

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
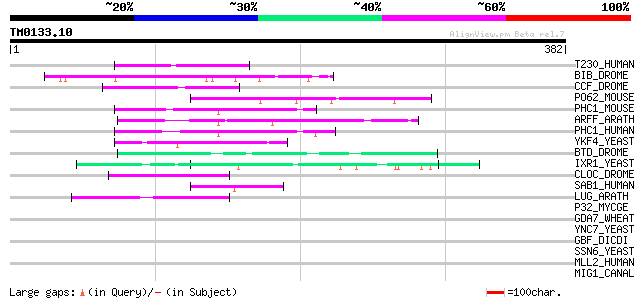
Query= TM0133.10
(382 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
T230_HUMAN (Q93074) Thyroid hormone receptor-associated protein ... 59 3e-08
BIB_DROME (P23645) Neurogenic protein big brain 53 1e-06
CCF_DROME (P41046) Centrosomal and chromosomal factor (CCF) (Chr... 47 6e-05
PO62_MOUSE (Q8BJI4) POU domain, class 6, transcription factor 2 47 8e-05
PHC1_MOUSE (Q64028) Polyhomeotic-like protein 1 (Early developme... 47 1e-04
ARFF_ARATH (Q9ZTX8) Auxin response factor 6 47 1e-04
PHC1_HUMAN (P78364) Polyhomeotic-like protein 1 (Early developme... 46 1e-04
YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4... 46 2e-04
BTD_DROME (Q24266) Transcription factor BTD (Buttonhead protein) 45 3e-04
IXR1_YEAST (P33417) Intrastrand crosslink recognition protein (S... 45 4e-04
CLOC_DROME (O61735) Circadian locomoter output cycles Kaput prot... 45 4e-04
SAB1_HUMAN (Q01826) DNA-binding protein SATB1 (Special AT-rich s... 44 7e-04
LUG_ARATH (Q9FUY2) Transcriptional corepressor LEUNIG 44 7e-04
P32_MYCGE (Q49417) P32 adhesin (Cytadhesin P32) 43 0.001
GDA7_WHEAT (P04727) Alpha/beta-gliadin clone PW8142 precursor (P... 43 0.001
YNC7_YEAST (P53968) Hypothetical 76.3 kDa zinc finger protein in... 43 0.002
GBF_DICDI (P36417) G-box binding factor (GBF) 43 0.002
SSN6_YEAST (P14922) Glucose repression mediator protein 42 0.002
MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia p... 42 0.002
MIG1_CANAL (Q9Y7G2) Regulatory protein MIG1 42 0.002
>T230_HUMAN (Q93074) Thyroid hormone receptor-associated protein
complex 230 kDa component (Trap230) (Activator-recruited
cofactor 240 KDa component) (ARC240) (CAG repeat protein
45) (OPA-containing protein) (Trinucleotide repeat
containing 11)
Length = 2212
Score = 58.5 bits (140), Expect = 3e-08
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 2093 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 2149
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 2150 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2182
Score = 39.3 bits (90), Expect = 0.017
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 2127 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2186
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2187 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2212
Score = 32.7 bits (73), Expect = 1.6
Identities = 16/31 (51%), Positives = 19/31 (60%)
Query: 121 ASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
A + E +QQQQQQ QQQQQQ+ Q Q
Sbjct: 2081 AILPEQQQQQQQQQQQQQQQQQQQQQQQQQQ 2111
Score = 32.0 bits (71), Expect = 2.7
Identities = 15/28 (53%), Positives = 17/28 (60%)
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSF 155
EQQQQQQ QQQQQQ+ Q Q +
Sbjct: 2085 EQQQQQQQQQQQQQQQQQQQQQQQQQQY 2112
Score = 31.6 bits (70), Expect = 3.5
Identities = 24/91 (26%), Positives = 37/91 (40%), Gaps = 13/91 (14%)
Query: 51 VNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARR 110
V+ A ++G LT STQ+ Q + T P + +
Sbjct: 2034 VHQQAPTYGHGLT-------------STQRFSHQTLQQTPMISTMTPMSAQGVQAGVRST 2080
Query: 111 EIVTALKFHRASMKEASEQQQQQQLQQQQQQ 141
I+ + + ++ +QQQQQQ QQQQQQ
Sbjct: 2081 AILPEQQQQQQQQQQQQQQQQQQQQQQQQQQ 2111
Score = 30.8 bits (68), Expect = 6.0
Identities = 14/29 (48%), Positives = 20/29 (68%)
Query: 114 TALKFHRASMKEASEQQQQQQLQQQQQQR 142
TA+ + ++ +QQQQQQ QQQQQQ+
Sbjct: 2080 TAILPEQQQQQQQQQQQQQQQQQQQQQQQ 2108
>BIB_DROME (P23645) Neurogenic protein big brain
Length = 700
Score = 52.8 bits (125), Expect = 1e-06
Identities = 66/251 (26%), Positives = 105/251 (41%), Gaps = 58/251 (23%)
Query: 25 EEEPPFSGA---YMRS--LVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKA-----CQS 74
++EP + G Y RS L + Q S+ + + + + G + A ++
Sbjct: 379 QQEPLYGGTRSLYCRSPTLTRSNLNRSQSVYAKSNTAINRDIVPRPGPLVPAQSLYPMRT 438
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ QQ QQ ++QV +S + + N + R E + ++ ++ +QQQQQQ
Sbjct: 439 QQQQQQQQQQQQQVASAPQSSHLQNQNVQNQMQQRSESIYGMRGSMRGQQQPIQQQQQQQ 498
Query: 135 ---LQQQ------QQQRAPDSPQLSQNPS----------------FDQDGRYK-SRRNP- 167
LQQQ QQQ+ PQ+ +P DG +K RR+P
Sbjct: 499 QQQLQQQQPNMGVQQQQMQPPPQMMSDPQQQPQGFQPVYGTRTNPTPMDGNHKYDRRDPQ 558
Query: 168 RIY------------PSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPI---APPPLMAE 212
++Y S ++++ SY G + PPA P+ P P P+ APPP
Sbjct: 559 QMYGVTGPRNRGQSAQSDDSSYGSYHGS-AVTPPARHPSVEPSPPPPPMLMYAPPP---- 613
Query: 213 NPNFILPNQPL 223
PN P QP+
Sbjct: 614 QPNAAHP-QPI 623
Score = 32.7 bits (73), Expect = 1.6
Identities = 18/42 (42%), Positives = 21/42 (49%)
Query: 120 RASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRY 161
+ S A +QQQQQQ QQQQQQ+ Q Q Q Y
Sbjct: 651 QGSAVTAQQQQQQQQQQQQQQQQQQQQQQQQQMMMQQQQQHY 692
Score = 32.0 bits (71), Expect = 2.7
Identities = 25/92 (27%), Positives = 39/92 (42%), Gaps = 5/92 (5%)
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
Q + PQ + Q R++ S P +++ A T + + ++ +QQQQQQ
Sbjct: 614 QPNAAHPQPIRTQSERKV--SAPV---VVSQPAACAVTYTTSQGSAVTAQQQQQQQQQQQ 668
Query: 135 LQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRN 166
QQQQQQ+ Q+ G R N
Sbjct: 669 QQQQQQQQQQQQQQMMMQQQQQHYGMLPLRPN 700
>CCF_DROME (P41046) Centrosomal and chromosomal factor (CCF)
(Chromocentrosomin)
Length = 550
Score = 47.4 bits (111), Expect = 6e-05
Identities = 29/94 (30%), Positives = 45/94 (47%), Gaps = 4/94 (4%)
Query: 65 HGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMK 124
H + Q QQ QQH + + +TS R + A + ++ A+ A+ +
Sbjct: 54 HAAVAAHQQQLLQQQQQQQHHRHHHQPANTSSSSNSRHSHAAATQTQVAAAV----ANSR 109
Query: 125 EASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQD 158
+ +QQQQQQ QQQQQQ A + + PS +D
Sbjct: 110 QQQQQQQQQQQQQQQQQTASSNSNTAPAPSPQKD 143
Score = 38.1 bits (87), Expect = 0.038
Identities = 26/88 (29%), Positives = 42/88 (47%), Gaps = 9/88 (10%)
Query: 70 KACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQ 129
+A S Q QQ QQ +Q +++L ++ A ++ + ++ H Q
Sbjct: 237 RAATSAQQQQQQQQRYQQQQQQLRQQHQQMSQMSQQAHYPQQQSSLVRQH---------Q 287
Query: 130 QQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
QQQQQ QQQQ+ + S + SQ+ S Q
Sbjct: 288 QQQQQQQQQQRASSNQSQRQSQSQSQSQ 315
Score = 31.2 bits (69), Expect = 4.6
Identities = 19/50 (38%), Positives = 26/50 (52%), Gaps = 4/50 (8%)
Query: 108 ARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
ARR A + ++ QQQQQQL+QQ QQ + Q+SQ + Q
Sbjct: 232 ARRGARAATSAQQQQQQQQRYQQQQQQLRQQHQQMS----QMSQQAHYPQ 277
>PO62_MOUSE (Q8BJI4) POU domain, class 6, transcription factor 2
Length = 691
Score = 47.0 bits (110), Expect = 8e-05
Identities = 51/184 (27%), Positives = 76/184 (40%), Gaps = 19/184 (10%)
Query: 125 EASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP---SCNTNFSSYM 181
+ +QQQQQQ QQQQQQ+ P P SQ+P +S+ P P S
Sbjct: 180 QQQQQQQQQQQQQQQQQQQPPPPPTSQHPQPASQAPPQSQPTPPHQPPPASQQLPAPPAQ 239
Query: 182 GDFSSYPPAFVPNSY--------PVPVASPIAPPPLMAENPNFILPN-----QPLGLNLN 228
+ ++ P P+S+ P P +PP + +P LP+ PL L +N
Sbjct: 240 LEQATQPQQHQPHSHPQNQTQNQPSPTQQSSSPPQKPSPSPGHSLPSPLTPPNPLQL-VN 298
Query: 229 FHDFNNLDATIHLSNTSLSSNSFSSATSSSQEVPS--VELSQGDGISSLVNSTESNAASQ 286
+ A + SS +F +A SS Q V V +QG I ++ +SQ
Sbjct: 299 NPLASQAAAAAAAMGSIASSQAFGNALSSLQGVTGQLVTNAQGQIIGTIPLMPNPGPSSQ 358
Query: 287 VNAG 290
+G
Sbjct: 359 AASG 362
>PHC1_MOUSE (Q64028) Polyhomeotic-like protein 1 (Early development
regulator protein 1) (MPH1) (RAE-28)
Length = 1012
Score = 46.6 bits (109), Expect = 1e-04
Identities = 39/145 (26%), Positives = 63/145 (42%), Gaps = 13/145 (8%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q +Q Q + +H + +Q R + T L+ + ++ +QQQQ
Sbjct: 382 QQQQIHLQQKQVVIQQQIAIHHQQQFQHRQSQLLHT----ATHLQLAQQQQQQQQQQQQQ 437
Query: 133 QQLQQQQQQR------APDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSS 186
QQ QQQQQQ+ AP PQ+ + + + P ++ + + +S
Sbjct: 438 QQQQQQQQQQQGTTLTAPQPPQVPPTQQVPPSQSQQQAQTLVVQPMLQSSPLTLPPEPTS 497
Query: 187 YPPAFVPNSYPVPVASPIAPPPLMA 211
PP + + PV +PI PP L A
Sbjct: 498 KPPIPIQSKPPV---APIKPPQLGA 519
>ARFF_ARATH (Q9ZTX8) Auxin response factor 6
Length = 933
Score = 46.6 bits (109), Expect = 1e-04
Identities = 52/213 (24%), Positives = 96/213 (44%), Gaps = 29/213 (13%)
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ +QQ QQ +Q +++ S+ Q++L + ++ S+QQQQQ
Sbjct: 484 QQLSQQQQQLSQQQQQQQQLSQQQQQQLSQQQQ----------------QQLSQQQQQQL 527
Query: 135 LQQQQQQR---APDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSS--YMGDFSSYPP 189
QQQQQQ P++ Q Q+ + Q + S++ ++ + N + SS + S +
Sbjct: 528 SQQQQQQAYLGVPETHQ-PQSQAQSQSNNHLSQQQQQVVDNHNPSASSAAVVSAMSQFGS 586
Query: 190 AFVPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSN 249
A PN+ P+ + + ++ P PL L+ + +HL+ T N
Sbjct: 587 ASQPNTSPLQSMTSLCHQQSFSDTNGGNNPISPLHTLLSNFSQDESSQLLHLTRT----N 642
Query: 250 SFSSATSSSQEVPSVELS-QGDGISSLVNSTES 281
S +++ + P+V+ S Q G + N+T+S
Sbjct: 643 SAMTSSGWPSKRPAVDSSFQHSGAGN--NNTQS 673
Score = 30.8 bits (68), Expect = 6.0
Identities = 21/67 (31%), Positives = 32/67 (47%), Gaps = 2/67 (2%)
Query: 123 MKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMG 182
M + QQQQQL QQQQQ+ S Q Q S Q + ++ ++ S +Y+G
Sbjct: 481 MLQQQLSQQQQQLSQQQQQQQQLSQQQQQQLSQQQQQQLSQQQQQQL--SQQQQQQAYLG 538
Query: 183 DFSSYPP 189
++ P
Sbjct: 539 VPETHQP 545
>PHC1_HUMAN (P78364) Polyhomeotic-like protein 1 (Early development
regulator protein 1) (HPH1)
Length = 1004
Score = 46.2 bits (108), Expect = 1e-04
Identities = 44/165 (26%), Positives = 68/165 (40%), Gaps = 28/165 (16%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ Q +Q Q + +H + +Q R + H A+ + ++QQQQ
Sbjct: 382 QQQQIHLQQKQVVIQQQIAIHHQQQFQHRQSQL------------LHTATHLQLAQQQQQ 429
Query: 133 QQLQQQQQQR-------APDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFS 185
QQ QQQQQQ+ AP PQ+ + + + P ++ S D +
Sbjct: 430 QQQQQQQQQQPQATTLTAPQPPQVPPTQQVPPSQSQQQAQTLVVQPMLQSSPLSLPPDAA 489
Query: 186 SYPPAFVPNSYPVPVASPIAPPPL------MAENPNFILPNQPLG 224
PP + + PV +PI PP L A+ P +P Q +G
Sbjct: 490 PKPPIPIQSKPPV---APIKPPQLGAAKMSAAQQPPPHIPVQVVG 531
>YKF4_YEAST (P35732) Hypothetical 84.0 kDa protein in NUP120-CSE4
intergenic region
Length = 738
Score = 45.8 bits (107), Expect = 2e-04
Identities = 36/131 (27%), Positives = 61/131 (46%), Gaps = 16/131 (12%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVT------------ALKFHR 120
Q QQ QQPQQ ++Q++++ + Q+ + A+A+ E ++ A + H+
Sbjct: 406 QQQQQPQQPQQPQQQLQQQ---QQQQQQPVQAQAQAQEEQLSQNYYTQQQQQQYAQQQHQ 462
Query: 121 ASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSY 180
+ S+QQQ Q QQQ Q PQ Q+P + G + + +Y + + SY
Sbjct: 463 LQQQYLSQQQQYAQQQQQHPQPQSQQPQSQQSPQSQKQGNNVAAQQYYMYQNQFPGY-SY 521
Query: 181 MGDFSSYPPAF 191
G F S A+
Sbjct: 522 PGMFDSQGYAY 532
Score = 35.8 bits (81), Expect = 0.19
Identities = 23/90 (25%), Positives = 39/90 (42%), Gaps = 8/90 (8%)
Query: 76 QSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREI--------VTALKFHRASMKEAS 127
+ Q+ + K+QV+ T+ ++ N+A E+ ++A+ + +
Sbjct: 320 EEVQEEAEKKEQVKEEEQTAEELEQEQDNVAAPEEEVTVVEEKVEISAVISEPPEDQANT 379
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQ 157
Q QQQ QQ QQ + P PQ Q P Q
Sbjct: 380 VPQPQQQSQQPQQPQQPQQPQQPQQPQQQQ 409
Score = 30.4 bits (67), Expect = 7.8
Identities = 21/79 (26%), Positives = 32/79 (39%), Gaps = 4/79 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q Q + P++ V ++ S E E + V + ++ + QQ
Sbjct: 344 QEQDNVAAPEEEVTVVEEKVEISAVISEP----PEDQANTVPQPQQQSQQPQQPQQPQQP 399
Query: 133 QQLQQQQQQRAPDSPQLSQ 151
QQ QQ QQQ+ P PQ Q
Sbjct: 400 QQPQQPQQQQQPQQPQQPQ 418
>BTD_DROME (Q24266) Transcription factor BTD (Buttonhead protein)
Length = 644
Score = 45.1 bits (105), Expect = 3e-04
Identities = 51/220 (23%), Positives = 88/220 (39%), Gaps = 24/220 (10%)
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ Q QQH +Q + LH + L+ +A+++ + L + ++ +QQQQQQ
Sbjct: 20 QQQHAQHQQHAQQQQHHLHMQQAQHHLHLSHQQAQQQHMQHLTQQQQQQQQQQQQQQQQQ 79
Query: 135 LQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVPN 194
QQQQ PQ Q+ + P ++ SS PP +
Sbjct: 80 QQQQQ-------PQQQQHDFLSAAALLSA---PPSLSGSSSGSSSGSSPLYGKPPMKLEL 129
Query: 195 SYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSNSFSSA 254
YP ++ A +PN + + P +++ F + S +S S S +
Sbjct: 130 PYPQASSTGTA-------SPNSSIQSAPSSASVSPSIFPS-------PAQSFASISASPS 175
Query: 255 TSSSQEVPSVELSQGDGISSLVNSTESNAASQVNAGLHAA 294
T ++ P + G S +S+ S++A+ A AA
Sbjct: 176 TPTTTLAPPTTAAAGALAGSPTSSSPSSSAASAAAAAAAA 215
>IXR1_YEAST (P33417) Intrastrand crosslink recognition protein
(Structure-specific recognition protein) (SSRP)
Length = 597
Score = 44.7 bits (104), Expect = 4e-04
Identities = 62/232 (26%), Positives = 81/232 (34%), Gaps = 67/232 (28%)
Query: 125 EASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDF 184
+ +QQQQQQ QQQQQQ+AP Q+P Q Y
Sbjct: 56 QPQQQQQQQQQQQQQQQQAPYQGHFQQSPQQQQQNVY----------------------- 92
Query: 185 SSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDA------- 237
YP+P S P + Q N N ++ N++A
Sbjct: 93 -----------YPLPPQSLTQPTSQSQQQQQQQQQQQYANSNSNSNNNVNVNALPQDFGY 141
Query: 238 -----------TIHLSNTSLSSNSFSSATSSSQEVPSV--------ELSQGDGISSLVNS 278
TI+ S NSF S + Q PSV S + +SS NS
Sbjct: 142 MQQTGSGQNYPTINQQQFSEFYNSFLSHLTQKQTNPSVTGTGASSNNNSNNNNVSSGNNS 201
Query: 279 TESN----AASQVN---AGLHAAMDEEAIAEIRSLGEQYQMEWNDTMNLVKS 323
T SN AASQ+N A AA + A S Q Q + + MN + S
Sbjct: 202 TSSNPAQLAASQLNPATAAAAAANNAAGPASYLSQLPQVQRYYPNNMNALSS 253
Score = 43.9 bits (102), Expect = 7e-04
Identities = 65/262 (24%), Positives = 103/262 (38%), Gaps = 28/262 (10%)
Query: 47 QGCVVNSDASSHGQNLTKHGKIGK-ACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNM 105
QG N + T H + + Q QQ QQ QQ ++Q ++ PYQ
Sbjct: 29 QGLEHNDSQYRDASHQTPHQYLNQFQAQPQQQQQQQQQQQQQQQQA-----PYQGHFQQS 83
Query: 106 AEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFD---QD-GRY 161
+ +++ V S+ + + Q QQQQ QQQQQQ A + + N + + QD G
Sbjct: 84 PQQQQQNVY-YPLPPQSLTQPTSQSQQQQQQQQQQQYANSNSNSNNNVNVNALPQDFGYM 142
Query: 162 KSRRNPRIYPSCN-TNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPN 220
+ + + YP+ N FS + F S+ N V A + N N N
Sbjct: 143 QQTGSGQNYPTINQQQFSEFYNSFLSHLTQKQTNP---SVTGTGASSNNNSNNNNVSSGN 199
Query: 221 QPLGLN---LNFHDFNNLDATIHLSNTSLSSNSFSSATSSSQEVPSVEL---SQGDGISS 274
N L N A +N + S+ S ++P V+ + + +SS
Sbjct: 200 NSTSSNPAQLAASQLNPATAAAAAANNAAGPASYLS------QLPQVQRYYPNNMNALSS 253
Query: 275 LVN-STESNAASQVNAGLHAAM 295
L++ S+ NAA N H +
Sbjct: 254 LLDPSSAGNAAGNANTATHPGL 275
Score = 33.9 bits (76), Expect = 0.71
Identities = 29/118 (24%), Positives = 56/118 (46%), Gaps = 13/118 (11%)
Query: 80 QPQ--QHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQ 137
QPQ H++Q++++L + Q++L + +++ H+ ++ +QQ QQ
Sbjct: 281 QPQLTHHQQQMQQQLQLQQ--QQQLQQQQQLQQQ-------HQLQQQQQLQQQHHHLQQQ 331
Query: 138 QQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGD--FSSYPPAFVP 193
QQQQ+ P +LS S + + ++ P+ S FS + + +P A VP
Sbjct: 332 QQQQQHPVVKKLSSTQSRIERRKQLKKQGPKRPSSAYFLFSMSIRNELLQQFPEAKVP 389
>CLOC_DROME (O61735) Circadian locomoter output cycles Kaput protein
(dCLOCK) (dPAS1)
Length = 1027
Score = 44.7 bits (104), Expect = 4e-04
Identities = 27/84 (32%), Positives = 43/84 (51%), Gaps = 1/84 (1%)
Query: 69 GKACQSQQS-TQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEAS 127
G A S+Q QQ QQ Q ++ LHT + + + + ++ + T + + ++
Sbjct: 751 GFALNSEQMLNQQDQQMMMQQQQNLHTQHQHNLQQQHQSHSQLQQHTQQQHQQQQQQQQQ 810
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQ 151
+QQQQQQ QQQQQQ+ Q Q
Sbjct: 811 QQQQQQQQQQQQQQQQQQQQQQQQ 834
Score = 37.4 bits (85), Expect = 0.064
Identities = 27/112 (24%), Positives = 48/112 (42%), Gaps = 14/112 (12%)
Query: 41 QLRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQE 100
Q + N G +NS+ + Q+ + + +Q QQH+ + + HT + +Q+
Sbjct: 744 QPQFNQYGFALNSEQMLNQQDQQMMMQQQQNLHTQHQHNLQQQHQSHSQLQQHTQQQHQQ 803
Query: 101 RLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQN 152
+ + +++ +QQQQQQ QQQQQQ+ QN
Sbjct: 804 QQQQQQQQQQQ--------------QQQQQQQQQQQQQQQQQQQQLQLQQQN 841
>SAB1_HUMAN (Q01826) DNA-binding protein SATB1 (Special AT-rich
sequence binding protein 1)
Length = 763
Score = 43.9 bits (102), Expect = 7e-04
Identities = 28/81 (34%), Positives = 36/81 (43%), Gaps = 17/81 (20%)
Query: 125 EASEQQQQQQLQQQQQQRAPDSPQLSQNP-----------------SFDQDGRYKSRRNP 167
E +QQQQQQ QQQQQQ+AP PQ Q P D++ R K+R
Sbjct: 590 EQIQQQQQQQQQQQQQQQAPPPPQPQQQPQTGPRLPPRQPTVASPAESDEENRQKTRPRT 649
Query: 168 RIYPSCNTNFSSYMGDFSSYP 188
+I S++ D YP
Sbjct: 650 KISVEALGILQSFIQDVGLYP 670
>LUG_ARATH (Q9FUY2) Transcriptional corepressor LEUNIG
Length = 931
Score = 43.9 bits (102), Expect = 7e-04
Identities = 32/110 (29%), Positives = 52/110 (47%), Gaps = 9/110 (8%)
Query: 43 RTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERL 102
RTN++ V + + ++ ++ Q S QQ QQ ++Q++ Q+ L
Sbjct: 66 RTNEKHSEVAASYIETQMIKAREQQLQQSQHPQVSQQQQQQQQQQIQM--------QQLL 117
Query: 103 LNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQ-QRAPDSPQLSQ 151
L A+ +++ H ++ +QQQQQQ QQQQQ Q P S Q Q
Sbjct: 118 LQRAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQ 167
Score = 31.6 bits (70), Expect = 3.5
Identities = 20/74 (27%), Positives = 33/74 (44%), Gaps = 20/74 (27%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H ++P ++ ++++ Q QQ
Sbjct: 136 QQQQQQQQQQQQQQQQQQQQHQNQPPSQQ--------------------QQQQSTPQHQQ 175
Query: 133 QQLQQQQQQRAPDS 146
Q QQQ QR S
Sbjct: 176 QPTPQQQPQRRDGS 189
Score = 30.4 bits (67), Expect = 7.8
Identities = 26/134 (19%), Positives = 49/134 (36%), Gaps = 18/134 (13%)
Query: 20 LSRNLEEEPPFSGAYMRSLV-----KQLRTNDQGCVVNSDASSHGQNLTKHGKIGKACQS 74
++R E+ + +Y+ + + +QL+ + V Q + + + Q
Sbjct: 64 IARTNEKHSEVAASYIETQMIKAREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQ 123
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ QQ Q H Q +++ + Q++ + + +QQQQ
Sbjct: 124 QQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQ-------------QHQNQPPSQQQQQQST 170
Query: 135 LQQQQQQRAPDSPQ 148
Q QQQ PQ
Sbjct: 171 PQHQQQPTPQQQPQ 184
>P32_MYCGE (Q49417) P32 adhesin (Cytadhesin P32)
Length = 280
Score = 43.1 bits (100), Expect = 0.001
Identities = 42/140 (30%), Positives = 62/140 (44%), Gaps = 23/140 (16%)
Query: 99 QERLLNMAEARREIVTALKFHRASMKEASEQQQQQ--QLQQQQQQRAPDSPQLSQ--NPS 154
++RLL E + +I L+ S+QQ+QQ ++ QQ Q P PQ+ Q P
Sbjct: 99 EKRLLEEKERQEQIAEQLQ-------RISDQQEQQTVEIDPQQSQAQPSQPQVQQPLQPQ 151
Query: 155 FDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASP-IAPP---PLM 210
F Q R P + P+ N N G F+ F P++ P +P + P P M
Sbjct: 152 FQQ-------RVPLLRPAFNPNMQQRPG-FNQPNQQFQPHNNFNPRMNPNMQRPGFNPNM 203
Query: 211 AENPNFILPNQPLGLNLNFH 230
+ P F PNQ + NF+
Sbjct: 204 QQRPGFNQPNQQFQPHNNFN 223
>GDA7_WHEAT (P04727) Alpha/beta-gliadin clone PW8142 precursor
(Prolamin)
Length = 313
Score = 43.1 bits (100), Expect = 0.001
Identities = 29/81 (35%), Positives = 43/81 (52%), Gaps = 1/81 (1%)
Query: 77 STQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQ 136
S+Q QQ Q+ ++L + Q + +A +V A+ H+ ++ +QQQQQQLQ
Sbjct: 165 SSQVLQQSTYQLLQQLCCQQLLQIPEQSRCQAIHNVVHAIIMHQQEQQQQLQQQQQQQLQ 224
Query: 137 QQQQQRAPDSPQLSQNPSFDQ 157
QQQQQ+ Q S SF Q
Sbjct: 225 QQQQQQ-QQQQQPSSQVSFQQ 244
Score = 32.3 bits (72), Expect = 2.1
Identities = 14/21 (66%), Positives = 18/21 (85%)
Query: 122 SMKEASEQQQQQQLQQQQQQR 142
S ++A +QQQQQQ QQQQQQ+
Sbjct: 117 SQQQAQQQQQQQQQQQQQQQQ 137
Score = 31.6 bits (70), Expect = 3.5
Identities = 27/100 (27%), Positives = 39/100 (39%), Gaps = 11/100 (11%)
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGD 183
K S+QQ Q+Q+ QQQ+ P Q F Y P+ +PS +
Sbjct: 28 KNPSQQQPQEQVPLVQQQQFP-----GQQQQFPPQQPYP---QPQPFPSQQPYLQ--LQP 77
Query: 184 FSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPL 223
F P YP P + P P + P ++ P QP+
Sbjct: 78 FPQPQPFLPQLPYPQPQSFP-PQQPYPQQRPKYLQPQQPI 116
>YNC7_YEAST (P53968) Hypothetical 76.3 kDa zinc finger protein in
KTR5-UME3 intergenic region
Length = 678
Score = 42.7 bits (99), Expect = 0.002
Identities = 45/161 (27%), Positives = 67/161 (40%), Gaps = 30/161 (18%)
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNF------ 177
++ +QQQQQQ QQQQQQ+ + ++ Q+ +F D N PS F
Sbjct: 121 QQLQQQQQQQQQQQQQQQKQTPTLKVEQSDTFQWDDILTPADNQH-RPSLTNQFLSPRSN 179
Query: 178 -----------SSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNF------ILPN 220
S+Y S+Y P YP + S A L A N +F N
Sbjct: 180 YDGTTRSSGIDSNYSDTESNY---HTPYLYPQDLVSSPAMSHLTANNDDFDDLLSVASMN 236
Query: 221 QPLGLNLNFHDF---NNLDATIHLSNTSLSSNSFSSATSSS 258
L +N H + +NLD L + + S N+ SA+++S
Sbjct: 237 SNYLLPVNSHGYKHISNLDELDDLLSLTYSDNNLLSASNNS 277
>GBF_DICDI (P36417) G-box binding factor (GBF)
Length = 708
Score = 42.7 bits (99), Expect = 0.002
Identities = 22/80 (27%), Positives = 39/80 (48%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q Q QQ QH +Q++++ H + Q++ + +++ ++ H+ + QQQQ
Sbjct: 138 QHHQQQQQQPQHHQQMQQQQHHQQMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQ 197
Query: 133 QQLQQQQQQRAPDSPQLSQN 152
QQ Q QQQ Q Q+
Sbjct: 198 QQQQHQQQHHQQQQQQQQQH 217
Score = 42.0 bits (97), Expect = 0.003
Identities = 32/142 (22%), Positives = 57/142 (39%), Gaps = 12/142 (8%)
Query: 26 EEPPFSGAYMRSLVKQLRTNDQGCVVNSDASSHGQNLTK--HGKIGKACQSQQSTQQPQQ 83
+ P ++ Y ++Q +Q N H Q + H + + Q Q QQ QQ
Sbjct: 108 QNPHYNYQYQLMFMQQQAQQNQPPQQNQQQQHHQQQQQQPQHHQQMQQQQHHQQMQQQQQ 167
Query: 84 HKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRA 143
H +Q++++ H + +L H+ ++ QQQ Q QQQQQQ+
Sbjct: 168 HHQQMQQQQHHQQMQHHQLQQHQHQ----------HQQQQQQQQHQQQHHQQQQQQQQQH 217
Query: 144 PDSPQLSQNPSFDQDGRYKSRR 165
Q Q+ Q ++ ++
Sbjct: 218 HQQQQHHQHSQPQQQHQHNQQQ 239
Score = 38.1 bits (87), Expect = 0.038
Identities = 29/124 (23%), Positives = 53/124 (42%), Gaps = 7/124 (5%)
Query: 58 HGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALK 117
H Q + H Q Q Q QQ ++Q +++ H + Q++ + + + +
Sbjct: 177 HHQQMQHH-----QLQQHQHQHQQQQQQQQHQQQHHQQQQQQQQQHHQQQQHHQHSQPQQ 231
Query: 118 FHRASMKEASEQQQQQQLQQQQQ-QRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTN 176
H+ + ++ + QQQ QQQ Q Q P PQ N S + + + N + N N
Sbjct: 232 QHQHNQQQQHQHNQQQHQQQQNQIQMVPQQPQSLSN-SGNNNNNNNNNNNSNNNNNNNNN 290
Query: 177 FSSY 180
+S+
Sbjct: 291 NNSH 294
Score = 35.4 bits (80), Expect = 0.24
Identities = 21/101 (20%), Positives = 42/101 (40%), Gaps = 7/101 (6%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q Q Q PQQ+++Q + +P + + + +++ + H+ ++ QQ Q
Sbjct: 123 QQAQQNQPPQQNQQQQHHQQQQQQPQHHQQMQQQQHHQQMQQQQQHHQQMQQQQHHQQMQ 182
Query: 133 -------QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRN 166
Q QQQQQ+ Q Q Q ++ +++
Sbjct: 183 HHQLQQHQHQHQQQQQQQQHQQQHHQQQQQQQQQHHQQQQH 223
Score = 35.0 bits (79), Expect = 0.32
Identities = 23/94 (24%), Positives = 40/94 (42%), Gaps = 2/94 (2%)
Query: 58 HGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALK 117
H Q + + + + Q QQ QQ Q H+ Q + H + Q++ + + ++ +
Sbjct: 158 HHQQMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQQQQ--HQQQHHQQQQQQQQ 215
Query: 118 FHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQ 151
H + Q QQQ Q QQQ+ + Q Q
Sbjct: 216 QHHQQQQHHQHSQPQQQHQHNQQQQHQHNQQQHQ 249
>SSN6_YEAST (P14922) Glucose repression mediator protein
Length = 966
Score = 42.4 bits (98), Expect = 0.002
Identities = 33/125 (26%), Positives = 50/125 (39%), Gaps = 5/125 (4%)
Query: 37 SLVKQLRTNDQGCVVNSDASSHGQN----LTKHGKIGKACQSQQSTQQPQQHKKQVRRRL 92
S+V+Q Q +NS A+ + L + Q+Q Q Q + Q + +
Sbjct: 464 SMVQQQHPAQQ-TPINSSATMYSNGASPQLQAQAQAQAQAQAQAQAQAQAQAQAQAQAQA 522
Query: 93 HTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQN 152
Q + A A+ + K + +A +QQQQQQ QQQQQQ+ Q Q
Sbjct: 523 QAQAQAQAQAQAQAHAQAQAQAQAKAQAQAQAQAQQQQQQQQQQQQQQQQQQQQQQQQQQ 582
Query: 153 PSFDQ 157
Q
Sbjct: 583 QQQQQ 587
Score = 39.3 bits (90), Expect = 0.017
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 7/98 (7%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q+Q Q Q + Q + + H Q + A+A+ + + ++ +QQQQ
Sbjct: 517 QAQAQAQAQAQAQAQAQAQAHAQAQAQAQAKAQAQAQAQAQ-----QQQQQQQQQQQQQQ 571
Query: 133 QQLQQQQQQRAPDSPQLSQNP--SFDQDGRYKSRRNPR 168
QQ QQQQQQ+ QL P Q G NP+
Sbjct: 572 QQQQQQQQQQQQQQQQLQPLPRQQLQQKGVSVQMLNPQ 609
Score = 34.7 bits (78), Expect = 0.42
Identities = 16/31 (51%), Positives = 22/31 (70%)
Query: 123 MKEASEQQQQQQLQQQQQQRAPDSPQLSQNP 153
M++ ++QQQQQQ QQQQQQ+ PQ +P
Sbjct: 10 MEQPAQQQQQQQQQQQQQQQQAAVPQQPLDP 40
Score = 34.3 bits (77), Expect = 0.54
Identities = 34/152 (22%), Positives = 58/152 (37%), Gaps = 22/152 (14%)
Query: 42 LRTNDQGCVVNSDASSHGQNLT------KHG-------------KIGKACQSQQSTQQPQ 82
L+ NDQG +N+ S+ N T +H G + Q Q Q
Sbjct: 441 LQPNDQGNPLNTRISAQSANATASMVQQQHPAQQTPINSSATMYSNGASPQLQAQAQAQA 500
Query: 83 QHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEA---SEQQQQQQLQQQQ 139
Q + Q + + Q + A+A+ + + H + +A ++ Q Q Q QQQQ
Sbjct: 501 QAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAHAQAQAQAQAKAQAQAQAQAQQQQ 560
Query: 140 QQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ+ Q Q Q + + ++ + P
Sbjct: 561 QQQQQQQQQQQQQQQQQQQQQQQQQQQLQPLP 592
>MLL2_HUMAN (O14686) Myeloid/lymphoid or mixed-lineage leukemia
protein 2 (ALL1-related protein)
Length = 5262
Score = 42.4 bits (98), Expect = 0.002
Identities = 22/70 (31%), Positives = 40/70 (56%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++L + Q++ L + ++++ + ++ +QQQQ
Sbjct: 3626 QQQQQLQQQQQLQQQQQQQLQQQQQLQQQQLQQQQQQQQLQQQQQQQLQQQQQQLQQQQQ 3685
Query: 133 QQLQQQQQQR 142
QQ QQ QQQ+
Sbjct: 3686 QQQQQFQQQQ 3695
Score = 37.4 bits (85), Expect = 0.064
Identities = 25/86 (29%), Positives = 40/86 (46%), Gaps = 3/86 (3%)
Query: 66 GKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKE 125
G +G + Q QQ QQH++Q + ++ L + ++ +++
Sbjct: 3571 GLMGHRLVTAQQQQQQQQHQQQGSM---AGLSHLQQSLMSHSGQPKLSAQPMGSLQQLQQ 3627
Query: 126 ASEQQQQQQLQQQQQQRAPDSPQLSQ 151
+ QQQQQLQQQQQQ+ QL Q
Sbjct: 3628 QQQLQQQQQLQQQQQQQLQQQQQLQQ 3653
Score = 32.7 bits (73), Expect = 1.6
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 10/95 (10%)
Query: 65 HGKIGKACQSQQSTQQ------PQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKF 118
H K KA ++Q T + P+ ++++ Q++L + + ++E +
Sbjct: 3260 HRKSRKALCAKQRTAKKAGREFPEADAEKLKLVTEQQSKIQKQLDQVRKQQKEHTNLM-- 3317
Query: 119 HRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNP 153
A + +QQQQQQ QQQQQ A + SQ+P
Sbjct: 3318 --AEYRNKQQQQQQQQQQQQQQHSAVLALSPSQSP 3350
>MIG1_CANAL (Q9Y7G2) Regulatory protein MIG1
Length = 574
Score = 42.4 bits (98), Expect = 0.002
Identities = 67/277 (24%), Positives = 102/277 (36%), Gaps = 58/277 (20%)
Query: 29 PFSGAYMRSLVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQV 88
P + YM+ +V+ T V N+D H Q H + Q Q Q+ QH++Q
Sbjct: 156 PHANGYMQPMVQ---TQGVTVVPNTDQQQHQQQQQYHQQ-----QQQYQFQERPQHQQQQ 207
Query: 89 RRRLHTSRPYQER---------LLNMAEARR------EIVTALKFHRASMKEASEQQQQQ 133
+ + T+ Q+ MA A+ ++K S+ + QQQ
Sbjct: 208 QTNIGTNNHLQQNGSAVFSLPSSPTMASAQTPNSGTSSSSESIKLAPLSIPNNISRPQQQ 267
Query: 134 QLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVP 193
Q QQQQQQ+ P KS + +Y N FS P
Sbjct: 268 QQQQQQQQQIP-----------------KSESSTSLYSDGNKLFSR-------------P 297
Query: 194 NSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLN--FHDFNNLDATIHLSNTSLSSNSF 251
NS + S +P A P +P P NLN F +N + + + +S S +S
Sbjct: 298 NSTLHSLGS--SPENNNASGPLSGVPT-PSFSNLNDYFQQKSNSNNSRLFNASSSSLSSL 354
Query: 252 SSATSSSQEVPSVELSQGDGISSLVNSTESNAASQVN 288
S SS L + ++S N+T + S N
Sbjct: 355 SGKIRSSSSTNLAGLQRLTPLTSTTNNTNNTTTSNTN 391
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.312 0.126 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,747,391
Number of Sequences: 164201
Number of extensions: 1836357
Number of successful extensions: 18188
Number of sequences better than 10.0: 421
Number of HSP's better than 10.0 without gapping: 268
Number of HSP's successfully gapped in prelim test: 161
Number of HSP's that attempted gapping in prelim test: 9236
Number of HSP's gapped (non-prelim): 2824
length of query: 382
length of database: 59,974,054
effective HSP length: 112
effective length of query: 270
effective length of database: 41,583,542
effective search space: 11227556340
effective search space used: 11227556340
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0133.10


