

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0129.4
(1259 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
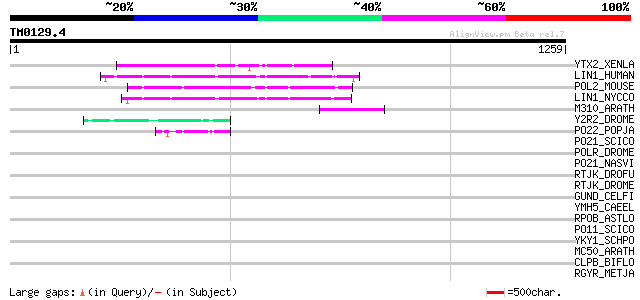
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 141 1e-32
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 133 3e-30
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 129 7e-29
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 126 3e-28
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 119 4e-26
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 53 6e-06
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 48 1e-04
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 45 0.002
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 44 0.002
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 42 0.008
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 42 0.011
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 42 0.014
GUND_CELFI (P50400) Endoglucanase D precursor (EC 3.2.1.4) (Endo... 40 0.052
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 39 0.12
RPOB_ASTLO (P27059) DNA-directed RNA polymerase beta chain (EC 2... 36 0.58
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 35 0.99
YKY1_SCHPO (O13796) Hypothetical protein C1142.01 in chromosome I 35 1.7
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 35 1.7
CLPB_BIFLO (Q8G4X4) Chaperone clpB 34 2.9
RGYR_METJA (Q58907) Reverse gyrase [Includes: Helicase (EC 3.6.1... 33 4.9
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 141 bits (355), Expect = 1e-32
Identities = 138/500 (27%), Positives = 230/500 (45%), Gaps = 26/500 (5%)
Query: 243 RSRASWLKHGDKNTRFFHQKANQRRKRNLIEVLKDDNGREVEDDADIARVLGDYFGGLFT 302
RSR L D+ +RFF+ ++ R I L ++G +ED I ++ LF+
Sbjct: 359 RSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARSFYQNLFS 418
Query: 303 SS--NPEGIEETTNLVAGRVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFY 360
+P+ EE + + VSE L P T +E+ +AL M K+PGLDG F+
Sbjct: 419 PDPISPDACEELWDGLPV-VSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFF 477
Query: 361 QKYWDIIGDDVSAFCLQVLQGVIPPGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKI 420
Q +WD +G D + + P + +L L+PK +RP+SL + +KI
Sbjct: 478 QFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKI 537
Query: 421 ITKTIANRLKIILPDIICQTQSAFVPGRLIIDNALVVYECFHFMKKRISGRNGMMTLKLD 480
+ K I+ RLK +L ++I QS VPGR I DN ++ + HF ++ +G + + L LD
Sbjct: 538 VAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARR--TGLS-LAFLSLD 594
Query: 481 MSKAYDRVEWSFLQGVLQKMGFPPSWVSLIMSVSPLLGFPLCLTEIPSRVLLLLEAYVRV 540
KA+DRV+ +L G LQ F P +V + + + CL +I + L A+ R
Sbjct: 595 QEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKT---MYASAECLVKINWSLTAPL-AFGRG 650
Query: 541 I-----LFRLIFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARAN 595
+ L ++ +E LL + + L+L K + V+S +ADD I+ A+ +
Sbjct: 651 VRQGCPLSGQLYSLAIEPFLCLLRKRLTGLVL---KEPDMRVVLS--AYADDVILVAQ-D 704
Query: 596 TQEAECVLNILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLG 655
+ E Y AS IN KS + ++ + + +++ KYLG
Sbjct: 705 LVDLERAQECQEVYAAASSARINWSKSSGLLEGSLKVDFLPPAFR--DISWESKIIKYLG 762
Query: 656 LPTMIGKFK-TRIFNFVKDRVWKKLKGWK--ESTLSRAGREVLIKSVVQAIPSYVMSCFI 712
+ ++ ++ F +++ V +L WK LS GR ++I +V + Y + C
Sbjct: 763 VYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLS 822
Query: 713 LPDSLCADIERMVSRFYWGG 732
A I+R + F W G
Sbjct: 823 PTQEFIAKIQRRLLDFLWIG 842
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 133 bits (334), Expect = 3e-30
Identities = 143/628 (22%), Positives = 277/628 (43%), Gaps = 71/628 (11%)
Query: 206 LQELQRKDQT------KEVLMAINVAERDLDSLLEQEEVWWKQRSRASWLKHGDKNTRFF 259
L+EL++++QT ++ ++ I +++++ +++ SR+ + + +K R
Sbjct: 320 LKELEKQEQTNSKASRRQEIIKIRAELKEIETQKTLQKI---NESRSWFFEKINKIDRPL 376
Query: 260 HQKANQRRKRNLIEVLKDDNGREVEDDADIARVLGDYFGGLFTSSNPEGIEETTNLV--- 316
+ ++R++N I+ +K+D G D +I + +Y+ L+ ++ E +EE +
Sbjct: 377 ARLIKKKREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLY-ANKLENLEEMDKFLDTY 435
Query: 317 -AGRVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFC 375
R+++ + L P T E+E + + K+PG +GF FYQ+Y +++ F
Sbjct: 436 TLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRY----KEELVPFL 491
Query: 376 LQVLQGV----IPPGMINQTLLVLIPKIMKPEHATQ-FRPISLCNVLFKIITKTIANRLK 430
L++ Q + I P + ++LIPK + + FRPISL N+ KI+ K +AN+++
Sbjct: 492 LKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQ 551
Query: 431 IILPDIICQTQSAFVP---GRLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDR 487
+ +I Q F+P G I ++ + + + K M + +D KA+D+
Sbjct: 552 QHIKKLIHHDQVGFIPAMQGWFNIRKSINIIQHINRTKD-----TNHMIISIDAEKAFDK 606
Query: 488 VEWSFLQGVLQKMGFPPSWVSLIMSVSPLLGFPLCLT----EIPSRVLLLLEAYVRVILF 543
++ F+ L K+G +++ +I ++ + L E P + L
Sbjct: 607 IQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLL 666
Query: 544 RLIFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVL 603
I L + R + + GI++ +S LFADD I++ A+ +L
Sbjct: 667 PNIVLEVLARAIRQEKE------IKGIQLGKEEVKLS--LFADDMIVYLENPIVSAQNLL 718
Query: 604 NILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLGLP---TMI 660
++S + + SG IN+ KSQ + N Q + +L A + KYLG+ +
Sbjct: 719 KLISNFSKVSGYKINVQKSQAFLYTNNRQTESQIMSELPFTIASKRI-KYLGIQLTRDVK 777
Query: 661 GKFKTR---IFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVV--QAIPSYVMSCFILPD 715
FK + N +K+ K WK S GR ++K + + I + LP
Sbjct: 778 DLFKENYKPLLNEIKEDTNK----WKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPM 833
Query: 716 SLCADIERMVSRFYWGGDVDKRGLQWASWKKLTRSKFDGGLGFRDFKSFNIALVAKNWWR 775
+ ++E+ +F W KR + L++ GG+ DFK + A V K W
Sbjct: 834 TFFTELEKTTLKFIWN---QKRA--HIAKSTLSQKNKAGGITLPDFKLYYKATVTKTAWY 888
Query: 776 IY----------TQPETLLAQVFKGVYF 793
Y T+P ++ ++ + F
Sbjct: 889 WYQNRDIDQWNRTEPSEIMPHIYNYLIF 916
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 129 bits (323), Expect = 7e-29
Identities = 128/528 (24%), Positives = 235/528 (44%), Gaps = 38/528 (7%)
Query: 267 RKRNLIEVLKDDNGREVEDDADIARVLGDYFGGLFTSSNPEGIEETTNLV----AGRVSE 322
R + LI ++++ G D +I + ++ L+ S+ E ++E + ++++
Sbjct: 411 RDKILINKIRNEKGDITTDPEEIQNTIRSFYKRLY-STKLENLDEMDKFLDRYQVPKLNQ 469
Query: 323 THLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYW-DIIGDDVSAFCLQVLQG 381
+ L P + +E+E + + K+PG DGF FYQ + D+I F ++G
Sbjct: 470 DQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEG 529
Query: 382 VIPPGMINQTLLVLIPKIMK-PEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQT 440
+P T+ LIPK K P FRPISL N+ KI+ K +ANR++ + II
Sbjct: 530 TLPNSFYEATI-TLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPD 588
Query: 441 QSAFVPGRLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKM 500
Q F+PG N H++ K + +N M+ + LD KA+D+++ F+ VL++
Sbjct: 589 QVGFIPGMQGWFNIRKSINVIHYINK-LKDKNHMI-ISLDAEKAFDKIQHPFMIKVLERS 646
Query: 501 GFPPSWVSLIMSV--SPLLGFPL---CLTEIPSRVLLLLEAYVRVILFRLIFLSYVERCS 555
G ++++I ++ P+ + L IP + + LF ++
Sbjct: 647 GIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVL-------- 698
Query: 556 QLLLRSV-WLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNILSTYERASG 614
++L R++ + GI++ IS L ADD I++ +LN+++++ G
Sbjct: 699 EVLARAIRQQKEIKGIQIGKEEVKIS--LLADDMIVYISDPKNSTRELLNLINSFGEVVG 756
Query: 615 QVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVESFDKYLG--LPTMIGKFKTRIFNFVK 672
IN +KS + Q E+++ + V + KYLG L + + F +K
Sbjct: 757 YKINSNKSMAFLYTKNKQ-AEKEIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLK 815
Query: 673 DRVWKKLKGWKESTLSRAGREVLIKSVV--QAIPSYVMSCFILPDSLCADIERMVSRFYW 730
+ + L+ WK+ S GR ++K + +AI + +P ++E + +F W
Sbjct: 816 KEIKEDLRRWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVW 875
Query: 731 GGDVDKRGLQWASWKKLTRSK-FDGGLGFRDFKSFNIALVAKNWWRIY 777
+ K L + K GG+ D K + A+V K W Y
Sbjct: 876 NNKKPRIA------KSLLKDKRTSGGITMPDLKLYYRAIVIKTAWYWY 917
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 126 bits (317), Expect = 3e-28
Identities = 125/549 (22%), Positives = 238/549 (42%), Gaps = 47/549 (8%)
Query: 253 DKNTRFFHQKANQ-----------RRKRNLIEVLKDDNGREVEDDADIARVLGDYFGGLF 301
+K+ +F +K N+ +R ++LI +++ N D ++I ++L +Y+ L+
Sbjct: 359 NKSKSWFFEKINKIDKPLANLTRKKRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLY 418
Query: 302 TSSNPEGIEETTNLVAG----RVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPT 357
S E ++E + R+S+ + +L P + E+ + + K+PG DGF +
Sbjct: 419 -SHKYENLKEIDQYLEACHLPRLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTS 477
Query: 358 LFYQKYWDIIGDDVSAFCLQVLQGVIPPGMINQTL----LVLIPKIMK-PEHATQFRPIS 412
FYQ + +++ L + Q + G++ T + LIPK K P +RPIS
Sbjct: 478 EFYQTF----KEELVPILLNLFQNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPIS 533
Query: 413 LCNVLFKIITKTIANRLKIILPDIICQTQSAFVPGR---LIIDNALVVYECFHFMKKRIS 469
L N+ KI+ K + NR++ + II Q F+PG I ++ V + + +K +
Sbjct: 534 LMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLKNK-- 591
Query: 470 GRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPPSWVSLIMSVSPLLGFPLCLTEIPSR 529
M L +D KA+D ++ F+ L+K+G +++ LI ++ + L + +
Sbjct: 592 ---DHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLK 648
Query: 530 VLLLLEAYVRVILFRLIFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDSI 589
L + + + V + +R + GI + + +S LFADD I
Sbjct: 649 SFPLRSGTRQGCPLSPLLFNIVMEVLAIAIREE--KAIKGIHIGSEEIKLS--LFADDMI 704
Query: 590 IFARANTQEAECVLNILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLLNVNAVES 649
++ +L ++ Y SG IN KS + N Q +K + V
Sbjct: 705 VYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTNNNQ-AEKTVKDSIPFTVVPK 763
Query: 650 FDKYLG--LPTMIGKFKTRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVV--QAIPS 705
KYLG L + + ++ + + + WK S GR ++K + +AI +
Sbjct: 764 KMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKAIYN 823
Query: 706 YVMSCFILPDSLCADIERMVSRFYWGGDVDKRGLQWASWKKLTRSKFDGGLGFRDFKSFN 765
+ P S D+E+++ F W +++ Q A ++K GG+ D + +
Sbjct: 824 FNAIPIKAPLSYFKDLEKIILHFIW----NQKKPQIAKTLLSNKNK-AGGITLPDLRLYY 878
Query: 766 IALVAKNWW 774
++V K W
Sbjct: 879 KSIVIKTAW 887
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 119 bits (299), Expect = 4e-26
Identities = 60/149 (40%), Positives = 84/149 (56%), Gaps = 1/149 (0%)
Query: 702 AIPSYVMSCFILPDSLCADIERMVSRFYWGGDVDKRGLQWASWKKLTRSKFD-GGLGFRD 760
A+P Y MSCF L LC + ++ F+W +KR + W +W+KL +SK D GGLGFRD
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWSSCENKRKISWVAWQKLCKSKEDDGGLGFRD 61
Query: 761 FKSFNIALVAKNWWRIYTQPETLLAQVFKGVYFAQGDLLGAKRGYRPSYAWSSILQSSWI 820
FN AL+AK +RI QP TLL+++ + YF ++ G RPSYAW SI+ +
Sbjct: 62 LGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGREL 121
Query: 821 FQEGGRWKIGDGSQVDILHDQWLPQGVPV 849
G IGDG + D+W+ P+
Sbjct: 122 LSRGLLRTIGDGIHTKVWLDRWIMDETPL 150
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 52.8 bits (125), Expect = 6e-06
Identities = 83/346 (23%), Positives = 130/346 (36%), Gaps = 49/346 (14%)
Query: 167 GRSLTEVPDRLSDVSGLLGRWGKEQFGDLPRKISDGQALLQELQRKDQTKEVLMAINVAE 226
GR LT P R + RW R++ + LLQ+ +R+D V + +
Sbjct: 292 GRKLTRSPSRRA-------RWWTADLCAARREVRRLRRLLQDGRRRDDDAAVELVVVELR 344
Query: 227 RDLDSLLEQEEVWWKQRSRAS---WLK----HGDKNTRFFHQKANQRRKRNLIEVLKDDN 279
R + K RA W + H D ++ RRK I L+ N
Sbjct: 345 R-------ASAYYKKLIGRAKMDDWKRFVGDHADDPWGRVYKICRGRRKCTEIGCLRV-N 396
Query: 280 GREVEDDADIARVLGDYFGGLFTSSNPEGIEETTNLVAGRVSETHLTVLGEPFTREEVEE 339
G + D D ARVL F + S P I E E F EV+
Sbjct: 397 GELITDWGDCARVLLRNFFPVAESEAPTAIAEEVPPAL------------EVF---EVDT 441
Query: 340 ALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDV-SAFCLQVLQGVIPPGMINQTLLVLIPK 398
+ ++ ++PGLDG + W I + + S F + G P ++ L+
Sbjct: 442 CVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVSLLKG 501
Query: 399 IMKPE-HATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVPGRLIIDNALVV 457
K + + +R I L V K++ + NR++ +LP+ C+ Q F GR + D
Sbjct: 502 PDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPE-GCRWQFGFRQGRCVED----- 555
Query: 458 YECFHFMKKRISGRNGMMTLK--LDMSKAYDRVEWSFLQGVLQKMG 501
+ +K + L +D A+D VEWS L +G
Sbjct: 556 --AWRHVKSSVGASAAQYVLGTFVDFKGAFDNVEWSAALSRLADLG 599
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 48.1 bits (113), Expect = 1e-04
Identities = 50/182 (27%), Positives = 84/182 (45%), Gaps = 33/182 (18%)
Query: 331 PFTREEVEEALFQMHPTKAPGLDGF----------PTLFYQKYWDIIGDDVSAFCLQVLQ 380
P REE++ A+ P+ APG DG P F Q L +L+
Sbjct: 5 PIAREEIQCAIKGWKPS-APGSDGLTVQAITRTRLPRNFVQ--------------LHLLR 49
Query: 381 GVIP-PGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQ 439
G +P P +T LIPK E+ + +RPI++ + L +++ + +A RL+ + Q
Sbjct: 50 GHVPTPWTAMRT--TLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVELHPAQ 107
Query: 440 TQSAFVPGRLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQK 499
A + G L+ N+L++ ++ R R + LD+ KA+D V S + LQ+
Sbjct: 108 KGYARIDGTLV--NSLLLDT---YISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQR 162
Query: 500 MG 501
+G
Sbjct: 163 LG 164
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 44.7 bits (104), Expect = 0.002
Identities = 68/274 (24%), Positives = 114/274 (40%), Gaps = 39/274 (14%)
Query: 256 TRFFHQKANQRRKRN--LIEVL--KDDNG--REVEDDADI-------ARVLGDYFGGLFT 302
T +H Q+RK+ L++ L KD + R V D+ D R + DY+ +F
Sbjct: 74 TTIYHGTNKQKRKQRYALVQSLYKKDISAAARVVLDENDKIATKIPPVRHMFDYWKDVFA 133
Query: 303 SSNPEGIEETTNLVAGRVSETHLTVLGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQK 362
+ G TN+ + H+ L +P + E++ A + K G DG +
Sbjct: 134 TG---GGSAATNINRAPPAP-HMETLWDPVSLIEIKSA--RASNEKGAGPDGVTP----R 183
Query: 363 YWDIIGDDVSA--FCLQVLQGVIPPGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKI 420
W+ + D + + V G +P I + V PKI FRP+S+C+V+ +
Sbjct: 184 SWNALDDRYKRLLYNIFVFYGRVP-SPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILRE 242
Query: 421 ITKTIANRLKIILPDIICQT----QSAFVPGRLIIDNALVVYECFHFMKKRISGRNGMMT 476
K +A R + C T Q+A++P + N ++ K+ R +
Sbjct: 243 FNKILARRF------VSCYTYDERQTAYLPIDGVCINVSMLTAIIAEAKRL---RKELHI 293
Query: 477 LKLDMSKAYDRVEWSFLQGVLQKMGFPPSWVSLI 510
LD+ KA++ V S L + + G PP V I
Sbjct: 294 AILDLVKAFNSVYHSALIDAITEAGCPPGVVDYI 327
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 44.3 bits (103), Expect = 0.002
Identities = 60/243 (24%), Positives = 99/243 (40%), Gaps = 25/243 (10%)
Query: 385 PGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAF 444
P I V IPK + + FRPIS+ +VL + + +A RL + Q F
Sbjct: 392 PHSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSIN--WDPRQRGF 449
Query: 445 VPGRLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPP 504
+P DNA +V K LD+SKA+D + + + L+ G P
Sbjct: 450 LPTDGCADNATIVDLVLRHSHKHF---RSCYIANLDVSKAFDSLSHASIYDTLRAYGAPK 506
Query: 505 SWVSLIMSV-----SPLLGFPLCLTE-IPSRVLLLLEAYVRVILFRLIFLSYVERCSQLL 558
+V + + + L G E +P+R + + + ILF L+ ++R + L
Sbjct: 507 GFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDP-LSPILFNLV----MDRLLRTL 561
Query: 559 LRSVWLLMLHGIKVANRAPVISHLLFADDSIIFARANTQEAECVLNILSTYERASGQVIN 618
+ G KV N + + FADD ++FA + +L+ + G +N
Sbjct: 562 PSEI------GAKVGN--AITNAAAFADDLVLFAETR-MGLQVLLDKTLDFLSIVGLKLN 612
Query: 619 LDK 621
DK
Sbjct: 613 ADK 615
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 42.4 bits (98), Expect = 0.008
Identities = 44/186 (23%), Positives = 78/186 (41%), Gaps = 11/186 (5%)
Query: 328 LGEPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQVLQGVIPPGM 387
L P + +E++E + A G DG T W+ I + + + ++ P
Sbjct: 315 LWRPISNDEIKEV--EACKRTAAGPDGMTTTA----WNSIDECIKSLFNMIMYHGQCPRR 368
Query: 388 INQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAFVPG 447
+ VLIPK FRP+S+ +V + + +ANR I ++ Q AF+
Sbjct: 369 YLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANR--IGEHGLLDTRQRAFIVA 426
Query: 448 RLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMGFPPSWV 507
+ +N ++ + +I G+ LD+ KA+D VE + L++ P
Sbjct: 427 DGVAENTSLLSAMIKEARMKI---KGLYIAILDVKKAFDSVEHRSILDALRRKKLPLEMR 483
Query: 508 SLIMSV 513
+ IM V
Sbjct: 484 NYIMWV 489
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 42.0 bits (97), Expect = 0.011
Identities = 38/165 (23%), Positives = 74/165 (44%), Gaps = 12/165 (7%)
Query: 330 EPFTREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQVLQ-----GVIP 384
+P T EEV+E + ++ P KAPG D L + ++ D + + + G P
Sbjct: 436 DPVTLEEVKELVSKLKPKKAPGED----LLDNRTIRLLPDQALLYLVLIFNSILRVGYFP 491
Query: 385 PGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAF 444
++++++ +P +RP SL L K++ + I NR I+ + + + F
Sbjct: 492 KARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNR--ILTSEEVTRAIPKF 549
Query: 445 VPG-RLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRV 488
G RL ++ +F + + + + LD+ +A+DRV
Sbjct: 550 QFGFRLQHGTPEQLHRVVNFALEALEKKEYAGSCFLDIQQAFDRV 594
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 41.6 bits (96), Expect = 0.014
Identities = 74/322 (22%), Positives = 120/322 (36%), Gaps = 63/322 (19%)
Query: 241 KQRSRASWLKHGDKNTRFFHQ-------KANQRRKRNLIEVLKDDNGREVEDDADIARVL 293
K+R R + + GD + H KA RRK+ I+ D+ G + + + R+
Sbjct: 309 KRRVRKEYARTGDPRMQQIHSRLANCLHKALARRKQAQIDTFLDNLGADASTNYSLWRIT 368
Query: 294 GDYFGGLFTSS---NPEGIEETTNLVAGRVSETHLT-------------------VLGEP 331
+ S NP G T+L V +L L P
Sbjct: 369 KRFKAQPTPKSAIKNPSGGWCRTSLEKTEVFANNLEQRFTPYNYAPESLCRQVEEYLESP 428
Query: 332 F---------TREEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQVLQGV 382
F T EEV+ + ++ KAPG D L + ++ D F + V
Sbjct: 429 FQMSLPLSAVTLEEVKNLIAKLPLKKAPGED----LLDNRTIRLLPDQALQFLALIFNSV 484
Query: 383 IPPGMI----NQTLLVLIPKIMK-PEHATQFRPISLCNVLFKIITKTIANRLKIILPDII 437
+ G +++I K K P +RP SL L KI+ + I NRL +
Sbjct: 485 LDVGYFPKAWKSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRL------LT 538
Query: 438 CQTQSAFVPG-----RLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSF 492
C+ + +P RL ++ +F + + + + LD+ +A+DRV W
Sbjct: 539 CKDVTKAIPKFQFGFRLQHGTPEQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-WH- 596
Query: 493 LQGVLQKMG--FPPSWVSLIMS 512
G+L K FPP ++ S
Sbjct: 597 -PGLLYKAKRLFPPQLYLVVKS 617
>GUND_CELFI (P50400) Endoglucanase D precursor (EC 3.2.1.4)
(Endo-1,4-beta-glucanase) (Cellulase)
Length = 747
Score = 39.7 bits (91), Expect = 0.052
Identities = 32/121 (26%), Positives = 54/121 (44%), Gaps = 10/121 (8%)
Query: 147 FEELWLQKNEECKEVIAETWGRSLTEVPDRLSDVSGLL-GRWGKEQFGDLPRKISDGQAL 205
F++ W QK+ + + A+ WG + + D D++ LL G WG + G PR+ A
Sbjct: 311 FDQKWFQKDFDKASLTADVWGPNWLFIHD--EDIAPLLIGEWG-GRLGQDPRQ-DKWMAA 366
Query: 206 LQELQRKDQTKEVLMAINVAERDLDSLLEQEEVWWKQRSRASWL-----KHGDKNTRFFH 260
L++L + + + +N D LL + W + ++ L KHG K H
Sbjct: 367 LRDLVAERRLSQTFWVLNPNSGDTGGLLLDDWKTWDEVKYSTMLEPTLWKHGGKYVGLDH 426
Query: 261 Q 261
Q
Sbjct: 427 Q 427
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 38.5 bits (88), Expect = 0.12
Identities = 27/117 (23%), Positives = 50/117 (42%), Gaps = 3/117 (2%)
Query: 385 PGMINQTLLVLIPKIMKPEHATQFRPISLCNVLFKIITKTIANRLKIILPDIICQTQSAF 444
P +++ IPK P + +RPISL + +I+ + I +R++ ++ Q F
Sbjct: 662 PNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGF 721
Query: 445 VPGRLIIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEWSFLQGVLQKMG 501
+ R + + +H + K + L D +KA+D+V L L G
Sbjct: 722 LNFRSCPSSLVRSISLYHSILK---NEKSLDILFFDFAKAFDKVSHPILLKKLALFG 775
>RPOB_ASTLO (P27059) DNA-directed RNA polymerase beta chain (EC
2.7.7.6) (PEP) (Plastid-encoded RNA polymerase beta
subunit) (RNA polymerase beta subunit)
Length = 1076
Score = 36.2 bits (82), Expect = 0.58
Identities = 24/128 (18%), Positives = 56/128 (43%), Gaps = 4/128 (3%)
Query: 584 FADDSIIFARANTQEAECVLNILSTYERASGQVINLDKSQLSVSRNVPQNGFNELKQLLN 643
F D II R ++ +++ ++ +IN + + +++N+P +K L N
Sbjct: 691 FEDAIIISERLRNEDILTSIHV----KKCKSFLINHKEKKEEITKNIPDIKLKNVKNLNN 746
Query: 644 VNAVESFDKYLGLPTMIGKFKTRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVVQAI 703
++ K G +IG K R+ N+ + +++ L ++ +S + L+ V
Sbjct: 747 NGIIKIGSKINGKEILIGIIKKRLINYEVELIYEILSEYERKNISVLSPKNLVGIVTNTK 806
Query: 704 PSYVMSCF 711
+ +C+
Sbjct: 807 IHKIKNCY 814
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 35.4 bits (80), Expect = 0.99
Identities = 38/161 (23%), Positives = 69/161 (42%), Gaps = 12/161 (7%)
Query: 335 EEVEEALFQMHPTKAPGLDGFPTLFYQKYWDIIGDDVSAFCLQ---VLQGVIPPGMINQT 391
+EV +++ + K+PG DG + W I + + FCL +L+ P +
Sbjct: 408 DEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYM--FCLYKQCLLESYFPQKWKIAS 465
Query: 392 LLVLIPKIMK-PEHATQFRPISLCNVLFKIITKTIANRLKIILPDI-ICQTQSAFVPGRL 449
L++L+ + + +RPI L + L K++ + RL L D+ + Q AF G+
Sbjct: 466 LVILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKS 525
Query: 450 IIDNALVVYECFHFMKKRISGRNGMMTLKLDMSKAYDRVEW 490
D + C S ++ L +D A+D + W
Sbjct: 526 TED----AWRCVQ-RHVECSEMKYVIGLNIDFQGAFDNLGW 561
>YKY1_SCHPO (O13796) Hypothetical protein C1142.01 in chromosome I
Length = 667
Score = 34.7 bits (78), Expect = 1.7
Identities = 34/130 (26%), Positives = 57/130 (43%), Gaps = 13/130 (10%)
Query: 191 QFGDLPRKISDGQALLQELQ-RKDQTKEVLMAINVAERDL--DSLLEQEEVWWKQRSR-- 245
QF +L +S LL E RK + +L ++ L D+ L + +V+ + SR
Sbjct: 523 QFAELTENLSRRVILLNEQSLRKFLPQRILQGTILSFDPLPPDTYLSESQVFGRDISRRI 582
Query: 246 ASWLKHGDKNTRFFHQKANQRRKRN--------LIEVLKDDNGREVEDDADIARVLGDYF 297
AS+L R ++ + L++ ++ + G E ED + L +F
Sbjct: 583 ASFLSDYLSRAREVNENEEEPPAHEFDLPPAEQLLQQIESEVGEESEDGTPVMTRLRSFF 642
Query: 298 GGLFTSSNPE 307
G LFTS+N E
Sbjct: 643 GSLFTSTNSE 652
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 34.7 bits (78), Expect = 1.7
Identities = 19/43 (44%), Positives = 27/43 (62%)
Query: 546 IFLSYVERCSQLLLRSVWLLMLHGIKVANRAPVISHLLFADDS 588
+F+ E S L R+ L GI+V+N +P I+HLLFADD+
Sbjct: 38 LFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHLLFADDT 80
>CLPB_BIFLO (Q8G4X4) Chaperone clpB
Length = 889
Score = 33.9 bits (76), Expect = 2.9
Identities = 26/79 (32%), Positives = 39/79 (48%), Gaps = 8/79 (10%)
Query: 132 DRRKRKQSKREYMFRFEELWLQKNEECKEVIAETWGRSLTEVPDRLSDVSGLLGRWGKEQ 191
D +RK ++ E EE+ L+K E+ E G+ E+ D +SGL RW EQ
Sbjct: 414 DELQRKVTRLE----MEEMQLKKAED--PASKERLGKLQAELADTREKLSGLKARWDAEQ 467
Query: 192 FGDLPRKISDGQALLQELQ 210
G K+ D +A L +L+
Sbjct: 468 AGH--NKVGDLRAKLDDLR 484
>RGYR_METJA (Q58907) Reverse gyrase [Includes: Helicase (EC
3.6.1.-); Topoisomerase (EC 5.99.1.3)] [Contains: Mja
r-Gyr intein]
Length = 1613
Score = 33.1 bits (74), Expect = 4.9
Identities = 33/120 (27%), Positives = 54/120 (44%), Gaps = 11/120 (9%)
Query: 585 ADDSIIFARANTQEAECVLNILSTYERASGQVINLDKS--QLSVSRNVPQNGFNELKQLL 642
A ++I R E + + ++T G+ I D + L +RN+ + GFNE+ +
Sbjct: 659 AKENIEIIREIADEVDAIF--IATDIDTEGEKIGYDIAINALPFNRNIYRVGFNEITKRA 716
Query: 643 NVNAVESFDKYLGLPTMIGKFKTRIFNFVKDRVWKKLKGWKESTLSRAGREVLIKSVVQA 702
+ AVESF K L K K ++ ++DR W LS+ EV K+ + A
Sbjct: 717 ILKAVESFKKGEELSLDENKVKGQVVRRIEDR-------WIGFRLSQKLWEVFNKNYLSA 769
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.139 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 154,075,927
Number of Sequences: 164201
Number of extensions: 6763700
Number of successful extensions: 18692
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 18642
Number of HSP's gapped (non-prelim): 51
length of query: 1259
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1137
effective length of database: 39,941,532
effective search space: 45413521884
effective search space used: 45413521884
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0129.4


