

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0152.2
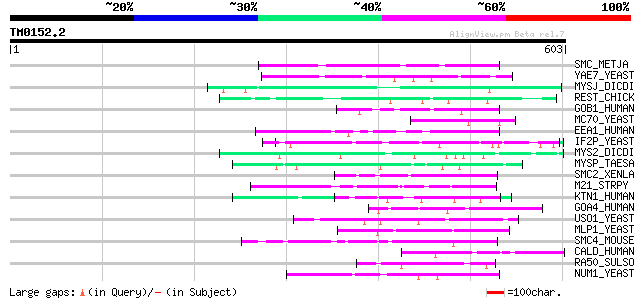
(603 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SMC_METJA (Q59037) Chromosome partition protein smc homolog 53 2e-06
YAE7_YEAST (P39723) Hypothetical 72.1 kDa protein in ACS1-GCV3 i... 50 1e-05
MYSJ_DICDI (P54697) Myosin IJ heavy chain 50 2e-05
REST_CHICK (O42184) Restin (Cytoplasmic linker protein-170) (CLI... 49 3e-05
GOB1_HUMAN (Q14789) Golgi autoantigen, golgin subfamily B member... 49 3e-05
MC70_YEAST (Q12411) Sporulation-specific protein 21 (Meiotic pla... 49 4e-05
EEA1_HUMAN (Q15075) Early endosome antigen 1 (Endosome-associate... 49 4e-05
IF2P_YEAST (P39730) Eukaryotic translation initiation factor 5B ... 49 5e-05
MYS2_DICDI (P08799) Myosin II heavy chain, non muscle 48 6e-05
MYSP_TAESA (Q8T305) Paramyosin 48 8e-05
SMC2_XENLA (P50533) Structural maintenance of chromosome 2 (Chro... 47 1e-04
M21_STRPY (P50468) M protein, serotype 2.1 precursor 47 1e-04
KTN1_HUMAN (Q86UP2) Kinectin (Kinesin receptor) (CG-1 antigen) 47 1e-04
GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member... 47 1e-04
USO1_YEAST (P25386) Intracellular protein transport protein USO1 47 2e-04
MLP1_YEAST (Q02455) MLP1 protein (Myosin-like protein 1) 47 2e-04
SMC4_MOUSE (Q8CG47) Structural maintenance of chromosomes 4-like... 46 2e-04
CALD_HUMAN (Q05682) Caldesmon (CDM) 46 2e-04
RA50_SULSO (Q97WH0) DNA double-strand break repair rad50 ATPase 46 3e-04
NUM1_YEAST (Q00402) Nuclear migration protein NUM1 46 3e-04
>SMC_METJA (Q59037) Chromosome partition protein smc homolog
Length = 1169
Score = 53.1 bits (126), Expect = 2e-06
Identities = 58/266 (21%), Positives = 128/266 (47%), Gaps = 24/266 (9%)
Query: 271 ILRFRREQQAAREKRNPAKTLLDA-DASHSGTESNRPQKKKRKNETPESAKGKDSSQPSM 329
I F +++ A E+ A+ L++ D S E+N + KK K + + K + + +
Sbjct: 171 IAEFDEKKKKAEEELKKARELIEMIDIRISEVENNLKKLKKEKEDAEKYIKLNEELKAAK 230
Query: 330 EKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSL-VETNL 388
++K K + + ++ +K EEL +E ++ + + ++ +L + N
Sbjct: 231 YALILK--------KVSYLNVLLENIQNDIKNLEELKNEFLSKVREIDVEIENLKLRLNN 282
Query: 389 VFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLE-EKEREIL 447
+ EK + L E ++I +EI K++D IN K++E E E +
Sbjct: 283 IINELNEKGNEEVL-ELHKSIKELEVEIENDKKVLD------SSINELKKVEVEIENKKK 335
Query: 448 KMKATMK-LLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLN 506
++K T K ++++ + + EK+ + + +++ +++LN ++ +E + +S++ I HL
Sbjct: 336 EIKETQKKIIENRDSIIEKEQ-----QIKEIEEKIKNLNYEKERLKEAIAESESIIKHLK 390
Query: 507 SSNAELKNENSKLHSEVSELKNSVLD 532
S E+ +E +K +E+ LK + D
Sbjct: 391 ESEMEIADEIAKNQNELYRLKKELND 416
Score = 35.4 bits (80), Expect = 0.43
Identities = 46/203 (22%), Positives = 94/203 (45%), Gaps = 22/203 (10%)
Query: 357 SLLKEFEELTSEEVTSLWDSKIDFNSLVETNL-VFEADREKVKKIGLKEACQAIMTKGLE 415
+++ E E +EEV L S + +E + V ++ ++KK+ ++ I K E
Sbjct: 282 NIINELNEKGNEEVLELHKSIKELEVEIENDKKVLDSSINELKKVEVE-----IENKKKE 336
Query: 416 IAAISKMIDLESAD--FDGINSAKQLEEKEREILKMKATMK--LLDSANKVNEKKAADLA 471
I K I +E+ D + K++EEK + + K +K + +S + + K +++
Sbjct: 337 IKETQKKI-IENRDSIIEKEQQIKEIEEKIKNLNYEKERLKEAIAESESIIKHLKESEME 395
Query: 472 LENE---------RLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSK-LHS 521
+ +E RLKK + DL+ K E+ K+ I L +++ ++K L+
Sbjct: 396 IADEIAKNQNELYRLKKELNDLDNLINRKNFEIEKNNEMIKKLKEELETVEDVDTKPLYL 455
Query: 522 EVSELKNSVLDQFEAGFAKAKEQ 544
E+ L N ++ + G + +E+
Sbjct: 456 ELENL-NVEIEFSKRGIKELEEK 477
Score = 34.7 bits (78), Expect = 0.74
Identities = 38/149 (25%), Positives = 70/149 (46%), Gaps = 33/149 (22%)
Query: 381 NSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLE 440
N + + + E++ K+K+ EI +SK++ +SAK++E
Sbjct: 679 NKIADEIIAIESELRKIKE---------------EIERLSKIVKR--------SSAKKME 715
Query: 441 -EKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSK 499
E EI+K K M+ + A K N K +L L+N+ + + +E+LN+ K EE++
Sbjct: 716 IENTLEIIK-KNEMRKREIAEK-NTIKIKELELKNKDILEELEELNL----KREEILN-- 767
Query: 500 AEITHLNSSNAELKNENSKLHSEVSELKN 528
I + S EL K+ +E+ E ++
Sbjct: 768 -RINEIESKINELIERREKIINELKEYES 795
>YAE7_YEAST (P39723) Hypothetical 72.1 kDa protein in ACS1-GCV3
intergenic region
Length = 622
Score = 50.4 bits (119), Expect = 1e-05
Identities = 67/288 (23%), Positives = 120/288 (41%), Gaps = 32/288 (11%)
Query: 274 FRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFM 333
F + A K++ + L A +S +G++ K E + D S
Sbjct: 209 FLHSKSRAESKQDKEEFLSLAQSSPAGSQLESRDSPSSKEENTDGGYQNDEIHDSNNHID 268
Query: 334 VKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKID--FNSLVETNLVFE 391
+ + A S+S P + +S ++ + E V + + D NS+ L FE
Sbjct: 269 TEN------VMANSTSLPISAVESRFEKTLDTQLEIVIEILHKEYDQFINSI---RLKFE 319
Query: 392 ADREKVKKIGLKEACQAIMTKGLEI-----AAISKMIDLESADFDGINSAK----QLEEK 442
++ K I K Q+ + LE+ + I K L S + I S LE+
Sbjct: 320 KSQKLEKIIASKLNEQSHLLDSLELEENSSSVIEKQDHLISQLKEKIESQSVLINNLEKL 379
Query: 443 EREILKMKATMKLL----DSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKS 498
+ +I+KMK K+L ++ K+N+ K + L+K + DL I K EE
Sbjct: 380 KEDIIKMKQNEKVLTKELETQTKINKLKENNWDSYINDLEKQINDLQID---KSEEFHVI 436
Query: 499 KAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQIL 546
+ ++ L+ N +LKN+ + L ++ +L Q+E+ F K + +L
Sbjct: 437 QNQLDKLDLENYQLKNQLNTLDNQ-----KLILSQYESNFIKFNQNLL 479
>MYSJ_DICDI (P54697) Myosin IJ heavy chain
Length = 2245
Score = 49.7 bits (117), Expect = 2e-05
Identities = 76/413 (18%), Positives = 153/413 (36%), Gaps = 54/413 (13%)
Query: 216 EAIIKLPDDSLTPEE------------MVDCTFLSSIDLNLTEFLDAHYEN---RLGEYV 260
E +IKL + L EE + D I L LTE + + +L EY
Sbjct: 1217 EQLIKLSSEKLGSEEEAKKQINQLELELTDHKSKLQIQLQLTEQSNEKIKKLKGKLEEYQ 1276
Query: 261 AKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAK 320
+ +L + + R ++ +Q+ +++N T L S S +K K T +S
Sbjct: 1277 DEKKQLQQE-LERIKQSKQSVEDEKNSLITQLTTVKFESTQVSTNVSHQKEKITTLKSTI 1335
Query: 321 GKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDF 380
+ + + K + + + KEF +L S++ S+I
Sbjct: 1336 EELNKSIGKLQAEQKNKDDEIRKIQFELNDQKQQFTRQTKEFSDLQSQQSIDRPKSEITI 1395
Query: 381 NSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLE 440
+SL TN ++D E+V++ L+ + D +
Sbjct: 1396 HSLERTNETLKSDFERVQQ------------------------SLKQQERDCQQYKDTIN 1431
Query: 441 EKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKA 500
E E+ ++ + ++ V +++ ++ E+ LK+ + Q E EL + K
Sbjct: 1432 RLENEVKQLTQLKERFENEFFVAKEQNSNQTQESVYLKEVTTQMQQNQSRIERELEEKKQ 1491
Query: 501 EITHLNSSNAELKNENSKL--------------HSEVSELKNSVLDQFEAGFAKAKEQIL 546
IT ++ ELK + ++L +E+ L+ L E G +K+Q
Sbjct: 1492 HITRIDDERDELKKQLTQLQQQHEQSSTQLLLAQNELERLRKKELKYKERGHETSKQQDQ 1551
Query: 547 FLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNEGE 599
F S+ + +D + D + D+ +++ ++ E++ + E
Sbjct: 1552 FNMEIQSLRITNNDQLKSLQDYEQEKKKLKDKLSSSKQEAQQQRESIIKMDAE 1604
Score = 36.6 bits (83), Expect = 0.19
Identities = 31/127 (24%), Positives = 61/127 (47%), Gaps = 17/127 (13%)
Query: 423 IDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVE 482
I L+ + D N + QL+ E + K++ + LD ++K+N+K DL+ +++ ++K
Sbjct: 1032 IQLKYQELDNSNQSSQLQLSEC-LSKLEEQTQQLDHSSKLNKKLEKDLSDQHDSIEKLQS 1090
Query: 483 DLNITQ------KAKEEELVKSKAEIT--------HLNSSNAELKNENSKLHSEVSELK- 527
N T+ K + EEL ++ T + + E +N+ E+ +LK
Sbjct: 1091 QFNETEQQLQQFKQQSEELSSKLSKTTQQLDFNKQEFDRLSQERDTDNTNNQLEIQQLKK 1150
Query: 528 -NSVLDQ 533
NS L++
Sbjct: 1151 ANSTLEE 1157
Score = 36.2 bits (82), Expect = 0.25
Identities = 40/174 (22%), Positives = 83/174 (46%), Gaps = 28/174 (16%)
Query: 433 INSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLAL-----ENERLKKHVEDLN-- 485
I S +++++K R I+ ++A ++ + + +A +L + +L++ +E+L
Sbjct: 934 ILSKREVDKKLRGIILIQARWRMKLAKRVYIQLRAEARSLRTVQEQKNKLQEKLEELQWR 993
Query: 486 ITQKAK-----EEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLD---QFEAG 537
+T +AK E++ VKS I+ L+S+N L+ + S++ + EL NS Q
Sbjct: 994 LTSEAKRKQQLEDQKVKSDTTISELSSNNDHLELQLSEIQLKYQELDNSNQSSQLQLSEC 1053
Query: 538 FAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDE 591
+K +EQ L+ +N K + D D+ D E+ +++ +E
Sbjct: 1054 LSKLEEQTQQLDHSSKLN-------------KKLEKDLSDQHDSIEKLQSQFNE 1094
Score = 35.4 bits (80), Expect = 0.43
Identities = 53/300 (17%), Positives = 124/300 (40%), Gaps = 38/300 (12%)
Query: 254 NRLGEYVAKMTRLSDDDILRFRREQQAAREKR-NPAKTLLDADASHSGTESNRPQKKKRK 312
NRL V ++T+L + RF E A+E+ N + + + + N+ + ++
Sbjct: 1431 NRLENEVKQLTQLKE----RFENEFFVAKEQNSNQTQESVYLKEVTTQMQQNQSRIEREL 1486
Query: 313 NETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTS 372
E + D + ++K + + QH A + KE + TS
Sbjct: 1487 EEKKQHITRIDDERDELKKQLTQLQQQHEQSSTQLLLAQNELERLRKKELKYKERGHETS 1546
Query: 373 ---------LWDSKIDFNSLVETNLVFEADREKVK------KIGLKEACQAIMTKGLEIA 417
+ +I N +++ +E +++K+K K ++ ++I+ E++
Sbjct: 1547 KQQDQFNMEIQSLRITNNDQLKSLQDYEQEKKKLKDKLSSSKQEAQQQRESIIKMDAELS 1606
Query: 418 AISKMIDLESADFDGINSAKQLEEKEREILKMKATMK--------LLDSANKVNEKKAAD 469
AI + NS ++++ +E+++ A K +DS K E + +
Sbjct: 1607 AIKQHSQWVE------NSFTDMKQRNQELIESSALYKQQLLQQTSTIDSTIKEKENEISK 1660
Query: 470 LALE----NERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSE 525
L + N++L + E+LN +++ + E + ++ L N +LK+ +++ ++ +
Sbjct: 1661 LQQQLETSNQQLHQLKEELNSMKQSNQLESTEQSKQLNQLIQENQQLKSVTNEISKQLDD 1720
Score = 32.0 bits (71), Expect = 4.8
Identities = 34/164 (20%), Positives = 69/164 (41%), Gaps = 14/164 (8%)
Query: 437 KQLEEKEREIL-KMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEEL 495
+Q +++ E+ K+ T + LD + ++ + + +N + ++ L EE+
Sbjct: 1100 QQFKQQSEELSSKLSKTTQQLDFNKQEFDRLSQERDTDNTNNQLEIQQLKKANSTLEEDY 1159
Query: 496 VKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSIN 555
+L EL++EN + + L QF++G A K+Q+ L + S
Sbjct: 1160 FSLSGIRDNLERQVLELRDENQLIKERLDSL-GQQSSQFQSGAALEKQQLEQLVQEQSEQ 1218
Query: 556 LAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNEGE 599
L KL S G EE E ++ N+ + + D++ +
Sbjct: 1219 LI-----------KLSSEKLGSEE-EAKKQINQLELELTDHKSK 1250
>REST_CHICK (O42184) Restin (Cytoplasmic linker protein-170)
(CLIP-170)
Length = 1433
Score = 49.3 bits (116), Expect = 3e-05
Identities = 84/386 (21%), Positives = 151/386 (38%), Gaps = 72/386 (18%)
Query: 229 EEMVDCTFLSSIDLNLTEFLDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPA 288
+++ D N + L A YE E + K D DI F++ A E
Sbjct: 990 DQLTDMKKQMETSQNQYKDLQAKYEKETSEMITK----HDADIKGFKQNLLDAEE----- 1040
Query: 289 KTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSS 348
L A + E+ + KK+ + + ++ Q +MEK + + H
Sbjct: 1041 -ALKAAQKKNDELETQAEELKKQAEQAKADKRAEEVLQ-TMEKVTKEKDAIHQ------- 1091
Query: 349 SAPPPSWKSLLKEFEELTSEEVTSLWDSKI--DFNSLVETNLVFEADREKVKKIGLKEAC 406
++ E L S E + + K+ + + L + NL E + K K++ E
Sbjct: 1092 -----------EKIETLASLENSRQTNEKLQNELDMLKQNNLKNEEELTKSKELLNLENK 1140
Query: 407 QAIMTK----GLEIAAISKMIDLESADFDGINSAKQLEEKEREIL---KMKATMKLLDSA 459
+ K L++AA K L + + + A++L E+ K++ +L+
Sbjct: 1141 KVEELKKEFEALKLAAAQKSQQLAALQEENVKLAEELGRSRDEVTSHQKLEEERSVLN-- 1198
Query: 460 NKVNEKKAADLALENE------RLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELK 513
N++ E K + L+ E L+K + D + K+EEL K + EIT L NA K
Sbjct: 1199 NQLLEMKKRESTLKKEIDEERASLQKSISDTSALITQKDEELEKLRNEITVLRGENASAK 1258
Query: 514 NENS---KLHSEVSELKNSVLDQFEAGFAKAKEQILFLNP--QVSINLAGSDPYARIVDG 568
S L S+ +L+ V + + AK+++ + +P ++ NL
Sbjct: 1259 TLQSVVKTLESDKLKLEEKVKNLEQKLKAKSEQPLTVTSPSGDIAANLL----------- 1307
Query: 569 KLISPDTGDEEDEGEEDKNEEDENVN 594
+DE EDK +E + +N
Sbjct: 1308 ----------QDESAEDKQQEIDFLN 1323
Score = 38.5 bits (88), Expect = 0.051
Identities = 59/312 (18%), Positives = 127/312 (39%), Gaps = 46/312 (14%)
Query: 242 LNLTEFLDAHYENRLG-EYVAKMTRLSDDDILRFRREQQA--AREKRNPAKTLLDADASH 298
L+L A+ E +L + +++ + ++ I E+ + +E + + LLD + +
Sbjct: 764 LDLAALQKANSEGKLEIQKLSEQLQAAEKQIQNLETEKVSNLTKELQGKEQKLLDLEKNL 823
Query: 299 SGTESNRPQKKKR----KNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPS 354
S + +K K + + G +++Q +M++ + K N + SS
Sbjct: 824 SAVNQVKDSLEKELQLLKEKFTSAVDGAENAQRAMQETINKLNQKEEQFALMSS------ 877
Query: 355 WKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGL 414
E E+L S +++ET L +RE+ + +A +
Sbjct: 878 ------ELEQLKSNL------------TVMETKLKEREEREQ-------QLTEAKVKLEN 912
Query: 415 EIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALEN 474
+IA I K SA +N +L+E++ E ++++ T K NEK
Sbjct: 913 DIAEIMKSSGDSSAQLMKMNDELRLKERQLEQIQLELT--------KANEKAVQLQKNVE 964
Query: 475 ERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQF 534
+ +K + T K +EEL K + ++T + +N+ L ++ + + ++ +
Sbjct: 965 QTAQKAEQSQQETLKTHQEELKKMQDQLTDMKKQMETSQNQYKDLQAKYEKETSEMITKH 1024
Query: 535 EAGFAKAKEQIL 546
+A K+ +L
Sbjct: 1025 DADIKGFKQNLL 1036
Score = 33.1 bits (74), Expect = 2.1
Identities = 39/176 (22%), Positives = 79/176 (44%), Gaps = 28/176 (15%)
Query: 384 VETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDL---ESADFDG-INSAKQL 439
+E +L+FE + + L++ A +++ I + + + L E A+ G + S+K +
Sbjct: 470 LEQSLLFEKTKADKLQRELEDTRVATVSEKSRIMELERDLALRVKEVAELRGRLESSKHI 529
Query: 440 EEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSK 499
++ + ++ LL + + EK AA K+H +++ ++ E +
Sbjct: 530 DDVD-------TSLSLLQEISSLQEKMAA-------AGKEHQREMSSLKEKFESSEEALR 575
Query: 500 AEITHLNSSNAELKNENSKL----------HSEVSELKNSVLDQFEAGFAKAKEQI 545
EI L++SN + EN L +S+V EL S L+ A +A E++
Sbjct: 576 KEIKTLSASNERMGKENESLKTKLDHANKENSDVIELWKSKLESAIASHQQAMEEL 631
>GOB1_HUMAN (Q14789) Golgi autoantigen, golgin subfamily B member 1
(Giantin) (Macrogolgin) (Golgi complex-associated
protein, 372-kDa) (GCP372)
Length = 3259
Score = 49.3 bits (116), Expect = 3e-05
Identities = 50/197 (25%), Positives = 90/197 (45%), Gaps = 35/197 (17%)
Query: 356 KSLLKEFEELTSEEV--TSLWDSKI-----DFNSLVET--NLVFEADREKVKKIGLKEAC 406
+ L+KE E L S ++ ++ W K ++ L+++ N+ EA+R ++
Sbjct: 1584 EKLVKEIESLKSSKIAESTEWQEKHKELQKEYEILLQSYENVSNEAER-------IQHVV 1636
Query: 407 QAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKK 466
+A+ + E+ L S + + + KQL+E E+E+ +MK M+ K ++K
Sbjct: 1637 EAVRQEKQELYG-----KLRSTEANKKETEKQLQEAEQEMEEMKEKMRKFA---KSKQQK 1688
Query: 467 AADLALENERLKKHVEDLNITQK-----------AKEEELVKSKAEITHLNSSNAELKNE 515
+L EN+RL+ V T K + +EEL + K E L+ L +E
Sbjct: 1689 ILELEEENDRLRAEVHPAGDTAKECMETLLSSNASMKEELERVKMEYETLSKKFQSLMSE 1748
Query: 516 NSKLHSEVSELKNSVLD 532
L EV +LK+ + D
Sbjct: 1749 KDSLSEEVQDLKHQIED 1765
Score = 38.9 bits (89), Expect = 0.039
Identities = 47/201 (23%), Positives = 99/201 (48%), Gaps = 12/201 (5%)
Query: 395 EKVKKIGLK--EACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKAT 452
E+V+ I K E QA+ K LEI + +++ + D + + + +EEK++++ ++ +
Sbjct: 852 ERVRHISSKVEELSQALSQKELEITKMDQLLLEKKRDVETLQ--QTIEEKDQQVTEISFS 909
Query: 453 MKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAEL 512
M + ++NE+K + L +E + LK+ + L+ ++AK+E+ V+ E++ N +
Sbjct: 910 M--TEKMVQLNEEKFS-LGVEIKTLKEQLNLLSRAEEAKKEQ-VEEDNEVSSGLKQNYDE 965
Query: 513 KNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAG--SDPYARIVD-GK 569
+ ++ E + + +L + E K K Q +N + + + A + D K
Sbjct: 966 MSPAGQISKEELQHEFDLLKK-ENEQRKRKLQAALINRKELLQRVSRLEEELANLKDESK 1024
Query: 570 LISPDTGDEEDEGEEDKNEED 590
P + E E EEDK ++
Sbjct: 1025 KEIPLSETERGEVEEDKENKE 1045
Score = 37.4 bits (85), Expect = 0.11
Identities = 26/100 (26%), Positives = 51/100 (51%), Gaps = 7/100 (7%)
Query: 437 KQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENER--LKKHVEDLNITQKAKEEE 494
+QL K+ ++L + + ++ DS N+V A +L+NER L +E +++ K+
Sbjct: 2808 QQLLSKDEQLLHLSSQLE--DSYNQVQSFSKAMASLQNERDHLWNELEKFRKSEEGKQRS 2865
Query: 495 LVK---SKAEITHLNSSNAELKNENSKLHSEVSELKNSVL 531
+ S AE+ L + + L+N+ +L E+ L+ L
Sbjct: 2866 AAQPSTSPAEVQSLKKAMSSLQNDRDRLLKELKNLQQQYL 2905
Score = 35.0 bits (79), Expect = 0.56
Identities = 58/239 (24%), Positives = 87/239 (36%), Gaps = 37/239 (15%)
Query: 286 NPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKA 345
N + + DA + ES KK +E E + Q EK V+ + L+
Sbjct: 1939 NYCQDVTDAQIKNELLESEMKNLKKCVSELEE-----EKQQLVKEKTKVESEIRKEYLEK 1993
Query: 346 GSSSAPPPSWKSLLKEFEELTSE---EVTSLWDSKIDFNSLVETNLVFEADREKVKKIGL 402
+ P KS KE +EL E EV L I + +KI
Sbjct: 1994 IQGAQKEPGNKSHAKELQELLKEKQQEVKQLQKDCIRYQ----------------EKISA 2037
Query: 403 KEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKV 462
E K LE DLE N A+ +E +++ ++ + LLD
Sbjct: 2038 LERT----VKALEFVQTESQKDLEITKE---NLAQAVEHRKKAQAELASFKVLLDDTQSE 2090
Query: 463 NEKKAADLALENERLKKHVEDLNITQKAKEEELVK--SKAEITHLNSSNAELKNENSKL 519
+ AD + L+ + E + K K+E+L + +AE HL E KN KL
Sbjct: 2091 AARVLADNLKLKKELQSNKESVKSQMKQKDEDLERRLEQAEEKHLK----EKKNMQEKL 2145
Score = 31.6 bits (70), Expect = 6.2
Identities = 43/193 (22%), Positives = 83/193 (42%), Gaps = 29/193 (15%)
Query: 363 EELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKM 422
E + ++ + +SK + L ET+L +A RE ++ + I K +I IS +
Sbjct: 2339 EGIIRQQEADIQNSKFSYEQL-ETDL--QASRELTSRLH-----EEINMKEQKI--ISLL 2388
Query: 423 IDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVE 482
E A I +Q +KE +K L++ E++ L EN++
Sbjct: 2389 SGKEEAIQVAIAELRQQHDKE---------IKELENLLSQEEEENIVLEEENKKAVDKTN 2439
Query: 483 DLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSEL----------KNSVLD 532
L T K ++E ++ KA++ S + L+N+ ++ + +L K+ ++
Sbjct: 2440 QLMETLKTIKKENIQQKAQLDSFVKSMSSLQNDRDRIVGDYQQLEERHLSIILEKDQLIQ 2499
Query: 533 QFEAGFAKAKEQI 545
+ A K KE+I
Sbjct: 2500 EAAAENNKLKEEI 2512
Score = 31.2 bits (69), Expect = 8.2
Identities = 42/150 (28%), Positives = 73/150 (48%), Gaps = 30/150 (20%)
Query: 404 EACQAIMTKGLEIAAISK-MIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKV 462
E QA+ + IA++ K +++LE+ + S+ +LEE + E K+ + + LL++ N+
Sbjct: 460 EGTQAVTEEN--IASLQKRVVELENEKGALLLSSIELEELKAENEKLSSQITLLEAQNRT 517
Query: 463 NEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSE 522
E AD + V +++I A + S AE S L+N S+ H E
Sbjct: 518 GE---AD---------REVSEISIVDIANKR---SSSAE----ESGQDVLENTFSQKHKE 558
Query: 523 VSELKNSVLDQFEAGFAKAKEQILFLNPQV 552
+S L +L+ E A+E+I FL Q+
Sbjct: 559 LSVL---LLEMKE-----AQEEIAFLKLQL 580
>MC70_YEAST (Q12411) Sporulation-specific protein 21 (Meiotic plaque
component protein 70)
Length = 609
Score = 48.9 bits (115), Expect = 4e-05
Identities = 32/124 (25%), Positives = 64/124 (50%), Gaps = 10/124 (8%)
Query: 436 AKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEEL 495
A +L+EKE ++ + L++ NK + K +++LEN+++K+ ++L+ +EEL
Sbjct: 290 ASKLKEKEAQLKSQNDKILKLETTNKAYKTKYKEVSLENKKIKEAFKELDNESYNHDEEL 349
Query: 496 VK----SKAEITHLNSSNAELKNENSKLHSEVSELKNSV------LDQFEAGFAKAKEQI 545
+K ++ + +N + ++N L V+EL+N V F+ +AK + I
Sbjct: 350 LKKYKYTRETLDRVNREQQLIIDQNEFLKKSVNELQNEVNATNFKFSLFKEKYAKLADSI 409
Query: 546 LFLN 549
LN
Sbjct: 410 TELN 413
Score = 37.4 bits (85), Expect = 0.11
Identities = 45/182 (24%), Positives = 76/182 (41%), Gaps = 18/182 (9%)
Query: 358 LLKEFEELTSEEVTSLWDSKIDFNS-LVETNLVFEADREKVKKIGLKEACQAIMTKGLEI 416
L++E +L + ++ D+ S L E ++ +K+ K L+ +A TK E+
Sbjct: 267 LMQEMYQLVKKNSSARRTKITDYASKLKEKEAQLKSQNDKILK--LETTNKAYKTKYKEV 324
Query: 417 AAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENER 476
+ +K I + D + E + K K T + LD N+ + + +NE
Sbjct: 325 SLENKKIKEAFKELDN----ESYNHDEELLKKYKYTRETLDRVNREQQL----IIDQNEF 376
Query: 477 LKKHVEDL-------NITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNS 529
LKK V +L N +E+ K IT LN+S + + L E +ELK
Sbjct: 377 LKKSVNELQNEVNATNFKFSLFKEKYAKLADSITELNTSTKKREALGENLTFECNELKEI 436
Query: 530 VL 531
L
Sbjct: 437 CL 438
>EEA1_HUMAN (Q15075) Early endosome antigen 1 (Endosome-associated
protein p162) (Zinc finger FYVE domain containing protein
2)
Length = 1411
Score = 48.9 bits (115), Expect = 4e-05
Identities = 59/269 (21%), Positives = 114/269 (41%), Gaps = 38/269 (14%)
Query: 268 DDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQP 327
+ ++L R++ ++ EK + A+ L ++ + G ++ Q+ K T E K Q
Sbjct: 1039 ESELLATRQDLKSVEEKLSLAQEDLISNRNQIGNQNKLIQELKTAKATLEQDSAKKEQQL 1098
Query: 328 SMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELT---SEEVTSLWDSKIDFNSLV 384
++ + +LK + KS L E EE+ +E+T L N +
Sbjct: 1099 QERCKALQDIQKEKSLKEKELV----NEKSKLAEIEEIKCRQEKEITKL-------NEEL 1147
Query: 385 ETNLVFEADREKVKKI-GLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKE 443
+++ + E +K+I LK+A Q ++ + LE+ + D + +A + E++
Sbjct: 1148 KSHKL-----ESIKEITNLKDAKQLLIQQKLELQGKA----------DSLKAAVEQEKRN 1192
Query: 444 REILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEIT 503
++ILK D K E+ + + +L +++ + K EE K +IT
Sbjct: 1193 QQILK--------DQVKKEEEELKKEFIEKEAKLHSEIKEKEVGMKKHEENEAKLTMQIT 1244
Query: 504 HLNSSNAELKNENSKLHSEVSELKNSVLD 532
LN + +K E VSEL+ D
Sbjct: 1245 ALNENLGTVKKEWQSSQRRVSELEKQTDD 1273
Score = 35.0 bits (79), Expect = 0.56
Identities = 40/180 (22%), Positives = 78/180 (43%), Gaps = 21/180 (11%)
Query: 368 EEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLES 427
E+V L + +L++ N+ E +RE K LK+ C+ + ++ A + E
Sbjct: 221 EDVAVLKKELVQVQTLMD-NMTLERERESEK---LKDECKKLQSQYASSEATISQLRSEL 276
Query: 428 ADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNIT 487
A +++ +E+ K+K++ VNE + L LKK + +
Sbjct: 277 A-----KGPQEVAVYVQELQKLKSS---------VNELTQKNQTLTENLLKKEQDYTKLE 322
Query: 488 QKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFA--KAKEQI 545
+K EE + K + T L+ + + + S+L + + L ++ E G A K KE++
Sbjct: 323 EKHNEESVSKKNIQAT-LHQKDLDCQQLQSRLSASETSLHRIHVELSEKGEATQKLKEEL 381
Score = 34.3 bits (77), Expect = 0.96
Identities = 37/157 (23%), Positives = 67/157 (42%), Gaps = 11/157 (7%)
Query: 384 VETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKE 443
+E +V + KK E+ + +TK E I K DF+ ++ +++ +E
Sbjct: 776 MEKEIVSSTRLDLQKKSEALESIKQKLTKQEEEKQILKQ------DFETLSQETKIQHEE 829
Query: 444 REILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEIT 503
+++ T+ L E +L+ ++L K + L ++ E+E K KA I
Sbjct: 830 LNN-RIQTTVTELQKVKMEKEALMTELSTVKDKLSKVSDSLKNSKSEFEKENQKGKAAIL 888
Query: 504 HLNSSNAELKN----ENSKLHSEVSELKNSVLDQFEA 536
L + ELK+ + E ELK S+ + EA
Sbjct: 889 DLEKTCKELKHQLQVQMENTLKEQKELKKSLEKEKEA 925
Score = 33.5 bits (75), Expect = 1.6
Identities = 24/115 (20%), Positives = 51/115 (43%)
Query: 413 GLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLAL 472
G E ++ DL+S + + + L +I ++ L +A E+ +A
Sbjct: 1037 GRESELLATRQDLKSVEEKLSLAQEDLISNRNQIGNQNKLIQELKTAKATLEQDSAKKEQ 1096
Query: 473 ENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELK 527
+ + K ++D+ + KE+ELV K+++ + + E +KL+ E+ K
Sbjct: 1097 QLQERCKALQDIQKEKSLKEKELVNEKSKLAEIEEIKCRQEKEITKLNEELKSHK 1151
Score = 32.7 bits (73), Expect = 2.8
Identities = 39/187 (20%), Positives = 77/187 (40%), Gaps = 25/187 (13%)
Query: 402 LKEACQAIMTKGLEIAAISKMIDL--ESADFDGINSAKQLEEKEREIL-----------K 448
LKE + TK + A K + E + G+ ++ + ++L +
Sbjct: 377 LKEELSEVETKYQHLKAEFKQLQQQREEKEQHGLQLQSEINQLHSKLLETERQLGEAHGR 436
Query: 449 MKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITH---- 504
+K +L E++ ADL L+ RL++ +++ + +L K+K +
Sbjct: 437 LKEQRQLSSEKLMDKEQQVADLQLKLSRLEEQLKEKVTNSTELQHQLDKTKQQHQEQQAL 496
Query: 505 LNSSNAELKNENSKLHSEVSEL--KNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPY 562
S+ A+L+ + L + ++ K+ + EA K+KE I L + D Y
Sbjct: 497 QQSTTAKLREAQNDLEQVLRQIGDKDQKIQNLEALLQKSKENISLLEKE------REDLY 550
Query: 563 ARIVDGK 569
A+I G+
Sbjct: 551 AKIQAGE 557
Score = 32.7 bits (73), Expect = 2.8
Identities = 26/116 (22%), Positives = 56/116 (47%), Gaps = 8/116 (6%)
Query: 417 AAISKMIDLESADFDGINSAKQLEEKEREI-LKMKATMKLLDSANKVNEKKAADLALENE 475
AA +++ LE++ + + + +EK ++ +++KA +LL SA + ADL
Sbjct: 611 AAQDRVLSLETSVNELNSQLNESKEKVSQLDIQIKAKTELLLSAEAAKTAQRADL----- 665
Query: 476 RLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVL 531
+ H++ + K++EL K ++ + + + + S+L S + E K L
Sbjct: 666 --QNHLDTAQNALQDKQQELNKITTQLDQVTAKLQDKQEHCSQLESHLKEYKEKYL 719
>IF2P_YEAST (P39730) Eukaryotic translation initiation factor 5B
(eIF-5B) (Translation initiation factor IF-2)
Length = 1002
Score = 48.5 bits (114), Expect = 5e-05
Identities = 78/335 (23%), Positives = 146/335 (43%), Gaps = 40/335 (11%)
Query: 275 RREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMV 334
+++++ E++ K +L S E + +K+K K + E A K + Q + ++
Sbjct: 71 KKQEKKVIEEKKDGKPILK-----SKKEKEKEKKEKEKQKKKEQAARKKAQQQAQKEKNK 125
Query: 335 KGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFN-SLVETNLVFEAD 393
+ N Q++ A +A S KS + + S + + K+ + + L +
Sbjct: 126 ELNKQNVEKAAAEKAAAEKSQKSKGESDKPSASAKKPA---KKVPAGLAALRRQLELKKQ 182
Query: 394 REKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATM 453
E+ +K+ +E + LE ++ + E + + K+ E+ +RE K KA
Sbjct: 183 LEEQEKLEREEE------ERLEKEEEERLANEEKMKEEAKAAKKEKEKAKRE--KRKAEG 234
Query: 454 KLLDSANKVNEK-----KAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSS 508
KLL K +K +AA L+ N ++ + K K+ K K T N+S
Sbjct: 235 KLLTRKQKEEKKLLERRRAALLSSGNVKVAGLAKKDGEENKPKKVVYSKKKKRTTQENAS 294
Query: 509 NAELKNENSKLHSEV---SELKNS---VLDQFEAGFAKAKEQILFLNPQVSINLAGSDPY 562
A +K++ SK SEV ELK S ++D +E A + N + + A +
Sbjct: 295 EA-IKSD-SKKDSEVVPDDELKESEDVLIDDWE-NLALGDDDEEGTNEETQESTASHENE 351
Query: 563 ARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNE 597
+ + G+EE+EGEE++ EE+E + +E
Sbjct: 352 DQ---------NQGEEEEEGEEEEEEEEERAHVHE 377
Score = 45.4 bits (106), Expect = 4e-04
Identities = 74/343 (21%), Positives = 135/343 (38%), Gaps = 54/343 (15%)
Query: 290 TLLDADASHSGTES---------NRPQKKKRKNETPESAKGKD--SSQPSMEKFMVKGNP 338
T +A AS G E+ + +KK+ K E GK S+ EK +
Sbjct: 44 TSREASASAEGAEAIEGDFMSTLKQSKKKQEKKVIEEKKDGKPILKSKKEKEKEKKEKEK 103
Query: 339 QHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVK 398
Q +A A + K KE + E+ + + E + + + +K
Sbjct: 104 QKKKEQAARKKAQQQAQKEKNKELNKQNVEKAAAE-------KAAAEKSQKSKGESDKPS 156
Query: 399 KIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDS 458
K A + + GL AA+ + ++L+ + ++LE +E E L+ + +L +
Sbjct: 157 ASAKKPAKK--VPAGL--AALRRQLELKKQ----LEEQEKLEREEEERLEKEEEERLANE 208
Query: 459 ANKVNEKKAADLALEN-ERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENS 517
E KAA E +R K+ E +T+K KEE+ + + L+S N ++ +
Sbjct: 209 EKMKEEAKAAKKEKEKAKREKRKAEGKLLTRKQKEEKKLLERRRAALLSSGNVKVAGL-A 267
Query: 518 KLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDT-- 575
K E ++ K V ++K K++ N +I + D +L +
Sbjct: 268 KKDGEENKPKKVV-------YSKKKKRTTQENASEAIKSDSKKDSEVVPDDELKESEDVL 320
Query: 576 ---------GDEEDEGEEDKNEE--------DENVNDNEGEGE 601
GD+++EG ++ +E D+N + E EGE
Sbjct: 321 IDDWENLALGDDDEEGTNEETQESTASHENEDQNQGEEEEEGE 363
>MYS2_DICDI (P08799) Myosin II heavy chain, non muscle
Length = 2116
Score = 48.1 bits (113), Expect = 6e-05
Identities = 91/421 (21%), Positives = 167/421 (39%), Gaps = 64/421 (15%)
Query: 229 EEMVDCTFLSSIDLNLTEFLDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPA 288
EE+ + S D + E ++ L + ++ + D R++++ E +
Sbjct: 985 EELTESFSEESKDKGVLEKTRVRLQSELDDLTVRLDSETKDKSELLRQKKKLEEELKQVQ 1044
Query: 289 KTL-------LDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHM 341
+ L L +A++ + + ++ N + + S+ ++E +V N + +
Sbjct: 1045 EALAAETAAKLAQEAANKKLQGEYTELNEKFNSEVTARSNVEKSKKTLESQLVAVNNE-L 1103
Query: 342 TLKAGSSSAPPPSWKSL------LKEFEELTSEEVTSLWDSKIDFNSLVET--NLVFEAD 393
+ + A K+L +K+ E T E SL+D K+ S +E N + E
Sbjct: 1104 DEEKKNRDALEKKKKALDAMLEEMKDQLESTGGEKKSLYDLKVKQESDMEALRNQISELQ 1163
Query: 394 REKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQ-----LEEKEREILK 448
K +K + E+A + ++ E + K+ LE+K ++ +
Sbjct: 1164 STIAKLEKIKSTLEG------EVARLQGELEAEQLAKSNVEKQKKKVELDLEDKSAQLAE 1217
Query: 449 MKATMKLLDSANKVNEKKAADLALE----------NERLKKHVE--------DLNITQKA 490
A + LD K E++ +++ + ++ KH+E +L QKA
Sbjct: 1218 ETAAKQALDKLKKKLEQELSEVQTQLSEANNKNVNSDSTNKHLETSFNNLKLELEAEQKA 1277
Query: 491 K---EEELVKSKAEITHLNSSNAELK-----NENSK--LHSEVSELKNSVLDQFEAGFAK 540
K E++ + ++E+ H+N E K NE K L EVSELK DQ E A
Sbjct: 1278 KQALEKKRLGLESELKHVNEQLEEEKKQKESNEKRKVDLEKEVSELK----DQIEEEVAS 1333
Query: 541 AKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNEGEG 600
K N + S YA +V S D E+ + + KNEE N + E EG
Sbjct: 1334 KKAVTEAKNKKESELDEIKRQYADVVS----SRDKSVEQLKTLQAKNEELRNTAE-EAEG 1388
Query: 601 E 601
+
Sbjct: 1389 Q 1389
Score = 41.2 bits (95), Expect = 0.008
Identities = 66/310 (21%), Positives = 126/310 (40%), Gaps = 45/310 (14%)
Query: 245 TEFLDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESN 304
T+ +A+ +N + K S +++ +Q A++ + L+++ H +
Sbjct: 1241 TQLSEANNKNVNSDSTNKHLETSFNNLKLELEAEQKAKQALEKKRLGLESELKHVNEQLE 1300
Query: 305 RPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEE 364
+K+K NE + K+ S+ +K + + + +S L E +
Sbjct: 1301 EEKKQKESNEKRKVDLEKEVSE-------LKDQIEEEVASKKAVTEAKNKKESELDEIKR 1353
Query: 365 LTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMID 424
++ V+S S VE +A E+++ + Q L+ A SK
Sbjct: 1354 QYADVVSSRDKS-------VEQLKTLQAKNEELRNTAEEAEGQ------LDRAERSK--- 1397
Query: 425 LESADFDGINSAKQLEEKEREILKMKATMKL-----------LDSANKVNEKKAADLALE 473
+ A+FD + K LEE+ + +K + MK LD A V+ ++ +
Sbjct: 1398 -KKAEFDLEEAVKNLEEETAKKVKAEKAMKKAETDYRSTKSELDDAKNVSSEQYVQIKRL 1456
Query: 474 NER-------LKKHVEDLNITQKAK---EEELVKSKAEITHLNSSNAELKNENSKLHSEV 523
NE L++ E N KAK E L K EI N++ A+ + ++ +L V
Sbjct: 1457 NEELSELRSVLEEADERCNSAIKAKKTAESALESLKDEIDAANNAKAKAERKSKELEVRV 1516
Query: 524 SELKNSVLDQ 533
+EL+ S+ D+
Sbjct: 1517 AELEESLEDK 1526
Score = 36.2 bits (82), Expect = 0.25
Identities = 27/105 (25%), Positives = 54/105 (50%), Gaps = 12/105 (11%)
Query: 435 SAKQLEEKER-----EILKMKATMKLLDSANKVNEKKAADLALENERLKKHVED------ 483
S+K +E+ R E+ +++A + +A +EKK L E + +K+ +ED
Sbjct: 1636 SSKAADEEIRKQVWQEVDELRAQLDSERAALNASEKKIKSLVAEVDEVKEQLEDEILAKD 1695
Query: 484 -LNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELK 527
L ++A E EL + + ++ S +EL++ +L +EV ++K
Sbjct: 1696 KLVKAKRALEVELEEVRDQLEEEEDSRSELEDSKRRLTTEVEDIK 1740
Score = 35.4 bits (80), Expect = 0.43
Identities = 92/403 (22%), Positives = 155/403 (37%), Gaps = 72/403 (17%)
Query: 258 EYVAKMTRLSDDDILRFRREQQAA----REKRNPAKTLLDADASHSGTESNRPQ----KK 309
E++ K DD R RE ++ +K+N K D +A + KK
Sbjct: 1533 EFIRKKDAEIDDLRARLDRETESRIKSDEDKKNTRKQFADLEAKVEEAQREVVTIDRLKK 1592
Query: 310 KRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEE 369
K +++ + + D+ S K TL A +A S K+ +E + +E
Sbjct: 1593 KLESDIIDLSTQLDTETKSRIKIEKSKKKLEQTL-AERRAAEEGSSKAADEEIRKQVWQE 1651
Query: 370 VTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESAD 429
V L +++D A +K+K + + E + EI A K++ + A
Sbjct: 1652 VDEL-RAQLD-----SERAALNASEKKIKSL-VAEVDEVKEQLEDEILAKDKLVKAKRAL 1704
Query: 430 FDGINSAK-QLEEKE-----REILKMKATMKLLDSANKVNEKKAADLALEN--------- 474
+ + QLEE+E E K + T ++ D K + + + L+
Sbjct: 1705 EVELEEVRDQLEEEEDSRSELEDSKRRLTTEVEDIKKKYDAEVEQNTKLDEAKKKLTDDV 1764
Query: 475 ERLKKHVED----LNITQKAK-------EEELVKSKAEITHLNSSNAELKNENSKLHSEV 523
+ LKK +ED LN +++AK E+ L K AE+ N S AE + K ++
Sbjct: 1765 DTLKKQLEDEKKKLNESERAKKRLESENEDFLAKLDAEVK--NRSRAE--KDRKKYEKDL 1820
Query: 524 SELKNSVLD------QFEAGFAKAKEQILFLN---PQVSINLAGSDPYARIVDGKLISPD 574
+ K + D Q E G AK ++QI L Q +D + ++G+ I
Sbjct: 1821 KDTKYKLNDEAATKTQTEIGAAKLEDQIDELRSKLEQEQAKATQADKSKKTLEGE-IDNL 1879
Query: 575 TGDEEDEGE----------------EDKNEEDENVNDNEGEGE 601
EDEG+ E+ E E D++ E E
Sbjct: 1880 RAQIEDEGKIKMRLEKEKRALEGELEELRETVEEAEDSKSEAE 1922
Score = 35.0 bits (79), Expect = 0.56
Identities = 43/180 (23%), Positives = 77/180 (41%), Gaps = 28/180 (15%)
Query: 434 NSAKQLEEKEREILKMKATM-------KLLDSANKVNEKKAAD----LALENERLKKHVE 482
N K+++EKEREIL++K+ + L+ + K E D L E E LK +
Sbjct: 825 NFEKEIKEKEREILELKSNLTDSTTQKDKLEKSLKDTESNVLDLQRQLKAEKETLKAMYD 884
Query: 483 ----------DLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLD 532
+L I + E EL + K + +L + ++ + L E+ E + + +
Sbjct: 885 SKDALEAQKRELEIRVEDMESELDEKKLALENLQNQKRSVEEKVRDLEEELQE-EQKLRN 943
Query: 533 QFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDEN 592
E K +E+ L +N SD +R+ + I + E +E E +EE ++
Sbjct: 944 TLEKLKKKYEEE---LEEMKRVNDGQSDTISRL---EKIKDELQKEVEELTESFSEESKD 997
Score = 35.0 bits (79), Expect = 0.56
Identities = 25/96 (26%), Positives = 46/96 (47%)
Query: 438 QLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVK 497
++E+ E E+ + K ++ L + + E+K DL E + +K L +K EEEL +
Sbjct: 899 RVEDMESELDEKKLALENLQNQKRSVEEKVRDLEEELQEEQKLRNTLEKLKKKYEEELEE 958
Query: 498 SKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQ 533
K + + + L+ +L EV EL S ++
Sbjct: 959 MKRVNDGQSDTISRLEKIKDELQKEVEELTESFSEE 994
>MYSP_TAESA (Q8T305) Paramyosin
Length = 863
Score = 47.8 bits (112), Expect = 8e-05
Identities = 87/377 (23%), Positives = 151/377 (39%), Gaps = 69/377 (18%)
Query: 243 NLTEFLDAHYENRLGEYVAKMTRLSDDDIL--RFRREQQAAREKRNPA----------KT 290
+L +D NRL + D D L R+ E +AA RN KT
Sbjct: 226 DLKRAMDEDARNRLNLQTQLSSLQMDYDNLQARYEEEAEAAGNLRNQVAKFNADMAALKT 285
Query: 291 LLDADASHSGTESNRPQKKK---RKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGS 347
L+ + + TE K+K R E + A+ + + ++EK VK + L+A +
Sbjct: 286 RLERELM-AKTEEFEELKRKLTVRITELEDMAEHERTRANNLEKTKVKLTLEIKDLQAEN 344
Query: 348 SSAPPPSWKSL--LKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVK-KIG--- 401
+ + + +K+ E L +E + + ++ N+L N EAD ++K ++G
Sbjct: 345 EALAAENGELTHRVKQAENLANELQRRIDEMTVEINTLNSANSALEADNMRLKGQVGDLT 404
Query: 402 ----------------LKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKERE 445
LKE A+ + + + A+ D + SA L + E
Sbjct: 405 DRIANLDRENRQLGDQLKETKSALRDANRRLTDLEALRSQLEAERDNLASA--LHDAEEA 462
Query: 446 ILKMKATMKLLDSANKVNEKKAA--------DLALENERLK--KHVEDLNIT-------- 487
+ +M+A K + S N +N K+ D LEN R + +E+L T
Sbjct: 463 LKEMEA--KYVASQNALNHLKSEMEQRLREKDEELENLRKSTTRTIEELTTTISEMEVRF 520
Query: 488 -------QKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAK 540
+K E + + + ++ N +NA L EN L V EL+ ++ D+ A +
Sbjct: 521 KSDMSRLKKKYEATISELEVQLDVANKANANLNRENKTLAQRVQELQAALEDERRA--RE 578
Query: 541 AKEQILFLNPQVSINLA 557
A E L ++ + I LA
Sbjct: 579 AAESNLQVSERKRIALA 595
>SMC2_XENLA (P50533) Structural maintenance of chromosome 2
(Chromosome-associated protein E) (Chromosome assembly
protein XCAP-E)
Length = 1203
Score = 47.4 bits (111), Expect = 1e-04
Identities = 46/179 (25%), Positives = 86/179 (47%), Gaps = 10/179 (5%)
Query: 354 SWKSLLKEFEEL--TSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMT 411
S K ++E EE ++EV + K F L EA+RE+ LKEA Q + T
Sbjct: 748 SLKQTIEESEETLKNTKEVQKKAEEK--FKVLEHKMKNAEAERERE----LKEAQQKLDT 801
Query: 412 KGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLA 471
+ A +K + + + D + +LEE +RE K ++ +D A K +++A +A
Sbjct: 802 AKKKADASNKKMKEKQQEVDAL--VLELEELKREQTTYKQQIETVDEAMKAYQEQADSMA 859
Query: 472 LENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSV 530
E + K+ V+ ++E ++ EI +S +L+ N+ L ++ EL++++
Sbjct: 860 SEVSKNKEAVKKAQDELAKQKEIIMGHDKEIKTKSSEAGKLRENNNDLQLKIKELEHNI 918
Score = 31.6 bits (70), Expect = 6.2
Identities = 37/178 (20%), Positives = 80/178 (44%), Gaps = 21/178 (11%)
Query: 360 KEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAI 419
+E + ++EE+ + DS + + N KVK++G KE + + E+
Sbjct: 243 EETKVRSAEELKEMQDSILKLQDTMAEN------ERKVKELG-KEIAELEKMRDQEVGGA 295
Query: 420 SKMID-----LESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALEN 474
+ ++ + AD + SA L+++ + + K +L+ S E+ A L +
Sbjct: 296 LRSLEEALSEAQRADTK-VQSALDLKKQNMKAEREKKRKELVKSM----EEDAKVLTAKE 350
Query: 475 ERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKN----ENSKLHSEVSELKN 528
+ +KK + L+ Q+A ++++ + H N+ +A L + E + L ++ KN
Sbjct: 351 KEVKKITDGLSSLQEASQKDVEAFTSAQQHFNAVSAGLSSNEDGEEATLAGQMMACKN 408
>M21_STRPY (P50468) M protein, serotype 2.1 precursor
Length = 407
Score = 47.0 bits (110), Expect = 1e-04
Identities = 63/272 (23%), Positives = 124/272 (45%), Gaps = 24/272 (8%)
Query: 262 KMTRLSDDDILRFRRE-QQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAK 320
K+ + S+D + R+ Q +E++ K L + + R Q++ +K + E K
Sbjct: 98 KLEKKSEDVERHYLRQLDQEYKEQQERQKNLEELERQSQREVEKRYQEQLQKQQQLEKEK 157
Query: 321 G-KDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKID 379
++S+ S+ + + L+A LKE ++++ SL + D
Sbjct: 158 QISEASRKSLRRDLEASRAAKKDLEAEHQK---------LKEEKQISEASRKSL---RRD 205
Query: 380 FNSLVETNLVFEADREKVKKIG-LKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQ 438
+ EA+ +K+K+ + EA + +++ LE + +K DLE A+ + KQ
Sbjct: 206 LEASRAAKKDLEAEHQKLKEEKQISEASRQGLSRDLEASRAAKK-DLE-AEHQKLKEEKQ 263
Query: 439 LEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKS 498
+ E R+ L L+++ + +K ADLA N +L+ +E LN + ++ K
Sbjct: 264 ISEASRQGLSRD-----LEASREAKKKVEADLAEANSKLQA-LEKLNKELEEGKKLSEKE 317
Query: 499 KAEI-THLNSSNAELKNENSKLHSEVSELKNS 529
KAE+ L + LK + +K E+++LK +
Sbjct: 318 KAELQAKLEAEAKALKEQLAKQAEELAKLKGN 349
Score = 33.9 bits (76), Expect = 1.3
Identities = 38/162 (23%), Positives = 71/162 (43%), Gaps = 13/162 (8%)
Query: 391 EADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMK 450
+ +R ++++ + Q K LE ++E + + +QLE KE++I +
Sbjct: 105 DVERHYLRQLDQEYKEQQERQKNLEELERQSQREVEKRYQEQLQKQQQLE-KEKQI--SE 161
Query: 451 ATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNA 510
A+ K L + + DL E+++LK+ + ++K+ +L S+A L + +
Sbjct: 162 ASRKSLRRDLEASRAAKKDLEAEHQKLKEEKQISEASRKSLRRDLEASRAAKKDLEAEHQ 221
Query: 511 ELKNENSKLHSEVS--------ELKNSVLDQFEAGFAKAKEQ 544
+LK E K SE S E + EA K KE+
Sbjct: 222 KLKEE--KQISEASRQGLSRDLEASRAAKKDLEAEHQKLKEE 261
Score = 32.3 bits (72), Expect = 3.7
Identities = 56/278 (20%), Positives = 107/278 (38%), Gaps = 25/278 (8%)
Query: 274 FRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFM 333
F + +NP +A S + ++ K E E D + +K
Sbjct: 33 FANQTTVKANSKNPVPVKKEAKLSEAELHDKIKNLEEEKAELFEKL---DKVEEEHKKVE 89
Query: 334 VKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWD----SKIDFNSLVETNLV 389
+ H L+ S + L +E++E E +L + S+ + + L
Sbjct: 90 EEHKKDHEKLEKKSEDVERHYLRQLDQEYKE-QQERQKNLEELERQSQREVEKRYQEQLQ 148
Query: 390 FEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILK- 448
+ EK K+I EA + + + LE + +K DLE A+ + KQ+ E R+ L+
Sbjct: 149 KQQQLEKEKQIS--EASRKSLRRDLEASRAAKK-DLE-AEHQKLKEEKQISEASRKSLRR 204
Query: 449 -----------MKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVK 497
++A + L +++E L+ + E + +DL + +EE
Sbjct: 205 DLEASRAAKKDLEAEHQKLKEEKQISEASRQGLSRDLEASRAAKKDLEAEHQKLKEEKQI 264
Query: 498 SKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFE 535
S+A L+ + K+ ++++E NS L E
Sbjct: 265 SEASRQGLSRDLEASREAKKKVEADLAE-ANSKLQALE 301
>KTN1_HUMAN (Q86UP2) Kinectin (Kinesin receptor) (CG-1 antigen)
Length = 1357
Score = 47.0 bits (110), Expect = 1e-04
Identities = 65/316 (20%), Positives = 124/316 (38%), Gaps = 19/316 (6%)
Query: 243 NLTEFLDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNP--AKTLLDADASHSG 300
N T L++ L + RL ++ + + QQ +K+N A T L +
Sbjct: 435 NTTNQLESKQSAELNKLRQDYARLVNELTEKTGKLQQEEVQKKNAEQAATQLKVQLQEAE 494
Query: 301 TESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNP-QHMTLKAGSSSAPPPSWKSLL 359
Q RK +A+ + + Q KF+ K N Q + K + + L
Sbjct: 495 RRWEEVQSYIRKR----TAEHEAAQQDLQSKFVAKENEVQSLHSKLTDTLVSKQQLEQRL 550
Query: 360 KEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAI 419
+ E + V ++ ++E N +A ++ + +++ + L
Sbjct: 551 MQLMESEQKRVNKEESLQMQVQDILEQNEALKAQIQQFHSQIAAQTSASVLAEELHKVIA 610
Query: 420 SKMIDLESADFDGINSAKQLEEKEREI-------LKMKATMKLLDSANKVNEKKAADLAL 472
K ++ + + +L KE E+ +KA ++ L + NE+ AA L
Sbjct: 611 EKDKQIKQTEDSLASERDRLTSKEEELKDIQNMNFLLKAEVQKLQAL--ANEQAAAAHEL 668
Query: 473 ENERLKKHVED--LNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKN-S 529
E + +V+D + + ++ + E+ E LN N LK+E KL + VSE N
Sbjct: 669 EKMQQSVYVKDDKIRLLEEQLQHEISNKMEEFKILNDQNKALKSEVQKLQTLVSEQPNKD 728
Query: 530 VLDQFEAGFAKAKEQI 545
V++Q E + E++
Sbjct: 729 VVEQMEKCIQEKDEKL 744
Score = 45.1 bits (105), Expect = 5e-04
Identities = 53/213 (24%), Positives = 94/213 (43%), Gaps = 49/213 (23%)
Query: 354 SWKSLLKEFEELTSEEVTSLWDSKI-DFNSLVETNLVFEADREKVKKIGLKEACQAI--- 409
S+ SL++E +++ E+ D KI L+E L+ A++EK + LK+ +A+
Sbjct: 791 SFASLVEELKKVIHEK-----DGKIKSVEELLEAELLKVANKEKTVQ-DLKQEIKALKEE 844
Query: 410 -----MTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNE 464
+ K +++ SK+ +L++ L+ KE ++ MKA ++ +
Sbjct: 845 IGNVQLEKAQQLSITSKVQELQNL----------LKGKEEQMNTMKAVLEEKEKDLANTG 894
Query: 465 KKAADLALENERLKKHV-----------------EDLNITQKAKEEELVKSKAEITHLNS 507
K DL ENE LK HV E+L I K KE EL + +A + S
Sbjct: 895 KWLQDLQEENESLKAHVQEVAQHNLKEASSASQFEELEIVLKEKENELKRLEAMLKERES 954
Query: 508 SNA-------ELKNENSKLHSEVSELKNSVLDQ 533
+ ++++EN S++ +LK Q
Sbjct: 955 DLSSKTQLLQDVQDENKLFKSQIEQLKQQNYQQ 987
Score = 40.8 bits (94), Expect = 0.010
Identities = 56/247 (22%), Positives = 99/247 (39%), Gaps = 35/247 (14%)
Query: 336 GNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDF-------NSLVETNL 388
GN Q + S ++ ++LLK EE + L + + D L E N
Sbjct: 846 GNVQLEKAQQLSITSKVQELQNLLKGKEEQMNTMKAVLEEKEKDLANTGKWLQDLQEENE 905
Query: 389 VFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKEREILK 448
+A ++V + LKEA A + LEI K +L+ + L+E+E ++
Sbjct: 906 SLKAHVQEVAQHNLKEASSASQFEELEIVLKEKENELKRLE-------AMLKERESDLSS 958
Query: 449 MKATMKLLDSANKVNEKKAADLALENER----LKKHVEDLNITQKAKEE------ELVKS 498
++ + NK+ + + L +N + H E L + + ++E EL
Sbjct: 959 KTQLLQDVQDENKLFKSQIEQLKQQNYQQASSFPPHEELLKVISEREKEISGLWNELDSL 1018
Query: 499 KAEITHLNSSNAELKNEN-----------SKLHSEVSELKNSVLDQFEAGFAKAKEQILF 547
K + H N +L+ +N L +V++ Q EA +AKE +
Sbjct: 1019 KDAVEHQRKKNNDLREKNWEAMEALASTEKMLQDKVNKTSKERQQQVEAVELEAKEVLKK 1078
Query: 548 LNPQVSI 554
L P+VS+
Sbjct: 1079 LFPKVSV 1085
Score = 38.1 bits (87), Expect = 0.067
Identities = 41/197 (20%), Positives = 85/197 (42%), Gaps = 23/197 (11%)
Query: 359 LKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAA 418
+K+ E+ + E L + + + N + +A+ +K++ + ++A AA
Sbjct: 616 IKQTEDSLASERDRLTSKEEELKDIQNMNFLLKAEVQKLQALANEQA-----------AA 664
Query: 419 ISKMIDLESADFDGINSAKQLEEK-EREILKMKATMKLLDSANKVNE---KKAADLALEN 474
++ ++ + + + + LEE+ + EI K+L+ NK + +K L E
Sbjct: 665 AHELEKMQQSVYVKDDKIRLLEEQLQHEISNKMEEFKILNDQNKALKSEVQKLQTLVSEQ 724
Query: 475 ------ERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKN 528
E+++K +++ + K EE L ++ ++ ENS L EV +LK
Sbjct: 725 PNKDVVEQMEKCIQEKDEKLKTVEELLETGLIQVATKEEELNAIRTENSSLTKEVQDLKA 784
Query: 529 SVLDQFEAGFAKAKEQI 545
DQ FA E++
Sbjct: 785 KQNDQ--VSFASLVEEL 799
Score = 34.7 bits (78), Expect = 0.74
Identities = 71/359 (19%), Positives = 133/359 (36%), Gaps = 50/359 (13%)
Query: 277 EQQAAREK---RNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQPSMEKFM 333
E+Q +E + P K + + T + K+K +S G D +E M
Sbjct: 126 EEQVIKESDASKIPGKKVEPVPVTKQPTPPSEAAASKKKPGQKKSKNGSDDQDKKVETLM 185
Query: 334 VKGN-----PQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEV------TSLWDSKIDFNS 382
V P H K S S + K E + L D+ D +
Sbjct: 186 VPSKRQEALPLHQETKQESGSGKKKASSKKQKTENVFVDEPLIHATTYIPLMDN-ADSSP 244
Query: 383 LVETNLVFEADR----EKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADF---DGINS 435
+V+ V + + E ++K G K+ + E+ ++ L++ F + +
Sbjct: 245 VVDKREVIDLLKPDQVEGIQKSGTKKLKTETDKENAEVKFKDFLLSLKTMMFSEDEALCV 304
Query: 436 AKQLEEKE---REILKMKATMKLLDSANKVNEKK------AADLALENERLKKHVEDLNI 486
L+EK ++ LK + +L +++ EK D A +R K+ +++
Sbjct: 305 VDLLKEKSGVIQDALKKSSKGELTTLIHQLQEKDKLLAAVKEDAAATKDRCKQLTQEMMT 364
Query: 487 TQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELK---NSVLDQFEAGFAKAKE 543
++ + + K I L + +N+ + E +++ V +Q EA A K+
Sbjct: 365 EKERSNVVITRMKDRIGTLEKEHNVFQNKIHVSYQETQQMQMKFQQVREQMEAEIAHLKQ 424
Query: 544 QILFLNPQVS-------------INLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEE 589
+ L VS +N D YAR+V+ ++ TG + E + KN E
Sbjct: 425 ENGILRDAVSNTTNQLESKQSAELNKLRQD-YARLVNE--LTEKTGKLQQEEVQKKNAE 480
>GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member 4
(Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (72.1
protein)
Length = 2230
Score = 47.0 bits (110), Expect = 1e-04
Identities = 58/208 (27%), Positives = 93/208 (43%), Gaps = 25/208 (12%)
Query: 391 EADREKVKK------IGLKEACQAIMTKGLEIAAISKMIDLESAD--FDGINSAKQLEEK 442
E D+ K++K LK IM LE K I++ES + N K +E K
Sbjct: 1503 EDDKSKMEKKESNLETELKSQTARIME--LEDHITQKTIEIESLNEVLKNYNQQKDIEHK 1560
Query: 443 EREILKMKATMKLLDSA-NKVNEKKAADLALEN---------ERLKKHVEDLNITQKAKE 492
E + K++ +L + N+V E + L LEN E KK +E +N++ K+KE
Sbjct: 1561 EL-VQKLQHFQELGEEKDNRVKEAEEKILTLENQVYSMKAELETKKKELEHVNLSVKSKE 1619
Query: 493 EELVKSKAEITHLNS-SNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAKEQILFLNPQ 551
EEL KA L S S A+L K +++ +K +L Q E + K+ +
Sbjct: 1620 EEL---KALEDRLESESAAKLAELKRKAEQKIAAIKKQLLSQMEEKEEQYKKGTESHLSE 1676
Query: 552 VSINLAGSDPYARIVDGKLISPDTGDEE 579
++ L + I++ KL S ++ E
Sbjct: 1677 LNTKLQEREREVHILEEKLKSVESSQSE 1704
Score = 42.4 bits (98), Expect = 0.004
Identities = 69/290 (23%), Positives = 116/290 (39%), Gaps = 59/290 (20%)
Query: 248 LDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQ 307
L+ YE + KM ++ +Q+A + K LLD +A
Sbjct: 941 LNEEYETKFKNQEKKMEKV----------KQKAKEMQETLKKKLLDQEAK---------L 981
Query: 308 KKKRKNETPE-SAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELT 366
KK+ +N E S K K + +E M + N AG S A + ++ E LT
Sbjct: 982 KKELENTALELSQKEKQFNAKMLE--MAQAN------SAGISDAVSRLETNQKEQIESLT 1033
Query: 367 S------EEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAIS 420
+V S+W+ K++ + + +++ +I L+E K E+A +
Sbjct: 1034 EVHRRELNDVISIWEKKLNQQA---------EELQEIHEIQLQE-------KEQEVAELK 1077
Query: 421 KMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKH 480
+ I L + + +N K++ + E +K T+ L K LA + +LK H
Sbjct: 1078 QKILLFGCEKEEMN--KEITWLKEEGVKQDTTLNELQEQLKQKSAHVNSLAQDETKLKAH 1135
Query: 481 VEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSV 530
+E L E +L KS E T L ELK + +VSEL + +
Sbjct: 1136 LEKL-------EVDLNKSLKENTFLQEQLVELKMLAEEDKRKVSELTSKL 1178
Score = 37.4 bits (85), Expect = 0.11
Identities = 38/119 (31%), Positives = 58/119 (47%), Gaps = 22/119 (18%)
Query: 438 QLEEKEREILKMKATMKLLDSANKVNEKKAAD---LALENE----RLKKHV-EDLNITQK 489
QLEEKE +I MKA ++ L + + +K+ + A E E +LKK + E++N
Sbjct: 1298 QLEEKENQIKSMKADIESLVTEKEALQKEGGNQQQAASEKESCITQLKKELSENINAVTL 1357
Query: 490 AKEEELVKSKAEITHLNSS----NAELKNE---------NSKLHSEVSELKNSVLDQFE 535
K EEL + K EI+ L+ N +L+N S L + E K +LDQ +
Sbjct: 1358 MK-EELKEKKVEISSLSKQLTDLNVQLQNSISLSEKEAAISSLRKQYDEEKCELLDQVQ 1415
Score = 33.9 bits (76), Expect = 1.3
Identities = 38/188 (20%), Positives = 84/188 (44%), Gaps = 27/188 (14%)
Query: 363 EELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKM 422
E+ E+ T L D +I F + +E E + + ++K+ +K+ E+ ++S
Sbjct: 647 EKCEQEKETLLKDKEIIFQAHIE-----EMNEKTLEKLDVKQT---------ELESLS-- 690
Query: 423 IDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVE 482
++ + A+ E+E +LK + + K++E+K + +K+H
Sbjct: 691 -----SELSEVLKARHKLEEELSVLKDQTDKMKQELEAKMDEQKNHHQQQVDSIIKEHEV 745
Query: 483 DLNITQKAKEEEL----VKSKAEITHLNSSNAELKNENSKLHSEVSELK--NSVLDQFEA 536
+ T+KA ++++ + K HL A ++N + + EL+ ++ LD F++
Sbjct: 746 SIQRTEKALKDQINQLELLLKERDKHLKEHQAHVENLEADIKRSEGELQQASAKLDVFQS 805
Query: 537 GFAKAKEQ 544
+ EQ
Sbjct: 806 YQSATHEQ 813
Score = 33.5 bits (75), Expect = 1.6
Identities = 50/223 (22%), Positives = 89/223 (39%), Gaps = 30/223 (13%)
Query: 331 KFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFE---ELTSEEVTSLWDSKIDFNSLVET- 386
K ++ + + TL A L+E E +L E T L D +L+E
Sbjct: 299 KETIQSHKEQCTLLTSEKEALQEQLDERLQELEKIKDLHMAEKTKLITQLRDAKNLIEQL 358
Query: 387 -----NLVFEADREKVKKIGLKEA--------CQAIMTKGLEIAAISKMIDLESADFDGI 433
++ E R+ + + +KE + + T+G E+ + E A F+ +
Sbjct: 359 EQDKGMVIAETKRQMHETLEMKEEEIAQLRSRIKQMTTQGEELREQKEKS--ERAAFEEL 416
Query: 434 NSAKQLEEKEREIL-KMKATM-KLLDSANKVNEKKAADLALENERLKKHV---------E 482
A +K E K+KA M + + + K +E++ L E R+K+ V E
Sbjct: 417 EKALSTAQKTEEARRKLKAEMDEQIKTIEKTSEEERISLQQELSRVKQEVVDVMKKSSEE 476
Query: 483 DLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSE 525
+ QK E+EL + + E+T + E K+ E S+
Sbjct: 477 QIAKLQKLHEKELARKEQELTKKLQTREREFQEQMKVALEKSQ 519
Score = 32.3 bits (72), Expect = 3.7
Identities = 56/311 (18%), Positives = 125/311 (40%), Gaps = 26/311 (8%)
Query: 248 LDAHYENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQ 307
L+A + + + ++ + + + +R ++A +++ N + LL H + ++
Sbjct: 721 LEAKMDEQKNHHQQQVDSIIKEHEVSIQRTEKALKDQINQLELLLKERDKH--LKEHQAH 778
Query: 308 KKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEE-LT 366
+ + + S + ++ F + H KA K L E E L
Sbjct: 779 VENLEADIKRSEGELQQASAKLDVFQSYQSATHEQTKAYEEQLAQLQQKLLDLETERILL 838
Query: 367 SEEVTSLWDSKIDFNSLVETNLVFEAD------------REKVKKIGLKEACQAIMTKGL 414
+++V + K D + ++ + + D +KVK L + ++ + G
Sbjct: 839 TKQVAEVEAQKKDVCTELDAHKIQVQDLMQQLEKQNSEMEQKVKS--LTQVYESKLEDGN 896
Query: 415 EIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALEN 474
+ +K I +E + I ++ ++KE EIL K + K DS + +NE+ +
Sbjct: 897 KEQEQTKQILVEKENM--ILQMREGQKKEIEILTQKLSAKE-DSIHILNEEYETKFKNQE 953
Query: 475 ERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQF 534
++++K + QKAKE + K + EL+N +L + + +L+
Sbjct: 954 KKMEK------VKQKAKEMQETLKKKLLDQEAKLKKELENTALELSQKEKQFNAKMLEMA 1007
Query: 535 EAGFAKAKEQI 545
+A A + +
Sbjct: 1008 QANSAGISDAV 1018
>USO1_YEAST (P25386) Intracellular protein transport protein USO1
Length = 1790
Score = 46.6 bits (109), Expect = 2e-04
Identities = 58/259 (22%), Positives = 110/259 (42%), Gaps = 27/259 (10%)
Query: 309 KKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSE 368
++ KNE+ + SM + ++ ++ GS K + + E+ E
Sbjct: 986 EESKNESSIQLSNLQNKIDSMSQ-----EKENFQIERGSIEKNIEQLKKTISDLEQTKEE 1040
Query: 369 EVTSLWDSKIDFNSLV-----ETNLVFEADREKVKKIG----LKEACQAIMT--KGLEIA 417
++ SK ++ S + + A+ E V KI +E +A + K L+
Sbjct: 1041 IISKSDSSKDEYESQISLLKEKLETATTANDENVNKISELTKTREELEAELAAYKNLKNE 1100
Query: 418 AISKMIDLESADFDGINSAKQLEEK----EREILKMKATMKLLDSANKVNEKKAADLALE 473
+K+ E A + + + L+E+ E+E + K + L + + EK+ DLA +
Sbjct: 1101 LETKLETSEKALKEVKENEEHLKEEKIQLEKEATETKQQLNSLRANLESLEKEHEDLAAQ 1160
Query: 474 NERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQ 533
LKK+ E + ++ EE+ + EIT N +K +N +L EV +K++ +Q
Sbjct: 1161 ---LKKYEEQIANKERQYNEEISQLNDEITSTQQENESIKKKNDELEGEVKAMKSTSEEQ 1217
Query: 534 FEAGFAKAKEQILFLNPQV 552
K +I LN Q+
Sbjct: 1218 SNL----KKSEIDALNLQI 1232
Score = 42.4 bits (98), Expect = 0.004
Identities = 83/419 (19%), Positives = 173/419 (40%), Gaps = 67/419 (15%)
Query: 216 EAIIKLPDDSLTPEEMVDCTF--LSSIDLNLTEFLDAHY------ENRLGEYVAKMTRLS 267
+ +I+L +++ + +D T L + L+ E L+ ++ + Y K+TR +
Sbjct: 1392 DELIRLQNENELKAKEIDNTRSELEKVSLSNDELLEEKQNTIKSLQDEILSYKDKITR-N 1450
Query: 268 DDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRKNETPESAKGKDSSQP 327
D+ +L R+ + R+ + + L A S + E KK + E+ + + S+
Sbjct: 1451 DEKLLSIERDNK--RDLESLKEQLRAAQESKAKVEEGL---KKLEEESSKEKAELEKSKE 1505
Query: 328 SMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETN 387
M+K LK+ + K L++ ++ E++ +L K D S + +
Sbjct: 1506 MMKKLESTIESNETELKSSMETIRKSDEK--LEQSKKSAEEDIKNLQHEKSDLISRINES 1563
Query: 388 LVFEADREKVK---KIGLKEACQAIMTKGLEIAAISKMIDLESADFDGINSAKQLEEKER 444
E D E++K +I K + + T E+ + I + + + + S +LE+ ER
Sbjct: 1564 ---EKDIEELKSKLRIEAKSGSE-LETVKQELNNAQEKIRINAEENTVLKS--KLEDIER 1617
Query: 445 EILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITH 504
E+ +A +K + EK+ L RLK+ ++L+ TQ+ ++ + +AE+
Sbjct: 1618 ELKDKQAEIK-----SNQEEKE-----LLTSRLKELEQELDSTQQKAQKSEEERRAEVRK 1667
Query: 505 LNSSNAELKNENSKLHSEVSELKN-------------SVLDQFEAGFAKAKEQILFLNPQ 551
++L + L ++ ++L N D K +++ L +
Sbjct: 1668 FQVEKSQLDEKAMLLETKYNDLVNKEQAWKRDEDTVKKTTDSQRQEIEKLAKELDNLKAE 1727
Query: 552 VSINLAGSDPYARIVDGKLISPD-------------------TGDEEDEGEEDKNEEDE 591
S ++ + I D L+ D + DEED+ E+D+ +E+E
Sbjct: 1728 NSKLKEANEDRSEIDDLMLLVTDLDEKNAKYRSKLKDLGVEISSDEEDDEEDDEEDEEE 1786
Score = 42.0 bits (97), Expect = 0.005
Identities = 69/375 (18%), Positives = 154/375 (40%), Gaps = 53/375 (14%)
Query: 254 NRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKK---- 309
N+ +Y ++++L+D+ I ++E ++ ++K + + + A S S +SN + +
Sbjct: 1170 NKERQYNEEISQLNDE-ITSTQQENESIKKKNDELEGEVKAMKSTSEEQSNLKKSEIDAL 1228
Query: 310 -------KRKNETPE-----SAKGKDSSQPSMEKFMVKGNPQHMTL-----KAGSSSAPP 352
K+KNET E S K +S +++ + N + + K +S
Sbjct: 1229 NLQIKELKKKNETNEASLLESIKSVESETVKIKELQDECNFKEKEVSELEDKLKASEDKN 1288
Query: 353 PSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTK 412
+ L KE E++ E + KI + + E ++ ++ K + +
Sbjct: 1289 SKYLELQKESEKIKEELDAKTTELKIQLEKITNLSKAKEKSESELSRLK-KTSSEERKNA 1347
Query: 413 GLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLAL 472
++ + I +++ F EKER++L ++ + + K+N + + L
Sbjct: 1348 EEQLEKLKNEIQIKNQAF----------EKERKLLNEGSSTITQEYSEKINTLEDELIRL 1397
Query: 473 ENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAEL----KNENSKLHSEVSELKN 528
+NE + K +E+ +++E+ ++ SN EL +N L E+ K+
Sbjct: 1398 QNE------------NELKAKEIDNTRSELEKVSLSNDELLEEKQNTIKSLQDEILSYKD 1445
Query: 529 SVLDQFEAGFAKAKEQILFLNPQVSINLAGSDPYARIVDG-KLISPDTGDEEDEGEEDK- 586
+ E + ++ L A + A++ +G K + ++ E+ E E+ K
Sbjct: 1446 KITRNDEKLLSIERDNKRDLESLKEQLRAAQESKAKVEEGLKKLEEESSKEKAELEKSKE 1505
Query: 587 --NEEDENVNDNEGE 599
+ + + NE E
Sbjct: 1506 MMKKLESTIESNETE 1520
Score = 34.3 bits (77), Expect = 0.96
Identities = 62/251 (24%), Positives = 109/251 (42%), Gaps = 26/251 (10%)
Query: 301 TESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVK----GNPQHMTLKAGSSSAPPPSWK 356
T S+ Q K+ +S+K + SQ S+ K ++ N +++ K + +
Sbjct: 1030 TISDLEQTKEEIISKSDSSKDEYESQISLLKEKLETATTANDENVN-KISELTKTREELE 1088
Query: 357 SLLKEFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEI 416
+ L ++ L +E T L S+ + E + K +KI L++ +A TK
Sbjct: 1089 AELAAYKNLKNELETKLETSEKALKEVKENE-----EHLKEEKIQLEK--EATETKQ--- 1138
Query: 417 AAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENER 476
S +LES + + + A QL++ E +I + + +++N++ ENE
Sbjct: 1139 QLNSLRANLESLEKEHEDLAAQLKKYEEQIANKERQYN--EEISQLNDE-ITSTQQENES 1195
Query: 477 LKKHVEDLNITQKA----KEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLD 532
+KK ++L KA EE+ K+EI LN ELK +N + + E SV
Sbjct: 1196 IKKKNDELEGEVKAMKSTSEEQSNLKKSEIDALNLQIKELKKKNETNEASLLESIKSV-- 1253
Query: 533 QFEAGFAKAKE 543
E+ K KE
Sbjct: 1254 --ESETVKIKE 1262
Score = 32.0 bits (71), Expect = 4.8
Identities = 40/217 (18%), Positives = 87/217 (39%), Gaps = 20/217 (9%)
Query: 330 EKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVTSLWDSKIDFNSLVETNLV 389
EK+ + N H +LK S K++ +E+T K + +L+E
Sbjct: 776 EKYQIL-NSSHSSLKENFSILET-ELKNVRDSLDEMTQLRDVLETKDKENQTALLEYKST 833
Query: 390 FEADREKVKKI--GL-------KEACQAIMTKGLEIAAISKMIDL---------ESADFD 431
+ +K + GL K+A I G ++ A+S+ + + D
Sbjct: 834 IHKQEDSIKTLEKGLETILSQKKKAEDGINKMGKDLFALSREMQAVEENCKNLQKEKDKS 893
Query: 432 GINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALENERLKKHVEDLNITQKAK 491
+N K+ + + +I +K ++ + + + +L+ E E + K + + ++
Sbjct: 894 NVNHQKETKSLKEDIAAKITEIKAINENLEEMKIQCNNLSKEKEHISKELVEYKSRFQSH 953
Query: 492 EEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKN 528
+ + K ++ L ++ +++ EN L V E KN
Sbjct: 954 DNLVAKLTEKLKSLANNYKDMQAENESLIKAVEESKN 990
>MLP1_YEAST (Q02455) MLP1 protein (Myosin-like protein 1)
Length = 1875
Score = 46.6 bits (109), Expect = 2e-04
Identities = 51/195 (26%), Positives = 91/195 (46%), Gaps = 11/195 (5%)
Query: 357 SLLKEFEEL--TSEEVTSLWDSKIDFNSLVETNLVFEADREKVK---KIGLKEACQAIMT 411
S +KE+++L T+ + +SK+D + TN + EK KI L + +
Sbjct: 921 SQIKEYKDLYETTSQSLQQTNSKLDESFKDFTNQIKNLTDEKTSLEDKISLLKEQMFNLN 980
Query: 412 KGLEIAAISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKA-ADL 470
L++ K ++ E ADF S Q KE E +K + KL N ++++ A+
Sbjct: 981 NELDLQ--KKGMEKEKADFKKRISILQNNNKEVEAVKSEYESKLSKIQNDLDQQTIYANT 1038
Query: 471 ALEN--ERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKN 528
A N + L+KH D++ T E+L K ++ LN S +L+N + S K
Sbjct: 1039 AQNNYEQELQKHA-DVSKTISELREQLHTYKGQVKTLNLSRDQLENALKENEKSWSSQKE 1097
Query: 529 SVLDQFEAGFAKAKE 543
S+L+Q + ++ ++
Sbjct: 1098 SLLEQLDLSNSRIED 1112
Score = 33.9 bits (76), Expect = 1.3
Identities = 32/129 (24%), Positives = 62/129 (47%), Gaps = 11/129 (8%)
Query: 427 SADFDGINSAKQLEEKEREILKMKATMKLLDS---ANKVN----EKKAADLALENERLKK 479
S + G+N+ +ER+IL K T+ D+ K++ E + A L+N R++K
Sbjct: 1137 STNGPGLNNILITLRRERDILDTKVTVAERDAKMLRQKISLMDVELQDARTKLDNSRVEK 1196
Query: 480 HVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFA 539
I Q +++++ ++ L SN L+NE +++ EL+ S LD+ + A
Sbjct: 1197 ENHSSIIQQ---HDDIMEKLNQLNLLRESNITLRNELENNNNKKKELQ-SELDKLKQNVA 1252
Query: 540 KAKEQILFL 548
+ ++ L
Sbjct: 1253 PIESELTAL 1261
>SMC4_MOUSE (Q8CG47) Structural maintenance of chromosomes 4-like 1
protein (Chromosome-associated polypeptide C) (XCAP-C
homolog)
Length = 1286
Score = 46.2 bits (108), Expect = 2e-04
Identities = 65/285 (22%), Positives = 118/285 (40%), Gaps = 32/285 (11%)
Query: 253 ENRLGEYVAKMTRLSDDDILRFRREQQAAREKRNPAKTLLDADASHSGTESNRPQKKKRK 312
+NR+ E + ++ +D + EK N + A S + + +KK
Sbjct: 329 QNRIAEITTQKEKIHEDT--------KEITEKSNVLSNEMKAKNS-----AVKDVEKKLN 375
Query: 313 NETPESAKGKDS-SQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLKEFEELTSEEVT 371
T + K+ +Q +E V+ +H T KA K L K+ E++ EE+
Sbjct: 376 KVTKFIEQNKEKFTQLDLEDVQVREKLKHATSKAKKLE------KQLQKDKEKV--EELK 427
Query: 372 SL-WDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMIDLESADF 430
S+ SK N N E +REK +K LKE ++ E + K +++ +
Sbjct: 428 SVPAKSKTVINETTTRNNSLEKEREKEEK-KLKEVMDSLKQ---ETQGLQKEKEIQEKEL 483
Query: 431 DGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLAL--ENERLKKH---VEDLN 485
G N + + E+ + + + L V++ A AL +E LK+ ++D+N
Sbjct: 484 MGFNKSVNEARSKMEVAQSELDIYLSRHNTAVSQLSKAKEALITASETLKERKAAIKDIN 543
Query: 486 ITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSV 530
++EL + + E+ L LK+ L +V E K+S+
Sbjct: 544 TKLPQTQQELKEKEKELQKLTQEEINLKSLVHDLFQKVEEAKSSL 588
>CALD_HUMAN (Q05682) Caldesmon (CDM)
Length = 793
Score = 46.2 bits (108), Expect = 2e-04
Identities = 48/180 (26%), Positives = 76/180 (41%), Gaps = 13/180 (7%)
Query: 426 ESADFDGINSAKQLEEKEREILKMKATMKLLDSANK---VNEKKAADLALENERLKKHVE 482
E A + KQLEEK+R + + K + ++ + VNEKKA + L+ LKK E
Sbjct: 379 EKAKVEEQKRNKQLEEKKRAMQETKIKGEKVEQKIEGKWVNEKKAQEDKLQTAVLKKQGE 438
Query: 483 DLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVSELKNSVLDQFEAGFAKAK 542
+ +AK E+L + K E+K+E K E E S +D+ + GF + K
Sbjct: 439 EKGTKVQAKREKLQEDKPTF-----KKEEIKDEKIKKDKEPKEEVKSFMDR-KKGFTEVK 492
Query: 543 EQILFLNPQVSINLAGSDPYARIVDGKLISPDTGDEEDEGEEDKNEEDENVNDNEGEGEN 602
Q N + + G S DT + E + + + E + GE E+
Sbjct: 493 SQ----NGEFMTHKLKHTENTFSRPGGRASVDTKEAEGAPQVEAGKRLEELRRRRGETES 548
>RA50_SULSO (Q97WH0) DNA double-strand break repair rad50 ATPase
Length = 864
Score = 45.8 bits (107), Expect = 3e-04
Identities = 47/173 (27%), Positives = 79/173 (45%), Gaps = 32/173 (18%)
Query: 378 IDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAISKMID------------- 424
ID + + T +V + + +K+ E Q IM K L++ I K+ID
Sbjct: 118 IDKDIALSTIIVRQGELDKIL-----ENFQEIMGKILKLELIEKLIDSRGPIVEFRKNLE 172
Query: 425 -----LESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAADLALE-NERLK 478
L+ + D N K +EEK +L++K + L+ K EK+ D+ + +E K
Sbjct: 173 NKLRELDRIEQDYNNFKKTVEEKRARVLELKKDKEKLEDEIKNLEKRIKDIKDQFDEYEK 232
Query: 479 KHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNEN---SKLHSEVSELKN 528
K + L +T K +E E+ LN S EL+ + +L E++EL+N
Sbjct: 233 KRNQYLKLTTTLKIKE-----GELNELNRSIEELRKQTENMDQLEKEINELEN 280
Score = 37.4 bits (85), Expect = 0.11
Identities = 41/201 (20%), Positives = 88/201 (43%), Gaps = 18/201 (8%)
Query: 358 LLKEFEELTS-----EEVTSLWDSK---IDFNSLVETNLVFEADREKVKKIGLKEACQAI 409
+L+ F+E+ E + L DS+ ++F +E N + E DR + K+ +
Sbjct: 137 ILENFQEIMGKILKLELIEKLIDSRGPIVEFRKNLE-NKLRELDRIEQDYNNFKKTVEEK 195
Query: 410 MTKGLEIAAISKMID-----LESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNE 464
+ LE+ + ++ LE D + + E+K + LK+ T+K+ +
Sbjct: 196 RARVLELKKDKEKLEDEIKNLEKRIKDIKDQFDEYEKKRNQYLKLTTTLKIKEGELNELN 255
Query: 465 KKAADLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLHSEVS 524
+ +L + E + + +++N + + +L K E+ L S+ E+ L E+
Sbjct: 256 RSIEELRKQTENMDQLEKEINELENLRNIKLKFEKYEV--LAKSHTEMSANVINLEKEIE 313
Query: 525 ELKNSV--LDQFEAGFAKAKE 543
E + ++ ++ E + K KE
Sbjct: 314 EYEKAIRRKEELEPKYLKYKE 334
Score = 31.6 bits (70), Expect = 6.2
Identities = 46/197 (23%), Positives = 78/197 (39%), Gaps = 38/197 (19%)
Query: 355 WKSLLKEFEELTSE-----EVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAI 409
+K L ++ EEL + ++ S DSK++ +E + E + K L++ +
Sbjct: 332 YKELERKLEELQPKYQQYLKLKSDLDSKLNLKERLEKD-ASELSNDIDKVNSLEQKVEET 390
Query: 410 MTKGLEIAA-ISKMIDLESADFDGINSAKQLEEKEREILKMKATMKLLDSANKVNEKKAA 468
K L + A ++K+ L S + IN+ Q+E + + + LD +K K A
Sbjct: 391 RKKQLNLRAQLAKVESLISEKNEIINNISQVEGETCPVCG-----RPLDEEHKQKIIKEA 445
Query: 469 DLALENERLKKHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKN-------------- 514
K ++ L + + EEEL K E+ + L N
Sbjct: 446 ---------KSYILQLELNKNELEEELKKITNELNKIEREYRRLSNNKASYDNVMRQLKK 496
Query: 515 ---ENSKLHSEVSELKN 528
E LHSE+ LKN
Sbjct: 497 LNEEIENLHSEIESLKN 513
>NUM1_YEAST (Q00402) Nuclear migration protein NUM1
Length = 2748
Score = 45.8 bits (107), Expect = 3e-04
Identities = 59/244 (24%), Positives = 99/244 (40%), Gaps = 21/244 (8%)
Query: 301 TESNRPQKKKRKNETPESAKGKDSSQPSMEKFMVKGNPQHMTLKAGSSSAPPPSWKSLLK 360
+ +NR +K K+ + + S E G + LKA + K+
Sbjct: 2 SHNNRHKKNNDKDSSAGQYANSIDNSLSQESVSTNGVTRMANLKADECGSGDEGDKTKRF 61
Query: 361 EFEELTSEEVTSLWDSKIDFNSLVETNLVFEADREKVKKIGLKEACQAIMTKGLEIAAIS 420
+ S+ T D +F N V A+ K L+ +++ L +
Sbjct: 62 SISSILSKRETK--DVLPEFAGSSSHNGVLTANSSKDMNFTLE------LSENL-LVECR 112
Query: 421 KMIDLESADFDGINSAKQLEEK-----EREILKMKATMKLLDSANKVN---EKKAADLAL 472
K+ A + I S KQ++E E + K+ MK LDS +N E K +L++
Sbjct: 113 KLQSSNEAKNEQIKSLKQIKESLSDKIEELTNQKKSFMKELDSTKDLNWDLESKLTNLSM 172
Query: 473 ENERLK--KHVEDLNITQKAKEEELVKSKAEITHLNSSNAELKNENSKLH--SEVSELKN 528
E +LK K + + + + +L+K+ EI L + E + KLH E+SELK
Sbjct: 173 ECRQLKELKKKTEKSWNDEKESLKLLKTDLEILTLTKNGMENDLSSQKLHYDKEISELKE 232
Query: 529 SVLD 532
+LD
Sbjct: 233 RILD 236
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 71,845,279
Number of Sequences: 164201
Number of extensions: 3216479
Number of successful extensions: 19774
Number of sequences better than 10.0: 767
Number of HSP's better than 10.0 without gapping: 149
Number of HSP's successfully gapped in prelim test: 631
Number of HSP's that attempted gapping in prelim test: 16277
Number of HSP's gapped (non-prelim): 2679
length of query: 603
length of database: 59,974,054
effective HSP length: 116
effective length of query: 487
effective length of database: 40,926,738
effective search space: 19931321406
effective search space used: 19931321406
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Lotus: description of TM0152.2


