236 (41%), Gaps = 9/236 (3%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
S++T +T S +S+ + + K + K V +S E +D E+ + +
Sbjct: 4189 SETTLFETLTSKVESKVEVLESSVKQVEEK---VQTSVKQAETTVTDSLEQLTKKSSEQL 4245
Query: 68 NEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGD 127
E ++++ ++ + V A + S ++ PD ++ EE + + T + ++ D
Sbjct: 4246 TEIKSVLDTNFEEVAKIVADVAKVLK-SDKDITDIIPDFDERQLEEKLKS-TADTEEESD 4303
Query: 128 KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTASEAPS 187
K + K +++ + +S S + + + +S A+L++ T++
Sbjct: 4304 KSTRDEKSLEISVKVEIESEKSSPDQKSGPISIEEKDKIEQSEKAQLRQGILTSSRPESV 4363
Query: 188 VPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSE-SDSDVEADVP 242
Q ++ PSP+ + + K +E +EK + + S+ S+ D++ P
Sbjct: 4364 ASQPESVPSPSQSAASHEH---KEVELSESHKAEKSSRPESVASQVSEKDMKTSRP 4416
>NSR1_YEAST (P27476) Nuclear localization sequence binding protein
(P67)
Length = 414
Score = 47.4 bits (111), Expect = 1e-04
Identities = 34/152 (22%), Positives = 63/152 (41%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
+K+T K K ++ K+A+ K S S SSS + S+ E E + ++
Sbjct: 2 AKTTKVKGNKKEVKASKQAKEEKAKAVSSSSSESSSSSSSSSESESESESESESSSSSSS 61
Query: 68 NEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKGD 127
++ E+ S D +AE + + S + + + E+ EET E+ SS
Sbjct: 62 SDSESSSSSSSDSESEAETKKEESKDSSSSSSDSSSDEEEEEEKEETKKEESKESSSSDS 121
Query: 128 KGSVSSKETHSDEPIQVLVQNNSDGDDSDDDD 159
S SS E + + D ++ +D++
Sbjct: 122 SSSSSSDSESEKEESNDKKRKSEDAEEEEDEE 153
Score = 42.4 bits (98), Expect = 0.003
Identities = 52/177 (29%), Positives = 75/177 (41%), Gaps = 24/177 (13%)
Query: 7 SSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPAN 66
SS S+SS +E S S S K +SK SSS D +SDEEEE +
Sbjct: 56 SSSSSSSDSESSSSSSSDSESEAETKKEESKDSSSSSS-----DSSSDEEEE---EEKEE 107
Query: 67 INEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSKKG 126
+EE+ S D+ +S+ D+ +E + ED EE + SS+KK
Sbjct: 108 TKKEESKESSS----SDSSSSSSSDSESEKEESNDKKRKSEDAEEEE----DEESSNKK- 158
Query: 127 DKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTAS 183
K ++EP + V S D D+ FE + +GAR+ +R T S
Sbjct: 159 ------QKNEETEEPATIFVGRLSWSID-DEWLKKEFEHIGGVIGARVIYERGTDRS 208
Score = 37.7 bits (86), Expect = 0.082
Identities = 26/130 (20%), Positives = 55/130 (42%)
Query: 111 AEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESV 170
A+E + SSS + S SS E+ S+ + ++S DS+ ++ E+
Sbjct: 20 AKEEKAKAVSSSSSESSSSSSSSSESESESESESESSSSSSSSDSESSSSSSSDSESEAE 79
Query: 171 GARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIV 230
+ + K +++S S +++ + T K + K+ +++ S SE +K+
Sbjct: 80 TKKEESKDSSSSSSDSSSDEEEEEEKEETKKEESKESSSSDSSSSSSSDSESEKEESNDK 139
Query: 231 SESDSDVEAD 240
D E +
Sbjct: 140 KRKSEDAEEE 149
Score = 35.8 bits (81), Expect = 0.31
Identities = 23/114 (20%), Positives = 48/114 (41%)
Query: 111 AEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESV 170
+E +E+ SSS D S SS + S+ + + + D S D E E
Sbjct: 47 SESESESESSSSSSSSDSESSSSSSSDSESEAETKKEESKDSSSSSSDSSSDEEEEEEKE 106
Query: 171 GARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKK 224
+ + + +++S++ S ++ KK+ + + E+S ++K+K
Sbjct: 107 ETKKEESKESSSSDSSSSSSSDSESEKEESNDKKRKSEDAEEEEDEESSNKKQK 160
>DMP1_MOUSE (O55188) Dentin matrix acidic phosphoprotein 1 precursor
(Dentin matrix protein-1) (DMP-1) (AG1)
Length = 503
Score = 47.4 bits (111), Expect = 1e-04
Identities = 39/234 (16%), Positives = 85/234 (35%), Gaps = 18/234 (7%)
Query: 2 GLKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFH 61
G++S SK T S + +RK ++ S EE +
Sbjct: 231 GIRSEESKGDREPTSTQDSDDSQSVEFSSRKSFRR----------------SRVSEEDYR 274
Query: 62 DAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPS 121
+ N ET S D E+ + + ++ ++P+ +D +E + PS
Sbjct: 275 GELTDSNSRETQSDSTEDTASKEESRSESQEDTAESQSQEDSPEGQDPSSESSEEAGEPS 334
Query: 122 SSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTT 181
+ + E+ D P D +DS+ + T S + + +
Sbjct: 335 QESSSESQEGVTSESRGDNPDN--TSQTGDQEDSESSEEDSLNTFSSSESQSTEEQADSE 392
Query: 182 ASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSESDS 235
++E+ S+ ++ + + S ++ L+ + +T +S + ++ +SDS
Sbjct: 393 SNESLSLSEESQESAQDGDSSSQEGLQSQSASTESRSQESQSEQDSRSEEDSDS 446
Score = 37.7 bits (86), Expect = 0.082
Identities = 49/248 (19%), Positives = 93/248 (36%), Gaps = 30/248 (12%)
Query: 4 KSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDA 63
++ S + SS++ + L R + R PA KS G +D D ++ F D
Sbjct: 60 QANSDHTHSSESGEELGYDRGQYR-PAGGLSKSTGTGADK-----DDDEDDSGDDTFGDE 113
Query: 64 PANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSS 123
+ EE G + D +++ DT S E D ++E + +TPS S
Sbjct: 114 DNGLGPEEGQWGGPSKLDSDEDSA---DTTQSSE----------DSTSQENSAQDTPSDS 160
Query: 124 KKGD-------KGSVSSKETHSDEPIQVL----VQNNSDGDDSDDDDVPLFETLPESVGA 172
K D + HS+ Q + +S GD S+ DD + PES +
Sbjct: 161 KDQDSEDDAHSRPDAGDSAQHSESEEQRVGGGSEGQSSHGDGSEFDDEGMQSDDPESTRS 220
Query: 173 RLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKKKKAPVIVSE 232
R ++A + +P+ T + ++ + + +S ++ +
Sbjct: 221 DRGHARMSSAGIRSEESKGDREPTSTQDSDDSQSVEFSSRKSFRRSRVSEEDYRGELTDS 280
Query: 233 SDSDVEAD 240
+ + ++D
Sbjct: 281 NSRETQSD 288
Score = 35.0 bits (79), Expect = 0.53
Identities = 30/159 (18%), Positives = 67/159 (41%), Gaps = 10/159 (6%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
S+ S + + S+S ++A P+++ GV+S D + +
Sbjct: 312 SQEDSPEGQDPSSESSEEAGEPSQESSSESQEGVTSES---RGDNPDNTSQTGDQEDSES 368
Query: 68 NEEETLMGSDRDVVPDAEASAPQDTNVS-------QENPIPENPDVEDVIAEETVSTETP 120
+EE++L E A ++N S QE+ + ++ + ++ STE+
Sbjct: 369 SEEDSLNTFSSSESQSTEEQADSESNESLSLSEESQESAQDGDSSSQEGLQSQSASTESR 428
Query: 121 SSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDD 159
S + ++ S S +++ S + + ++NS G S ++
Sbjct: 429 SQESQSEQDSRSEEDSDSQDSSRSKEESNSTGSASSSEE 467
Score = 35.0 bits (79), Expect = 0.53
Identities = 36/165 (21%), Positives = 65/165 (38%), Gaps = 14/165 (8%)
Query: 8 SKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANI 67
S+ +SS++++ ++ + + SS L +S E + A +
Sbjct: 334 SQESSSESQEGVTSESRGDNPDNTSQTGDQEDSESSEEDSLNTFSSSESQSTEEQADSES 393
Query: 68 NEEETLMGSDRDVVPDAEASAP---QDTNVSQENPIPENPDVEDVIAEETVSTETPSSSK 124
NE +L ++ D ++S+ Q + S E+ E+ +D +EE ++ S SK
Sbjct: 394 NESLSLSEESQESAQDGDSSSQEGLQSQSASTESRSQESQSEQDSRSEEDSDSQDSSRSK 453
Query: 125 K--GDKGSVSSKE---------THSDEPIQVLVQNNSDGDDSDDD 158
+ GS SS E S + I N GD D+D
Sbjct: 454 EESNSTGSASSSEEDIRPKNMEADSRKLIVDAYHNKPIGDQDDND 498
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 46.6 bits (109), Expect = 2e-04
Identities = 51/234 (21%), Positives = 94/234 (39%), Gaps = 18/234 (7%)
Query: 5 SGSSKSTSSKTEKSLSQSRKKARTPAR------------KYYKSKSHGVSSSHVLLEDVT 52
SGSS S+SS +++S S+S + T R K K++ SSS +
Sbjct: 103 SGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKETKKAKTEPESSSSSESSSSGSS 162
Query: 53 SDEEEEPFHDAPANINEEETLMGSDRDVVPDAEA-SAPQDTNVSQENPIPENPDVEDVIA 111
S E E ++ ++ + + S D D+E+ S ++ S ++ + D +
Sbjct: 163 SSSESESGSESDSDSSSSSS---SSSDSESDSESDSQSSSSSSSSDSSSDSDSSSSDSSS 219
Query: 112 EETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVG 171
+ S+ + SSS D S SS ++ S ++S D+S D ++ +S
Sbjct: 220 DSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSDSDSDSDSGS 279
Query: 172 ARLKRKRRTTA--SEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEKK 223
+ + TA S+A P + +P+ S + T E ++K
Sbjct: 280 SSELETKEATADESKAEETPASSNESTPSASSSSSANKLNIPAGTDEIKEGQRK 333
Score = 46.2 bits (108), Expect = 2e-04
Identities = 53/242 (21%), Positives = 95/242 (38%), Gaps = 11/242 (4%)
Query: 5 SGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFHDAP 64
S SS S+SS E S S S + + + S S SSS +S + E ++
Sbjct: 40 SSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSSSSSSSSSSSSSSDSESSSESD 99
Query: 65 ANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPSSSK 124
++ + + S D S + ++E+ +N D ++ +T E+ SSS+
Sbjct: 100 SSSSGSSSSSSSSSDESSSESESEDETKKRARES---DNEDAKETKKAKT-EPESSSSSE 155
Query: 125 KGDKGSVSSKETHS-DEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLKRKRRTTAS 183
GS SS E+ S E ++S DS+ D ++ S + +++S
Sbjct: 156 SSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSSS 215
Query: 184 EAPSVPQKKTKPSPTTPKS-----KKKDLKGKGKATAEKSMSEKKKKAPVIVSESDSDVE 238
++ S + S ++ S D G + + S S+ SDSD +
Sbjct: 216 DSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSS-SDSSSDESTSSDSSDSDSD 274
Query: 239 AD 240
+D
Sbjct: 275 SD 276
Score = 41.6 bits (96), Expect = 0.006
Identities = 39/208 (18%), Positives = 86/208 (40%), Gaps = 11/208 (5%)
Query: 35 KSKSHGVSSSHVLLEDVTSDEEEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNV 94
KS S SSS +S ++ ++ + + SD D+E+S+ ++
Sbjct: 24 KSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSSSSSSS 83
Query: 95 SQENPIPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDD 154
S + ++ + E+ S+ + SSS SS E+ S++ + + + D+
Sbjct: 84 SSSSSSSDSES-----SSESDSSSSGSSSSSSSSSDESSSESESEDETK---KRARESDN 135
Query: 155 SDDDDVPLFETLPESVGARLKRKRRTTA---SEAPSVPQKKTKPSPTTPKSKKKDLKGKG 211
D + +T PES + +++ SE+ S + S ++ + D +
Sbjct: 136 EDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDS 195
Query: 212 KATAEKSMSEKKKKAPVIVSESDSDVEA 239
++++ S S+ + S+S SD ++
Sbjct: 196 QSSSSSSSSDSSSDSDSSSSDSSSDSDS 223
Score = 38.5 bits (88), Expect = 0.048
Identities = 38/188 (20%), Positives = 73/188 (38%), Gaps = 13/188 (6%)
Query: 56 EEEPFHDAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETV 115
+E+ + ++ + + S + +S+ + ++ S + + D D E+
Sbjct: 17 KEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESS 76
Query: 116 STETPSSSKKGDKGSVSSKETHSDEPIQVLVQNNSDGDDSDDDDVPLFETLPESVGARLK 175
S+ + SSS S S E+ S+ ++S SD E+ ES
Sbjct: 77 SSSSSSSS---SSSSSSDSESSSESDSSSSGSSSSSSSSSD-------ESSSESESEDET 126
Query: 176 RKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATAEKSMSEK---KKKAPVIVSE 232
+KR + + KK K P + S + G ++ +S SE + S+
Sbjct: 127 KKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSD 186
Query: 233 SDSDVEAD 240
S+SD E+D
Sbjct: 187 SESDSESD 194
Score = 37.0 bits (84), Expect = 0.14
Identities = 31/135 (22%), Positives = 60/135 (43%), Gaps = 9/135 (6%)
Query: 2 GLKSGSSKSTSSKTEKSLSQSRKKARTPARKYYKSKSHGVSSSHVLLEDVTSDEEEEPFH 61
G S S + S+++ S S + + + S+S SSS D +SD +
Sbjct: 160 GSSSSSESESGSESD---SDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSS-S 215
Query: 62 DAPANINEEETLMGSDRDVVPDAEASAPQDTNVSQENPIPENPDVEDVIAEETVSTETPS 121
D+ ++ + + S D D+++S+ D++ S ++ D ++E+ S+++
Sbjct: 216 DSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSS-----DSSSSSDSSSDESTSSDSSD 270
Query: 122 SSKKGDKGSVSSKET 136
S D GS S ET
Sbjct: 271 SDSDSDSGSSSELET 285
Score = 35.0 bits (79), Expect = 0.53
Identities = 30/145 (20%), Positives = 59/145 (40%), Gaps = 4/145 (2%)
Query: 100 IPENPDVEDVIAEETVSTETPSSSKKGDKGSVSSKETHSDE----PIQVLVQNNSDGDDS 155
+P+ E I E++ S+ + SSS S SS + S E ++SD DS
Sbjct: 11 VPKLSVKEKEIEEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDS 70
Query: 156 DDDDVPLFETLPESVGARLKRKRRTTASEAPSVPQKKTKPSPTTPKSKKKDLKGKGKATA 215
D + + S + ++ S++ S + S + S + + + + K A
Sbjct: 71 SDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRA 130
Query: 216 EKSMSEKKKKAPVIVSESDSDVEAD 240
+S +E K+ +E +S ++
Sbjct: 131 RESDNEDAKETKKAKTEPESSSSSE 155
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.309 0.127 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,487,648
Number of Sequences: 164201
Number of extensions: 3024878
Number of successful extensions: 13995
Number of sequences better than 10.0: 520
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 474
Number of HSP's that attempted gapping in prelim test: 12080
Number of HSP's gapped (non-prelim): 1532
length of query: 577
length of database: 59,974,054
effective HSP length: 116
effective length of query: 461
effective length of database: 40,926,738
effective search space: 18867226218
effective search space used: 18867226218
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0280b.3


