

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0216.15
(500 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
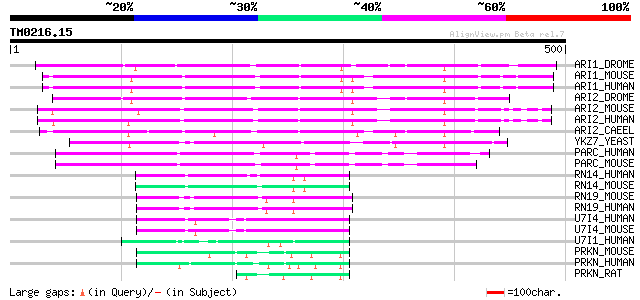
Score E
Sequences producing significant alignments: (bits) Value
ARI1_DROME (Q94981) Ariadne-1 protein (Ari-1) 183 1e-45
ARI1_MOUSE (Q9Z1K5) Ariadne-1 protein homolog (ARI-1) (Ubiquitin... 176 2e-43
ARI1_HUMAN (Q9Y4X5) Ariadne-1 protein homolog (ARI-1) (Ubiquitin... 176 2e-43
ARI2_DROME (O76924) Ariadne-2 protein (Ari-2) 170 9e-42
ARI2_MOUSE (Q9Z1K6) Ariadne-2 protein homolog (ARI-2) (Triad1 pr... 157 5e-38
ARI2_HUMAN (O95376) Ariadne-2 protein homolog (ARI-2) (Triad1 pr... 157 5e-38
ARI2_CAEEL (Q22431) Probable ariadne-2 protein (Ari-2) 154 4e-37
YKZ7_YEAST (P36113) Hypothetical 63.6 kDa protein in YPT52-GCN3 ... 134 4e-31
PARC_HUMAN (Q8IWT3) p53-associated parkin-like cytoplasmic prote... 132 2e-30
PARC_MOUSE (Q80TT8) p53-associated parkin-like cytoplasmic protein 130 6e-30
RN14_HUMAN (Q9UBS8) RING finger protein 14 (Androgen receptor-as... 93 2e-18
RN14_MOUSE (Q9JI90) RING finger protein 14 (Androgen receptor-as... 92 4e-18
RN19_MOUSE (P50636) RING finger protein 19 (XY body protein) (XY... 84 6e-16
RN19_HUMAN (Q9NV58) RING finger protein 19 (Dorfin) (Double ring... 84 6e-16
U7I4_HUMAN (P50876) Ubiquitin conjugating enzyme 7 interacting p... 68 5e-11
U7I4_MOUSE (Q925F3) Ubiquitin conjugating enzyme 7 interacting p... 65 3e-10
U7I1_HUMAN (Q9NWF9) Ubiquitin conjugating enzyme 7 interacting p... 59 2e-08
PRKN_MOUSE (Q9WVS6) Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) 59 3e-08
PRKN_HUMAN (O60260) Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRK... 57 8e-08
PRKN_RAT (Q9JK66) Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN) 56 2e-07
>ARI1_DROME (Q94981) Ariadne-1 protein (Ari-1)
Length = 503
Score = 183 bits (464), Expect = 1e-45
Identities = 135/485 (27%), Positives = 222/485 (44%), Gaps = 37/485 (7%)
Query: 24 NLPNTEEEEEDKRE-NFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTK 82
+LP++ + + D+ + + +L+ EI + Q + I++ +L +P + LL H W+ K
Sbjct: 33 DLPSSADRQMDQDDYQYKVLTTDEIVQHQREIIDEANLLLKLPTPTTRILLNHFKWDKEK 92
Query: 83 VLESWCEDE-EKVQKAAGLLNQPKVEIDWLK--------DLCQVCFRSSEKGKILSAGCA 133
+LE + +D ++ K A ++N P + +K + C++CF + C
Sbjct: 93 LLEKYFDDNTDEFFKCAHVIN-PFNATEAIKQKTSRSQCEECEICFSQLPPDSMAGLECG 151
Query: 134 HPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESS-KVMYEQFLL 192
H C CW +Y+ T I T+ C CD+ VD + L +++ +V Y+Q +
Sbjct: 152 HRFCMPCWHEYLSTKI-VAEGLGQTISCAAHGCDILVDDVTVANLVTDARVRVKYQQLIT 210
Query: 193 WSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCET 252
S+V+ +RWCP+ C Y V A V C C H FC+ CGE H PV C
Sbjct: 211 NSFVECNQLLRWCPSVDCTYAVKVP----YAEPRRVHCKCGHVFCFACGENWHDPVKCRW 266
Query: 253 AGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCK---CGFQFCWVCFHE 309
+W+KK D SE WI A+TK CPRC IEK+ G +M CK C +FCWVC
Sbjct: 267 LKKWIKKCD-DDSETSNWIAANTKECPRCSVTIEKDGGCNHMVCKNQNCKNEFCWVCLG- 324
Query: 310 QLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQK 369
E S + E + + + + + + + HYY + N S K
Sbjct: 325 --SWEPHGSSWYNCNRYDEDEAKTARDAQEKL-RSSLARYLHYYNRY-MNHMQSMKFENK 380
Query: 370 LIDLDRKYLSMSLQKRETYFD--FVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFS 427
L ++ + Q ++ + F+KKA + +CR+ L +T V+ Y+L KN ++
Sbjct: 381 LYASVKQKMEEMQQHNMSWIEVQFLKKAVDILCQCRQTLMYTYVFAYYLKKNNQS--MIF 438
Query: 428 EYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFE 487
E Q + E A E L + + + T E+L ++KV + LL+
Sbjct: 439 EDNQKDLESATEMLSEYLERDIT-------SENLADIKQKVQDKYRYCEKRCSVLLKHVH 491
Query: 488 DGLDK 492
+G DK
Sbjct: 492 EGYDK 496
>ARI1_MOUSE (Q9Z1K5) Ariadne-1 protein homolog (ARI-1)
(Ubiquitin-conjugating enzyme E2-binding protein 1)
(UbcH7-binding protein) (UbcM4-interacting protein 77)
Length = 555
Score = 176 bits (445), Expect = 2e-43
Identities = 130/480 (27%), Positives = 216/480 (44%), Gaps = 41/480 (8%)
Query: 30 EEEEDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCE 89
E+EED R + +L+ +I + + I +V V+ P LL H W+ K++E + +
Sbjct: 92 EQEEDYR--YEVLTAEQILQHMVECIREVNEVIQNPATITRILLSHFNWDKEKLMERYFD 149
Query: 90 DE-EKVQKAAGLLNQPKVEI-------DWLKDL-CQVCFRSSEKGKILSAGCAHPLCEAC 140
EK+ ++N K +D+ CQ+C+ + C H C C
Sbjct: 150 GNLEKLFAECHVINPSKKSRTRQMNTRSSAQDMPCQICYLNYPNSYFTGLECGHKFCMQC 209
Query: 141 WMDYIDTNI-NKGPSKCLTLRCPDPSCDVAVDRDMIRELASESS-KVMYEQFLLWSYVDS 198
W +Y+ T I +G + T+ CP CD+ VD + + L ++S K+ Y+ + S+V+
Sbjct: 210 WSEYLTTKIMEEGMGQ--TISCPAHGCDILVDDNTVMRLITDSKVKLKYQHLITNSFVEC 267
Query: 199 KAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMK 258
++WCPAP C +VV Q V C C FC+ CGE H PV C+ +W+K
Sbjct: 268 NRLLKWCPAPDCHHVVKVQYPDA----KPVRCKCGRQFCFNCGENWHDPVKCKWLKKWIK 323
Query: 259 KIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCK---CGFQFCWVCF--HEQLKC 313
K D SE WI A+TK CP+C IEK+ G +M C+ C +FCWVC E
Sbjct: 324 KCD-DDSETSNWIAANTKECPKCHVTIEKDGGCNHMVCRNQNCKAEFCWVCLGPWEPHGS 382
Query: 314 ESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYEC-WARNDFLSKSILQKLID 372
+C+ + D + E+ + + C N S KL
Sbjct: 383 AWYNCN--------RYNEDDAKAARDAQERSRAALQRYLFYCNRYMNHMQSLRFEHKLYA 434
Query: 373 LDRKYLSMSLQKRETYFD--FVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYI 430
++ + Q ++ + F+KKA + +CR L +T V+ ++L KN ++ E
Sbjct: 435 QVKQKMEEMQQHNMSWIEVQFLKKAVDVLCQCRATLMYTYVFAFYLKKNNQS--IIFENN 492
Query: 431 QVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGL 490
Q + E A E L +E + QD+ ++ K Q+K + ++ +++ +E L
Sbjct: 493 QADLENATEVLS--GYLERDISQDSL-QDIKQKVQDKYRYCESRRRVLLQHVHEGYEKDL 549
>ARI1_HUMAN (Q9Y4X5) Ariadne-1 protein homolog (ARI-1)
(Ubiquitin-conjugating enzyme E2-binding protein 1)
(UbcH7-binding protein) (UbcM4-interacting protein)
(HHARI) (H7-AP2) (HUSSY-27) (MOP-6)
Length = 557
Score = 176 bits (445), Expect = 2e-43
Identities = 130/480 (27%), Positives = 216/480 (44%), Gaps = 41/480 (8%)
Query: 30 EEEEDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCE 89
E+EED R + +L+ +I + + I +V V+ P LL H W+ K++E + +
Sbjct: 94 EQEEDYR--YEVLTAEQILQHMVECIREVNEVIQNPATITRILLSHFNWDKEKLMERYFD 151
Query: 90 DE-EKVQKAAGLLNQPKVEI-------DWLKDL-CQVCFRSSEKGKILSAGCAHPLCEAC 140
EK+ ++N K +D+ CQ+C+ + C H C C
Sbjct: 152 GNLEKLFAECHVINPSKKSRTRQMNTRSSAQDMPCQICYLNYPNSYFTGLECGHKFCMQC 211
Query: 141 WMDYIDTNI-NKGPSKCLTLRCPDPSCDVAVDRDMIRELASESS-KVMYEQFLLWSYVDS 198
W +Y+ T I +G + T+ CP CD+ VD + + L ++S K+ Y+ + S+V+
Sbjct: 212 WSEYLTTKIMEEGMGQ--TISCPAHGCDILVDDNTVMRLITDSKVKLKYQHLITNSFVEC 269
Query: 199 KAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHSPVDCETAGRWMK 258
++WCPAP C +VV Q V C C FC+ CGE H PV C+ +W+K
Sbjct: 270 NRLLKWCPAPDCHHVVKVQYPDA----KPVRCKCGRQFCFNCGENWHDPVKCKWLKKWIK 325
Query: 259 KIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTCK---CGFQFCWVCF--HEQLKC 313
K D SE WI A+TK CP+C IEK+ G +M C+ C +FCWVC E
Sbjct: 326 KCD-DDSETSNWIAANTKECPKCHVTIEKDGGCNHMVCRNQNCKAEFCWVCLGPWEPHGS 384
Query: 314 ESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYEC-WARNDFLSKSILQKLID 372
+C+ + D + E+ + + C N S KL
Sbjct: 385 AWYNCN--------RYNEDDAKAARDAQERSRAALQRYLFYCNRYMNHMQSLRFEHKLYA 436
Query: 373 LDRKYLSMSLQKRETYFD--FVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYI 430
++ + Q ++ + F+KKA + +CR L +T V+ ++L KN ++ E
Sbjct: 437 QVKQKMEEMQQHNMSWIEVQFLKKAVDVLCQCRATLMYTYVFAFYLKKNNQS--IIFENN 494
Query: 431 QVEAEVALEKLQQCAKVEETMFQDNYPEESLNKFQEKVIHLTDVTKIYFENLLRAFEDGL 490
Q + E A E L +E + QD+ ++ K Q+K + ++ +++ +E L
Sbjct: 495 QADLENATEVLS--GYLERDISQDSL-QDIKQKVQDKYRYCESRRRVLLQHVHEGYEKDL 551
>ARI2_DROME (O76924) Ariadne-2 protein (Ari-2)
Length = 509
Score = 170 bits (430), Expect = 9e-42
Identities = 123/455 (27%), Positives = 194/455 (42%), Gaps = 64/455 (14%)
Query: 39 FTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAA 98
+ L+ +I++L + + K+ ++L I + A LL + WN V+E + +D + A
Sbjct: 47 YECLTVEDIEKLLNERVEKLNTILQITPSLAKVLLLEHQWNNVAVVEKYRQDANALLVTA 106
Query: 99 GLLNQPKVEI-------------------------------DWLKDLCQVCFRSSEKGKI 127
+ P V + + +C VC S K
Sbjct: 107 RI-KPPSVAVTDTASTSAAAASAQLLRLGSSGYKTTASATPQYRSQMCPVCASSQLGDKF 165
Query: 128 LSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASES-SKVM 186
S C H C+ CW Y +T I +G S + C C+V V D++ L + +
Sbjct: 166 YSLACGHSFCKDCWTIYFETQIFQGIST--QIGCMAQMCNVRVPEDLVLTLVTRPVMRDK 223
Query: 187 YEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLCYHSFCWRCGEESHS 246
Y+QF YV S ++R+CP P C +V + + C+ FC+RCG + H+
Sbjct: 224 YQQFAFKDYVKSHPELRFCPGPNCQIIV---QSSEISAKRAICKACHTGFCFRCGMDYHA 280
Query: 247 PVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTC-KCGFQFCWV 305
P DC+ +W+ K SE +I AHTK CP+C IEKN G +M C C FCW+
Sbjct: 281 PTDCQVIKKWLTKC-ADDSETANYISAHTKDCPKCHICIEKNGGCNHMQCFNCKHDFCWM 339
Query: 306 CF-------HEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKDNPKKCNHYYECWAR 358
C E +C +P I + Q ++ KK HYYE W
Sbjct: 340 CLGDWKTHGSEYYECSRYKDNPNIANESVHVQA-----------REALKKYLHYYERW-E 387
Query: 359 NDFLSKSILQKLID-LDRKYLSMSLQKRETYFD--FVKKAWQQIAECRRILKWTNVYGYF 415
N S + Q+ ID L ++ S + T+ D ++ A +A+CR L++T Y Y+
Sbjct: 388 NHSKSLKLEQQTIDRLRQRINSKVMNGSGTWIDWQYLFNAAALLAKCRYTLQYTYPYAYY 447
Query: 416 LPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEET 450
+ ++K EY Q + E +E L + ET
Sbjct: 448 MEAG--SRKNLFEYQQAQLEAEIENLSWKIERAET 480
>ARI2_MOUSE (Q9Z1K6) Ariadne-2 protein homolog (ARI-2) (Triad1
protein) (UbcM4-interacting protein 48)
Length = 492
Score = 157 bits (398), Expect = 5e-38
Identities = 127/505 (25%), Positives = 209/505 (41%), Gaps = 74/505 (14%)
Query: 26 PNTEEEEEDKRE----------------------------NFTILSELEIKRLQEDAINK 57
PN EEEEE++ + FT L+ E + + +
Sbjct: 19 PNCEEEEEEEEDPGDIEDYYVGVASDVEQQGADAFDPEEYQFTCLTYKESEGALHEHMTS 78
Query: 58 VASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLLNQPKVEIDWLKDL--C 115
+ASVL + + A +L + W V+++L+ + + ++ A + P + C
Sbjct: 79 LASVLKVSHSVAKLILVNFHWQVSEILDRYRSNSAQLLVEARVQPNPSKHVPTAHPPHHC 138
Query: 116 QVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMI 175
VC + K +LS C H C +CW + + G + + C C + D +
Sbjct: 139 AVCMQFVRKENLLSLACQHQFCRSCWEQHCSVLVKDGVG--VGISCMAQDCPLRTPEDFV 196
Query: 176 RELA-SESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTC-LCY 233
L +E + Y ++L YV+S +++ CP C V+ Q R V C C
Sbjct: 197 FPLLPNEELRDKYRRYLFRDYVESHFQLQLCPGADCPMVIRVQEP----RARRVQCNRCS 252
Query: 234 HSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKY 293
FC++C + H+P DC T +W+ K SE +I AHTK CP+C IEKN G +
Sbjct: 253 EVFCFKCRQMYHAPTDCATIRKWLTKC-ADDSETANYISAHTKDCPKCNICIEKNGGCNH 311
Query: 294 MTC-KCGFQFCWVCF-------HEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKDN 345
M C KC FCW+C E +C +P I + Q ++
Sbjct: 312 MQCSKCKHDFCWMCLGDWKTHGSEYYECSRYKENPDIVNQSQQAQA-----------REA 360
Query: 346 PKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQKRETYFD--FVKKAWQQIAECR 403
KK Y+E W ++ + Q + K + T+ D +++ A + +A+CR
Sbjct: 361 LKKYLFYFERWENHNKSLQLEAQTYERIHEKIQERVMNNLGTWIDWQYLQNAAKLLAKCR 420
Query: 404 RILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLNK 463
L++T Y Y++ +K+ EY Q + E +E L KVE D+Y L
Sbjct: 421 YTLQYTYPYAYYMESG--PRKKLFEYQQAQLEAEIENLSW--KVERA---DSYDRGDL-- 471
Query: 464 FQEKVIHLTDVTKIYFENLLRAFED 488
E +H+ + + LL+ F D
Sbjct: 472 --ENQMHIAEQRR---RTLLKDFHD 491
>ARI2_HUMAN (O95376) Ariadne-2 protein homolog (ARI-2) (Triad1
protein) (HT005)
Length = 493
Score = 157 bits (398), Expect = 5e-38
Identities = 128/506 (25%), Positives = 210/506 (41%), Gaps = 75/506 (14%)
Query: 26 PNTEEEEEDKREN-----------------------------FTILSELEIKRLQEDAIN 56
PN EEEEE++ ++ FT L+ E + + +
Sbjct: 19 PNCEEEEEEEEDDPGDIEDYYVGVASDVEQQGADAFDPEEYQFTCLTYKESEGALNEHMT 78
Query: 57 KVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLLNQPK--VEIDWLKDL 114
+ASVL + + A +L + W V+++L+ + + ++ A + P V
Sbjct: 79 SLASVLKVSHSVAKLILVNFHWQVSEILDRYKSNSAQLLVEARVQPNPSKHVPTSHPPHH 138
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDM 174
C VC + K +LS C H C +CW + + G + + C C + D
Sbjct: 139 CAVCMQFVRKENLLSLACQHQFCRSCWEQHCSVLVKDGVG--VGVSCMAQDCPLRTPEDF 196
Query: 175 IRELA-SESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTC-LC 232
+ L +E + Y ++L YV+S +++ CP C V+ Q R V C C
Sbjct: 197 VFPLLPNEELREKYRRYLFRDYVESHYQLQLCPGADCPMVIRVQEP----RARRVQCNRC 252
Query: 233 YHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFK 292
FC++C + H+P DC T +W+ K SE +I AHTK CP+C IEKN G
Sbjct: 253 NEVFCFKCRQMYHAPTDCATIRKWLTKC-ADDSETANYISAHTKDCPKCNICIEKNGGCN 311
Query: 293 YMTC-KCGFQFCWVCF-------HEQLKCESRSCSPFIRMHIMEFQGTDGEEVEINMEKD 344
+M C KC FCW+C E +C +P I + Q ++
Sbjct: 312 HMQCSKCKHDFCWMCLGDWKTHGSEYYECSRYKENPDIVNQSQQAQA-----------RE 360
Query: 345 NPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQKRETYFD--FVKKAWQQIAEC 402
KK Y+E W ++ + Q + K + T+ D +++ A + +A+C
Sbjct: 361 ALKKYLFYFERWENHNKSLQLEAQTYQRIHEKIQERVMNNLGTWIDWQYLQNAAKLLAKC 420
Query: 403 RRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAKVEETMFQDNYPEESLN 462
R L++T Y Y++ +K+ EY Q + E +E L KVE D+Y L
Sbjct: 421 RYTLQYTYPYAYYMESG--PRKKLFEYQQAQLEAEIENLSW--KVERA---DSYDRGDL- 472
Query: 463 KFQEKVIHLTDVTKIYFENLLRAFED 488
E +H+ + + LL+ F D
Sbjct: 473 ---ENQMHIAEQRR---RTLLKDFHD 492
>ARI2_CAEEL (Q22431) Probable ariadne-2 protein (Ari-2)
Length = 482
Score = 154 bits (390), Expect = 4e-37
Identities = 117/432 (27%), Positives = 196/432 (45%), Gaps = 41/432 (9%)
Query: 28 TEEEEEDKRENFTILSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESW 87
T EE D + LS +++R+ D +N + S + I + A LL N W+V K+
Sbjct: 39 THSEEAD----YECLSVNQVERVFIDGVNSLVSRISINEKFARILLQANHWDVDKIARLV 94
Query: 88 CEDEEKVQKAAGLLNQPK-------VEIDWLKDLCQVCFRSSEKGKILSAGCAHPLCEAC 140
D + + +P+ + K C VC ++ C H CE C
Sbjct: 95 RNDRNDFLRKCHIDAKPEPKRKLSSTQSVLAKGYCSVCAMDGYT-ELPHLTCGHCFCEHC 153
Query: 141 WMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDMIRELASESS--KVMYEQFLLWSYVDS 198
W ++++ +++G + + C + C+V + + + S K+ YE+FLL V+S
Sbjct: 154 WKSHVESRLSEGVAS--RIECMESECEVYAPSEFVLSIIKNSPVIKLKYERFLLRDMVNS 211
Query: 199 KAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCL-CYHSFCWRCGEESHSPVDCETAGRWM 257
+++C C ++ + VTC+ C+ SFC +CG + H+P CET +WM
Sbjct: 212 HPHLKFCVGNECPVIIRSTE----VKPKRVTCMQCHTSFCVKCGADYHAPTSCETIKQWM 267
Query: 258 KKIDKSTSENGKWILAHTKPCPRCKSLIEKNQGFKYMTC-KCGFQFCWVCFHEQLK--CE 314
K SE +I AHTK CP+C S IEK G ++ C +C FCW+CF + E
Sbjct: 268 TKC-ADDSETANYISAHTKDCPQCHSCIEKAGGCNHIQCTRCRHHFCWMCFGDWKSHGSE 326
Query: 315 SRSCSPFIRMHIMEFQGTDGEEVEINMEKDNP--KKCNHYYECWARNDFLSKSILQKLID 372
CS ++ E N K +K HY+E + N S + ++L D
Sbjct: 327 YYECS--------RYKENPSVAAEANHVKARRALEKYLHYFERF-ENHSKSLKMEEELRD 377
Query: 373 LDRKYLSMSLQKRE-TYFD--FVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEY 429
RK + + + T+ D ++ K+ + +CR L++T + YFL + +K EY
Sbjct: 378 KIRKKIDDKVNEHNGTWIDWQYLHKSVSLLTKCRYTLQYTYPFAYFL--SATPRKNLFEY 435
Query: 430 IQVEAEVALEKL 441
Q + E +E+L
Sbjct: 436 QQAQLEKEVEEL 447
>YKZ7_YEAST (P36113) Hypothetical 63.6 kDa protein in YPT52-GCN3
intergenic region
Length = 551
Score = 134 bits (338), Expect = 4e-31
Identities = 105/422 (24%), Positives = 176/422 (40%), Gaps = 48/422 (11%)
Query: 55 INKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLLNQPKV-------- 106
++ + + IP A LL H WN ++LE W E +++ GL +
Sbjct: 107 VDHLQPIFAIPSADILILLQHYDWNEERLLEVWTEKMDELLVELGLSTTANIKKDNDYNS 166
Query: 107 ---EIDWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPD 163
E+++ D + + + + C H C C+ YI +++G + C D
Sbjct: 167 HFREVEFKNDFTCIICCDKKDTETFALECGHEYCINCYRHYIKDKLHEGN----IITCMD 222
Query: 164 PSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAK-MRWCPAPGCDYVVCYQGDGGC 222
C +A+ + I ++ S + S+V + +WCP C +V +
Sbjct: 223 --CSLALKNEDIDKVMGHPSSSKLMDSSIKSFVQKHNRNYKWCPFADCKSIVHLRDTSSL 280
Query: 223 ARDLD------VTCLCYHSFCWRCGEESHSPVDCETAGRWMKKIDKSTSENGKWILAHTK 276
V C +H FC+ CG E HSP DC+ W+KK K SE W+L+HTK
Sbjct: 281 PEYTRLHYSPFVKCNSFHRFCFNCGFEVHSPADCKITTAWVKKARKE-SEILNWVLSHTK 339
Query: 277 PCPRCKSLIEKNQGFKYMTC-KCGFQFCWVCFHEQLKCESRSCSPFIRMHIMEFQGTDGE 335
CP+C IEKN G +M C C ++FCW+C P+ FQ T +
Sbjct: 340 ECPKCSVNIEKNGGCNHMVCSSCKYEFCWICE-----------GPWAPHGKNFFQCTMYK 388
Query: 336 EVEINMEKDNP-------KKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQKRETY 388
E N K NP KK YY + ++ +K L K ++ + ++
Sbjct: 389 NNEDNKSK-NPQDANKTLKKYTFYYRLFNEHEVSAKLDWNLGQTLGTKVHALQERIGISW 447
Query: 389 FD--FVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQVEAEVALEKLQQCAK 446
D F+ ++ + + E R +LKW+ Y+ + K F + + A A+E L + +
Sbjct: 448 IDGQFLSESLKVLNEGRTVLKWSFAVAYYSDASHNLTKIFVDNQMLLAN-AVESLSELLQ 506
Query: 447 VE 448
++
Sbjct: 507 IK 508
>PARC_HUMAN (Q8IWT3) p53-associated parkin-like cytoplasmic protein
(UbcH7 associated protein 1)
Length = 2517
Score = 132 bits (332), Expect = 2e-30
Identities = 105/405 (25%), Positives = 173/405 (41%), Gaps = 51/405 (12%)
Query: 42 LSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLL 101
+S E++ L + + +V L + A LL H+ W ++L+S+ ED E + AAGL
Sbjct: 1997 MSPQEVEGLMKQTVRQVQETLNLEPDVAQHLLAHSHWGAEQLLQSYSEDPEPLLLAAGLC 2056
Query: 102 NQPKVEIDWLKDLCQVCFRS-SEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLR 160
+ D C VC + S C H C++CW +Y+ T I + + L
Sbjct: 2057 VHQAQAVPVRPDHCPVCVSPLGCDDDLPSLCCMHYCCKSCWNEYLTTRIEQ--NLVLNCT 2114
Query: 161 CPDPSCDVAVDRDMIRELASESSKV-MYEQFLLWSYVDSKAKMRWCPAP-GCDYVVCYQG 218
CP C IR + S + YE+ LL YV+S + + WC P GCD ++C QG
Sbjct: 2115 CPIADCPAQPTGAFIRAIVSSPEVISKYEKALLRGYVESCSNLTWCTNPQGCDRILCRQG 2174
Query: 219 DGGCARDLDVTC-LCYHSFCWRCG-EESHSPVDCETAGRWM--------KKIDKSTSENG 268
G TC C + C+ C E+H P C +W+ ++ +
Sbjct: 2175 LG-----CGTTCSKCGWASCFNCSFPEAHYPASCGHMSQWVDDGGYYDGMSVEAQSKHLA 2229
Query: 269 KWILAHTKPCPRCKSLIEKNQGFKYMTC-KCGFQFCWVCFHEQLKCESRSCSPFIRMHIM 327
K I +K CP C++ IEKN+G +MTC KC FCW C LK + + M
Sbjct: 2230 KLI---SKRCPSCQAPIEKNEGCLHMTCAKCNHGFCWRC----LKSWKPNHKDYYNCSAM 2282
Query: 328 EFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQKRET 387
+ E+ +D ++C +++ + ++L + ++
Sbjct: 2283 VSKAARQEK----RFQDYNERCTFHHQA-----------REFAVNLRNRVSAIHEVPPPR 2327
Query: 388 YFDFVKKAWQQIAECRRILKWTNVYGYFLPKNEKAKKQFSEYIQV 432
F F+ A Q + + R++L + VY ++ Q +EY+ V
Sbjct: 2328 SFTFLNDACQGLEQARKVLAYACVYSFY--------SQDAEYMDV 2364
>PARC_MOUSE (Q80TT8) p53-associated parkin-like cytoplasmic protein
Length = 1865
Score = 130 bits (328), Expect = 6e-30
Identities = 101/393 (25%), Positives = 168/393 (42%), Gaps = 43/393 (10%)
Query: 42 LSELEIKRLQEDAINKVASVLCIPDASACFLLCHNLWNVTKVLESWCEDEEKVQKAAGLL 101
+S E++ L E + +V L + A LL H+ W ++L+S+ +D E + AAGL
Sbjct: 1336 MSPQEVEGLMEQTVRQVQETLNLEPDVAQHLLAHSHWGTEQLLQSYSDDPEPLLLAAGLR 1395
Query: 102 NQPKVEIDWLKDLCQVCFRS-SEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLR 160
+ D C VC S C H C++CW +Y+ T I + + L
Sbjct: 1396 VPQAQVVPTRPDQCPVCVTPLGPHDDSPSLCCLHCCCKSCWNEYLTTRIEQ--NFVLNCT 1453
Query: 161 CPDPSCDVAVDRDMIRELASESSKV-MYEQFLLWSYVDSKAKMRWCPAP-GCDYVVCYQG 218
CP C IR + S + YE+ LL YV+S + + WC P GCD ++C QG
Sbjct: 1454 CPIADCPAQPTGAFIRNIVSSPEVISKYEKALLRGYVESCSNLTWCTNPQGCDRILCRQG 1513
Query: 219 DGGCARDLDVTC-LCYHSFCWRCG-EESHSPVDCETAGRWM--------KKIDKSTSENG 268
G TC C + C+ C E+H P C +W+ ++ +
Sbjct: 1514 LGS-----GTTCSKCGWASCFSCSFPEAHYPASCGHMSQWVDDGGYYDGMSVEAQSKHLA 1568
Query: 269 KWILAHTKPCPRCKSLIEKNQGFKYMTC-KCGFQFCWVCFHEQLKCESRSCSPFIRMHIM 327
K I +K CP C++ IEKN+G +MTC +C FCW C LK S + M
Sbjct: 1569 KLI---SKRCPSCQAPIEKNEGCLHMTCARCNHGFCWRC----LKSWKPSHKDYYNCSAM 1621
Query: 328 EFQGTDGEEVEINMEKDNPKKCNHYYECWARNDFLSKSILQKLIDLDRKYLSMSLQKRET 387
+ E+ +D ++C +++ + ++L + ++
Sbjct: 1622 VSKAARQEK----RFQDYNERCTFHHQA-----------REFAVNLRNQASAIQEVPPPK 1666
Query: 388 YFDFVKKAWQQIAECRRILKWTNVYGYFLPKNE 420
F F++ A + + + R++L + VY ++ E
Sbjct: 1667 SFTFLQDACRALEQARKVLAYACVYSFYSQDTE 1699
>RN14_HUMAN (Q9UBS8) RING finger protein 14 (Androgen
receptor-associated protein 54) (Triad2 protein) (HFB30)
(HRIHFB2038)
Length = 474
Score = 92.8 bits (229), Expect = 2e-18
Identities = 60/221 (27%), Positives = 90/221 (40%), Gaps = 34/221 (15%)
Query: 114 LCQVCFRSSEKGKILS-AGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDR 172
LC +CF + + C H C+AC DY + I G +CL CP+P C
Sbjct: 219 LCSICFCEKLGSECMYFLECRHVYCKACLKDYFEIQIRDGQVQCLN--CPEPKCPSVATP 276
Query: 173 DMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLC 232
++EL Y++ LL S +D A + +CP P C V + GC + + C
Sbjct: 277 GQVKELVEAELFARYDRLLLQSSLDLMADVVYCPRPCCQLPV--MQEPGCT--MGICSSC 332
Query: 233 YHSFCWRCGEESHSPVDCETAG-----------------------RWMKKIDKST---SE 266
+FC C H C+ R+ K++ + E
Sbjct: 333 NFAFCTLCRLTYHGVSPCKVTAEKLMDLRNEYLQADEANKRLLDQRYGKRVIQKALEEME 392
Query: 267 NGKWILAHTKPCPRCKSLIEKNQGFKYMTCK-CGFQFCWVC 306
+ +W+ ++K CP C + IEK G MTC C FCW+C
Sbjct: 393 SKEWLEKNSKSCPCCGTPIEKLDGCNKMTCTGCMQYFCWIC 433
>RN14_MOUSE (Q9JI90) RING finger protein 14 (Androgen
receptor-associated protein 54) (Triad2 protein)
Length = 485
Score = 91.7 bits (226), Expect = 4e-18
Identities = 60/221 (27%), Positives = 87/221 (39%), Gaps = 34/221 (15%)
Query: 114 LCQVCFRSSEKGKILS-AGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDR 172
LC +CF + C H C+AC DY + I G KCL CP+P C
Sbjct: 220 LCSICFCEKLGSDCMYFLECKHVYCKACLKDYFEIQIKDGQVKCLN--CPEPQCPSVATP 277
Query: 173 DMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTCLC 232
++EL Y++ LL S +D A + +CP P C V + G A + C
Sbjct: 278 GQVKELVEADLFARYDRLLLQSTLDLMADVVYCPRPCCQLPVMQEPGGTMA----ICSSC 333
Query: 233 YHSFCWRCGEESHSPVDCETAG-----------------------RWMKKIDKST---SE 266
+FC C H C+ R+ K++ + E
Sbjct: 334 NFAFCTLCRLTYHGLSPCKVTAEKLIDLRNEYLQADEATKRFLEQRYGKRVIQKALEEME 393
Query: 267 NGKWILAHTKPCPRCKSLIEKNQGFKYMTCK-CGFQFCWVC 306
+ W+ ++K CP C + I+K G MTC C FCW+C
Sbjct: 394 SKDWLEKNSKSCPCCGTPIQKLDGCNKMTCTGCMQYFCWIC 434
>RN19_MOUSE (P50636) RING finger protein 19 (XY body protein) (XYbp)
(Gametogenesis expressed protein GEG-154)
(UBCM4-interacting protein 117) (UIP117)
Length = 840
Score = 84.3 bits (207), Expect = 6e-16
Identities = 61/212 (28%), Positives = 92/212 (42%), Gaps = 25/212 (11%)
Query: 115 CQVCFRSSEKGKILSA-GCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRD 173
C +C K + C H C C Y+ I++ + + CP+ C +
Sbjct: 132 CPLCLLRHSKDRFPDIMTCHHRSCVDCLRQYLRIEISESR---VNISCPE--CTERFNPH 186
Query: 174 MIRELASESSKV-MYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTC-- 230
IR + S+ + YE+F+L ++ + RWCPAP C Y V G C + +TC
Sbjct: 187 DIRLILSDDVLMEKYEEFMLRRWLVADPDCRWCPAPDCGYAVIAFGCASCPK---LTCGR 243
Query: 231 -LCYHSFCWRCGEESHSPVDCETAG---------RWMKKIDKSTSENGKWILAHTKPCPR 280
C FC+ C + H C+ A R ++ S S+ KPCPR
Sbjct: 244 EGCGTEFCYHCKQIWHPNQTCDAARQERAQSLRLRTIRSSSISYSQESGAAADDIKPCPR 303
Query: 281 CKS-LIEKNQG-FKYMTCK-CGFQFCWVCFHE 309
C + +I+ N G +MTC CG +FCW+C E
Sbjct: 304 CAAYIIKMNDGSCNHMTCAVCGCEFCWLCMKE 335
>RN19_HUMAN (Q9NV58) RING finger protein 19 (Dorfin) (Double
ring-finger protein) (p38 protein)
Length = 838
Score = 84.3 bits (207), Expect = 6e-16
Identities = 61/212 (28%), Positives = 92/212 (42%), Gaps = 25/212 (11%)
Query: 115 CQVCFRSSEKGKILSA-GCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRD 173
C +C K + C H C C Y+ I++ + + CP+ C +
Sbjct: 132 CPLCLLRHSKDRFPDIMTCHHRSCVDCLRQYLRIEISESR---VNISCPE--CTERFNPH 186
Query: 174 MIRELASESSKV-MYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDVTC-- 230
IR + S+ + YE+F+L ++ + RWCPAP C Y V G C + +TC
Sbjct: 187 DIRLILSDDVLMEKYEEFMLRRWLVADPDCRWCPAPDCGYAVIAFGCASCPK---LTCGR 243
Query: 231 -LCYHSFCWRCGEESHSPVDCETAG---------RWMKKIDKSTSENGKWILAHTKPCPR 280
C FC+ C + H C+ A R ++ S S+ KPCPR
Sbjct: 244 EGCGTEFCYHCKQIWHPNQTCDAARQERAQSLRLRTIRSSSISYSQESGAAADDIKPCPR 303
Query: 281 CKS-LIEKNQG-FKYMTCK-CGFQFCWVCFHE 309
C + +I+ N G +MTC CG +FCW+C E
Sbjct: 304 CAAYIIKMNDGSCNHMTCAVCGCEFCWLCMKE 335
>U7I4_HUMAN (P50876) Ubiquitin conjugating enzyme 7 interacting
protein 4 (UbcM4-interacting protein 4) (RING finger
protein 144)
Length = 292
Score = 68.2 bits (165), Expect = 5e-11
Identities = 54/203 (26%), Positives = 83/203 (40%), Gaps = 19/203 (9%)
Query: 115 CQVCFRSSEKGKILS-AGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSC------- 166
C++C ++ + A C C C Y++ I +G ++ CPD +C
Sbjct: 20 CKLCLGEYPVEQMTTIAQCQCIFCTLCLKQYVELLIKEGLETAIS--CPDAACPKQGHLQ 77
Query: 167 DVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDL 226
+ ++ + E+ K+ +E+ +L+ + WCPA C VC D G
Sbjct: 78 ENEIECMVAAEIMQRYKKLQFEREVLFDPCRT-----WCPASTCQ-AVCQLQDVGLQTPQ 131
Query: 227 DVTC-LCYHSFCWRCGEESHSPVDC-ETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSL 284
V C C FC C H C ET + S + + A K CP+CK
Sbjct: 132 PVQCKACRMEFCSTCKASWHPGQGCPETMPITFLPGETSAAFKMEEDDAPIKRCPKCKVY 191
Query: 285 IEKNQGFKYMTCK-CGFQFCWVC 306
IE+++G M CK C FCW C
Sbjct: 192 IERDEGCAQMMCKNCKHAFCWYC 214
>U7I4_MOUSE (Q925F3) Ubiquitin conjugating enzyme 7 interacting
protein 4 (UbcM4-interacting protein 4) (RING finger
protein 144)
Length = 292
Score = 65.5 bits (158), Expect = 3e-10
Identities = 53/203 (26%), Positives = 84/203 (41%), Gaps = 19/203 (9%)
Query: 115 CQVCFRSSEKGKILS-AGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSC------- 166
C++C ++ + A C C C Y++ I +G ++ CPD +C
Sbjct: 20 CKLCLGEYPAEQMTTIAQCQCIFCTLCLKQYVELLIKEGLETAIS--CPDAACPKQGHLQ 77
Query: 167 DVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDL 226
+ ++ + E+ K+ +E+ +L+ + WCPA C VC D G
Sbjct: 78 ENEIECMVAAEIMQRYKKLQFEREVLFDPCRT-----WCPASTCQ-AVCQLQDIGLQTPQ 131
Query: 227 DVTC-LCYHSFCWRCGEESHSPVDC-ETAGRWMKKIDKSTSENGKWILAHTKPCPRCKSL 284
V C C FC C H C ET + S++ + A K CP+C+
Sbjct: 132 LVQCKACDMEFCSACKARWHPGQGCPETMPITFLPGETSSAFKMEEGDAPIKRCPKCRVY 191
Query: 285 IEKNQGFKYMTCK-CGFQFCWVC 306
IE+++G M CK C FCW C
Sbjct: 192 IERDEGCAQMMCKNCKHAFCWYC 214
>U7I1_HUMAN (Q9NWF9) Ubiquitin conjugating enzyme 7 interacting
protein 1 (Triad3 protein) (Zinc finger protein
inhibiting NF-kappa-B)
Length = 488
Score = 59.3 bits (142), Expect = 2e-08
Identities = 53/214 (24%), Positives = 86/214 (39%), Gaps = 19/214 (8%)
Query: 101 LNQPKVEIDWLKDLCQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLR 160
+N+ + + D C+ C+ ++ AH C+ C + Y + G K L L
Sbjct: 123 MNEEQYQKDGQLIECRCCYGEFPFEELTQCADAHLFCKECLIRYAQEAVF-GSGK-LELS 180
Query: 161 CPDPSCDVAVDRDMIRELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDG 220
C + SC + SE KV+ Q +L+ Y + KA+ A + V C
Sbjct: 181 CMEGSCTCSFP-------TSELEKVL-PQTILYKYYERKAEEEVAAAYADELVRCPSCSF 232
Query: 221 GCARDLDVTCL------CYHSFCWRCGE--ESHSPVDCETAGRWMKKIDKSTSENGKWIL 272
D DV C C +C + H+ + CE I TS K
Sbjct: 233 PALLDSDVKRFSCPNPHCRKETCRKCQGLWKEHNGLTCEELAE-KDDIKYRTSIEEKMTA 291
Query: 273 AHTKPCPRCKSLIEKNQGFKYMTCKCGFQFCWVC 306
A + C +C + + K++G M+C+CG Q C++C
Sbjct: 292 ARIRKCHKCGTGLIKSEGCNRMSCRCGAQMCYLC 325
>PRKN_MOUSE (Q9WVS6) Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN)
Length = 464
Score = 58.9 bits (141), Expect = 3e-08
Identities = 58/224 (25%), Positives = 78/224 (33%), Gaps = 44/224 (19%)
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINKGPSKCLTLRCPDPSCDVAVDRDM 174
C C + H +C C+ Y T +N C +
Sbjct: 237 CIACTDVRSPVLVFQCNHRHVICLDCFHLYCVTRLNDRQFVHDAQLGYSLPCVAGCPNSL 296
Query: 175 IREL-----ASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDY----------VVCYQGD 219
I+EL E Y+Q+ V + CP PGC V C G+
Sbjct: 297 IKELHHFRILGEEQYTRYQQYGAEECVLQMGGVL-CPRPGCGAGLLPEQGQRKVTCEGGN 355
Query: 220 G-GCARDLDVTCLCYHSFCWRCGEESHSPVDCET------AGRWMKKIDKSTSENGKW-- 270
G GC FC C E H DC++ A ++DK +E +W
Sbjct: 356 GLGCG----------FVFCRDCKEAYHEG-DCDSLLEPSGATSQAYRVDKRAAEQARWEE 404
Query: 271 -----ILAHTKPCPRCKSLIEKNQGFKYMTC---KCGFQFCWVC 306
I TKPCPRC IEKN G +M C +C ++CW C
Sbjct: 405 ASKETIKKTTKPCPRCNVPIEKNGGCMHMKCPQPQCKLEWCWNC 448
>PRKN_HUMAN (O60260) Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN)
(Parkinson juvenile disease protein 2) (Parkinson
disease protein 2)
Length = 465
Score = 57.4 bits (137), Expect = 8e-08
Identities = 55/220 (25%), Positives = 85/220 (38%), Gaps = 36/220 (16%)
Query: 115 CQVCFRSSEKGKILSAGCAHPLCEACWMDYIDTNINK-----GPSKCLTLRCPDPSCDVA 169
C C + H +C C+ Y T +N P +L C C +
Sbjct: 238 CITCTDVRSPVLVFQCNSRHVICLDCFHLYCVTRLNDRQFVHDPQLGYSLPCV-AGCPNS 296
Query: 170 VDRDMIR-ELASESSKVMYEQFLLWSYVDSKAKMRWCPAPGCDYVVCYQGDGGCARDLDV 228
+ +++ + E Y+Q+ V + CP PGC + + D V
Sbjct: 297 LIKELHHFRILGEEQYNRYQQYGAEECVLQMGGVL-CPRPGCGAGLLPEPD-----QRKV 350
Query: 229 TCL------CYHSFCWRCGEESHSPVDC----ETAGRWMK--KIDKSTSENGKWILAH-- 274
TC C +FC C E H +C E +G + ++D+ +E +W A
Sbjct: 351 TCEGGNGLGCGFAFCRECKEAYHEG-ECSAVFEASGTTTQAYRVDERAAEQARWEAASKE 409
Query: 275 -----TKPCPRCKSLIEKNQGFKYMTC---KCGFQFCWVC 306
TKPCPRC +EKN G +M C +C ++CW C
Sbjct: 410 TIKKTTKPCPRCHVPVEKNGGCMHMKCPQPQCRLEWCWNC 449
>PRKN_RAT (Q9JK66) Parkin (EC 6.3.2.-) (Ubiquitin E3 ligase PRKN)
Length = 465
Score = 56.2 bits (134), Expect = 2e-07
Identities = 40/128 (31%), Positives = 51/128 (39%), Gaps = 36/128 (28%)
Query: 205 CPAPGCDY----------VVCYQGDG-GCARDLDVTCLCYHSFCWRCGEESH-----SPV 248
CP PGC V C G+G GC FC C E H S
Sbjct: 332 CPRPGCGAGLLPEQGQKKVTCEGGNGLGCG----------FVFCRDCKEAYHEGECDSMF 381
Query: 249 DCETAGRWMKKIDKSTSENGKW-------ILAHTKPCPRCKSLIEKNQGFKYMTC---KC 298
+ A ++D+ +E +W I TKPCPRC IEKN G +M C +C
Sbjct: 382 EASGATSQAYRVDQRAAEQARWEEASKETIKKTTKPCPRCNVPIEKNGGCMHMKCPQPQC 441
Query: 299 GFQFCWVC 306
++CW C
Sbjct: 442 KLEWCWNC 449
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,918,035
Number of Sequences: 164201
Number of extensions: 2684468
Number of successful extensions: 7973
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 7819
Number of HSP's gapped (non-prelim): 101
length of query: 500
length of database: 59,974,054
effective HSP length: 114
effective length of query: 386
effective length of database: 41,255,140
effective search space: 15924484040
effective search space used: 15924484040
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0216.15


