

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.9
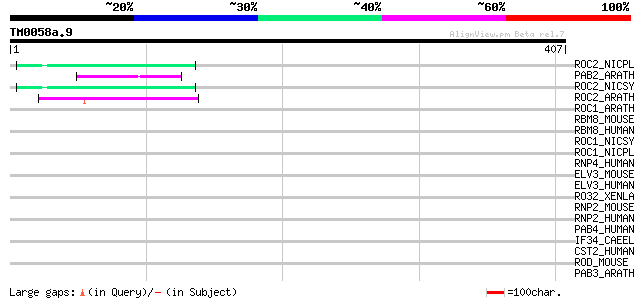
(407 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ROC2_NICPL (P49314) 31 kDa ribonucleoprotein, chloroplast precur... 46 1e-04
PAB2_ARATH (P42731) Polyadenylate-binding protein 2 (Poly(A)-bin... 46 1e-04
ROC2_NICSY (Q08937) 29 kDa ribonucleoprotein B, chloroplast prec... 46 2e-04
ROC2_ARATH (Q43349) 29 kDa ribonucleoprotein, chloroplast precur... 45 4e-04
ROC1_ARATH (Q9ZUU4) Putative ribonucleoprotein At2g37220, chloro... 42 0.002
RBM8_MOUSE (Q9CWZ3) RNA-binding protein 8A (RNA binding motif pr... 42 0.003
RBM8_HUMAN (Q9Y5S9) RNA-binding protein 8A (RNA binding motif pr... 42 0.003
ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A, chloroplast prec... 42 0.004
ROC1_NICPL (P49313) 30 kDa ribonucleoprotein, chloroplast precur... 41 0.005
RNP4_HUMAN (Q86U06) RNA-binding region containing protein 4 (Spl... 41 0.006
ELV3_MOUSE (Q60900) ELAV-like protein 3 (Hu-antigen C) (HuC) 41 0.006
ELV3_HUMAN (Q14576) ELAV-like protein 3 (Hu-antigen C) (HuC) (Pa... 41 0.006
RO32_XENLA (P51992) Heterogeneous nuclear ribonucleoprotein A3 h... 40 0.008
RNP2_MOUSE (Q8VH51) RNA-binding region containing protein 2 (Coa... 40 0.008
RNP2_HUMAN (Q14498) RNA-binding region containing protein 2 (Hep... 40 0.008
PAB4_HUMAN (Q13310) Polyadenylate-binding protein 4 (Poly(A)-bin... 40 0.011
IF34_CAEEL (Q19706) Probable eukaryotic translation initiation f... 40 0.011
CST2_HUMAN (P33240) Cleavage stimulation factor, 64 kDa subunit ... 40 0.011
ROD_MOUSE (Q60668) Heterogeneous nuclear ribonucleoprotein D0 (h... 40 0.014
PAB3_ARATH (O64380) Polyadenylate-binding protein 3 (Poly(A)-bin... 40 0.014
>ROC2_NICPL (P49314) 31 kDa ribonucleoprotein, chloroplast precursor
(CP-RBP31)
Length = 292
Score = 46.2 bits (108), Expect = 1e-04
Identities = 40/131 (30%), Positives = 53/131 (39%), Gaps = 3/131 (2%)
Query: 6 RASRSFAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFID 65
RA R A + A R+ S G G GG DG + G S V++
Sbjct: 157 RAIRVNAGPAPAKRENSSF---GGGRGGNSSYGGGRDGNSSFGGARGGRSVDSSNRVYVG 213
Query: 66 GIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFR 125
+ D L L+++ S+ G VV+ V R SGR FGF Y + AI LNG
Sbjct: 214 NLSWGVDDLALKELFSEQGNVVDAKVVYDRDSGRSRGFGFVTYSSAKEVNDAIDSLNGID 273
Query: 126 LGSAPLSASLA 136
L + S A
Sbjct: 274 LDGRSIRVSAA 284
>PAB2_ARATH (P42731) Polyadenylate-binding protein 2
(Poly(A)-binding protein 2) (PABP 2)
Length = 629
Score = 46.2 bits (108), Expect = 1e-04
Identities = 28/77 (36%), Positives = 40/77 (51%), Gaps = 1/77 (1%)
Query: 50 RSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYG 109
R S R ++FI ++ES D+ L D S FG +V+ V SG+ +GF +Y
Sbjct: 114 RDPSVRRSGAGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVD-SSGQSKGYGFVQYA 172
Query: 110 LRDQALKAIQGLNGFRL 126
+ A KAI+ LNG L
Sbjct: 173 NEESAQKAIEKLNGMLL 189
Score = 35.8 bits (81), Expect = 0.20
Identities = 19/76 (25%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query: 50 RSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYG 109
R +A + +V++ + ES L++ ++GK+ + V + + G+ FGF +
Sbjct: 205 RDSTANKTKFTNVYVKNLAESTTDDDLKNAFGEYGKITSAVVMKDGE-GKSKGFGFVNFE 263
Query: 110 LRDQALKAIQGLNGFR 125
D A +A++ LNG +
Sbjct: 264 NADDAARAVESLNGHK 279
Score = 32.0 bits (71), Expect = 2.9
Identities = 20/76 (26%), Positives = 39/76 (51%), Gaps = 1/76 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++++ ++ S L++I S FG V + V R +G GF + ++A +A+
Sbjct: 319 NLYVKNLDPSISDEKLKEIFSPFGTVTSSKVMRD-PNGTSKGSGFVAFATPEEATEAMSQ 377
Query: 121 LNGFRLGSAPLSASLA 136
L+G + S PL ++A
Sbjct: 378 LSGKMIESKPLYVAIA 393
>ROC2_NICSY (Q08937) 29 kDa ribonucleoprotein B, chloroplast
precursor (CP29B)
Length = 291
Score = 45.8 bits (107), Expect = 2e-04
Identities = 40/131 (30%), Positives = 53/131 (39%), Gaps = 3/131 (2%)
Query: 6 RASRSFAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFID 65
RA R A + A R+ S G G GG DG + G S V++
Sbjct: 156 RAIRVNAGPAPAKRENSSF---GGGRGGNSSYGGGRDGNSSFGGARGGRSVDSSNRVYVG 212
Query: 66 GIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFR 125
+ D L L+++ S+ G VV+ V R SGR FGF Y + AI LNG
Sbjct: 213 NLSWGVDDLALKELFSEQGNVVDAKVVYDRDSGRSRGFGFVTYSSSKEVNDAIDSLNGVD 272
Query: 126 LGSAPLSASLA 136
L + S A
Sbjct: 273 LDGRSIRVSAA 283
>ROC2_ARATH (Q43349) 29 kDa ribonucleoprotein, chloroplast precursor
(RNA-binding protein cp29)
Length = 342
Score = 44.7 bits (104), Expect = 4e-04
Identities = 37/125 (29%), Positives = 51/125 (40%), Gaps = 8/125 (6%)
Query: 22 GSTTARGVGDVAGEEEGGELDGEADGGSRSCS--------AGRRSCFSVFIDGIEESFDY 73
GS G G G G + G GGS+ S +G S +++ + D
Sbjct: 211 GSERGGGYGSERGGGYGSQRSGGGYGGSQRSSYGSGSGSGSGSGSGNRLYVGNLSWGVDD 270
Query: 74 LHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSA 133
+ L ++ ++ GKVV V R SGR FGF + KAI LNG L +
Sbjct: 271 MALENLFNEQGKVVEARVIYDRDSGRSKGFGFVTLSSSQEVQKAINSLNGADLDGRQIRV 330
Query: 134 SLAWA 138
S A A
Sbjct: 331 SEAEA 335
>ROC1_ARATH (Q9ZUU4) Putative ribonucleoprotein At2g37220,
chloroplast precursor
Length = 289
Score = 42.4 bits (98), Expect = 0.002
Identities = 31/101 (30%), Positives = 42/101 (40%), Gaps = 4/101 (3%)
Query: 38 GGELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKS 97
G G GG +G R V++ + D + L + S+ GKVV V R S
Sbjct: 186 GSSGSGYGGGGGSGAGSGNR----VYVGNLSWGVDDMALESLFSEQGKVVEARVIYDRDS 241
Query: 98 GRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWA 138
GR FGF Y + AI+ L+G L + S A A
Sbjct: 242 GRSKGFGFVTYDSSQEVQNAIKSLDGADLDGRQIRVSEAEA 282
Score = 32.3 bits (72), Expect = 2.2
Identities = 43/176 (24%), Positives = 65/176 (36%), Gaps = 23/176 (13%)
Query: 7 ASRSFAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDG 66
A++ +P S AR + T+ V EE G D A +S SA + +F+
Sbjct: 49 AAKWNSPASRFARNVAITSEFEV------EEDGFAD-VAPPKEQSFSADLK----LFVGN 97
Query: 67 IEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRL 126
+ + D L + G V + V + +GR FGF + A Q NG+ L
Sbjct: 98 LPFNVDSAQLAQLFESAGNVEMVEVIYDKITGRSRGFGFVTMSSVSEVEAAAQQFNGYEL 157
Query: 127 GSAPLSASLAWAP------------WVCGSPRRGFGLCSQASSAQGQRLAARVLSW 170
PL + P GS G+G + + G R+ LSW
Sbjct: 158 DGRPLRVNAGPPPPKREDGFSRGPRSSFGSSGSGYGGGGGSGAGSGNRVYVGNLSW 213
>RBM8_MOUSE (Q9CWZ3) RNA-binding protein 8A (RNA binding motif
protein 8A) (Ribonucleoprotein RBM8A)
Length = 174
Score = 42.0 bits (97), Expect = 0.003
Identities = 27/109 (24%), Positives = 48/109 (43%), Gaps = 5/109 (4%)
Query: 40 ELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGR 99
E DG+ G RS + +F+ G+ E + D +++G++ N+ + R++G
Sbjct: 58 EQDGDEPGPQRSVEG-----WILFVTGVHEEATEEDIHDKFAEYGEIKNIHLNLDRRTGY 112
Query: 100 FFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAPWVCGSPRRG 148
+ Y +A A++GLNG L P+S + RRG
Sbjct: 113 LKGYTLVEYETYKEAQAAMEGLNGQDLMGQPISVDWCFVRGPPKGKRRG 161
>RBM8_HUMAN (Q9Y5S9) RNA-binding protein 8A (RNA binding motif
protein 8A) (Ribonucleoprotein RBM8A) (RNA-binding
protein Y14) (Binder of OVCA1-1) (BOV-1)
Length = 174
Score = 42.0 bits (97), Expect = 0.003
Identities = 27/109 (24%), Positives = 48/109 (43%), Gaps = 5/109 (4%)
Query: 40 ELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGR 99
E DG+ G RS + +F+ G+ E + D +++G++ N+ + R++G
Sbjct: 58 EQDGDEPGPQRSVEG-----WILFVTGVHEEATEEDIHDKFAEYGEIKNIHLNLDRRTGY 112
Query: 100 FFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAPWVCGSPRRG 148
+ Y +A A++GLNG L P+S + RRG
Sbjct: 113 LKGYTLVEYETYKEAQAAMEGLNGQDLMGQPISVDWCFVRGPPKGKRRG 161
>ROC1_NICSY (Q08935) 29 kDa ribonucleoprotein A, chloroplast
precursor (CP29A)
Length = 273
Score = 41.6 bits (96), Expect = 0.004
Identities = 34/115 (29%), Positives = 48/115 (41%), Gaps = 9/115 (7%)
Query: 35 EEEGGELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQ 94
+ E G + GG S+ R V++ + D L + S+ GKVV+ V
Sbjct: 168 KRENSSFRGGSRGGGSFDSSNR-----VYVGNLAWGVDQDALETLFSEQGKVVDAKVVYD 222
Query: 95 RKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAPWVCGSPRRGF 149
R SGR FGF Y ++ AI+ L+G L + S P PRR F
Sbjct: 223 RDSGRSRGFGFVTYSSAEEVNNAIESLDGVDLNGRAIRVS----PAEARPPRRQF 273
>ROC1_NICPL (P49313) 30 kDa ribonucleoprotein, chloroplast precursor
(CP-RBP30)
Length = 279
Score = 41.2 bits (95), Expect = 0.005
Identities = 34/113 (30%), Positives = 47/113 (41%), Gaps = 9/113 (7%)
Query: 37 EGGELDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRK 96
E G + GG S+ R V++ + D L + S+ GKVV+ V R
Sbjct: 176 ENSSFRGGSRGGGSFDSSNR-----VYVGNLAWGVDQDALETLFSEQGKVVDAKVVYDRD 230
Query: 97 SGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSASLAWAPWVCGSPRRGF 149
SGR FGF Y ++ AI+ L+G L + S P PRR F
Sbjct: 231 SGRSRGFGFVTYSSAEEVNNAIESLDGVDLNGRAIRVS----PAEARPPRRQF 279
>RNP4_HUMAN (Q86U06) RNA-binding region containing protein 4
(Splicing factor SF2) (PP239)
Length = 439
Score = 40.8 bits (94), Expect = 0.006
Identities = 19/56 (33%), Positives = 31/56 (54%)
Query: 76 LRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAPL 131
LR I FGK+ N+ + + +GR +GF + + A +A++ LNGF L P+
Sbjct: 279 LRGIFEPFGKIDNIVLMKDSDTGRSKGYGFITFSDSECARRALEQLNGFELAGRPM 334
>ELV3_MOUSE (Q60900) ELAV-like protein 3 (Hu-antigen C) (HuC)
Length = 367
Score = 40.8 bits (94), Expect = 0.006
Identities = 30/94 (31%), Positives = 41/94 (42%), Gaps = 2/94 (2%)
Query: 41 LDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRF 100
+ G A G +AG C +F+ + D L + FG V N+ V R + +
Sbjct: 267 MSGLAGVGLSGGAAGAGWC--IFVYNLSPEADESVLWQLFGPFGAVTNVKVIRDFTTNKC 324
Query: 101 FRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSAS 134
FGF D+A AI LNG+RLG L S
Sbjct: 325 KGFGFVTMTNYDEAAMAIASLNGYRLGERVLQVS 358
Score = 40.0 bits (92), Expect = 0.011
Identities = 30/118 (25%), Positives = 60/118 (50%), Gaps = 7/118 (5%)
Query: 49 SRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRY 108
+R SA R ++++ G+ ++ + + S++G+++ + + +G GF R+
Sbjct: 115 ARPSSASIRDA-NLYVSGLPKTMSQKEMEQLFSQYGRIITSRILLDQATGVSRGVGFIRF 173
Query: 109 GLRDQALKAIQGLNGFR-LGSA-PLSASLAWAPWVCGSPRRGFGLCSQASSAQGQRLA 164
R +A +AI+GLNG + LG+A P++ A P S + G L + + +R A
Sbjct: 174 DKRIEAEEAIKGLNGQKPLGAAEPITVKFANNP----SQKTGQALLTHLYQSSARRYA 227
Score = 30.8 bits (68), Expect = 6.5
Identities = 16/76 (21%), Positives = 36/76 (47%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++ ++ + ++ + + G + + + R + +G+ +GF Y + A KAI
Sbjct: 40 NLIVNYLPQNMTQDEFKSLFGSIGDIESCKLVRDKITGQSLGYGFVNYSDPNDADKAINT 99
Query: 121 LNGFRLGSAPLSASLA 136
LNG +L + + S A
Sbjct: 100 LNGLKLQTKTIKVSYA 115
>ELV3_HUMAN (Q14576) ELAV-like protein 3 (Hu-antigen C) (HuC)
(Paraneoplastic cerebellar degeneration-associated
antigen) (Paraneoplastic limbic encephalitis antigen 21)
Length = 367
Score = 40.8 bits (94), Expect = 0.006
Identities = 30/94 (31%), Positives = 41/94 (42%), Gaps = 2/94 (2%)
Query: 41 LDGEADGGSRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRF 100
+ G A G +AG C +F+ + D L + FG V N+ V R + +
Sbjct: 267 MSGLAGVGLSGGAAGAGWC--IFVYNLSPEADESVLWQLFGPFGAVTNVKVIRDFTTNKC 324
Query: 101 FRFGFFRYGLRDQALKAIQGLNGFRLGSAPLSAS 134
FGF D+A AI LNG+RLG L S
Sbjct: 325 KGFGFVTMTNYDEAAMAIASLNGYRLGERVLQVS 358
Score = 39.3 bits (90), Expect = 0.018
Identities = 30/118 (25%), Positives = 60/118 (50%), Gaps = 7/118 (5%)
Query: 49 SRSCSAGRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRY 108
+R SA R ++++ G+ ++ + + S++G+++ + + +G GF R+
Sbjct: 115 ARPSSASIRDA-NLYVSGLPKTMSQKEMEQLFSQYGRIITSRILVDQVTGVSRGVGFIRF 173
Query: 109 GLRDQALKAIQGLNGFR-LGSA-PLSASLAWAPWVCGSPRRGFGLCSQASSAQGQRLA 164
R +A +AI+GLNG + LG+A P++ A P S + G L + + +R A
Sbjct: 174 DKRIEAEEAIKGLNGQKPLGAAEPITVKFANNP----SQKTGQALLTHLYQSSARRYA 227
Score = 30.8 bits (68), Expect = 6.5
Identities = 16/76 (21%), Positives = 36/76 (47%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++ ++ + ++ + + G + + + R + +G+ +GF Y + A KAI
Sbjct: 40 NLIVNYLPQNMTQDEFKSLFGSIGDIESCKLVRDKITGQSLGYGFVNYSDPNDADKAINT 99
Query: 121 LNGFRLGSAPLSASLA 136
LNG +L + + S A
Sbjct: 100 LNGLKLQTKTIKVSYA 115
>RO32_XENLA (P51992) Heterogeneous nuclear ribonucleoprotein A3
homolog 2 (hnRNP A3(B))
Length = 385
Score = 40.4 bits (93), Expect = 0.008
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 1/90 (1%)
Query: 62 VFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALK-AIQG 120
+F+ GI+E + HLRD +GK+ + V R+SG+ F F + D K +Q
Sbjct: 120 IFVGGIKEDTEEYHLRDYFEGYGKIETIEVMEDRQSGKKRGFAFVTFDDHDTVDKIVVQK 179
Query: 121 LNGFRLGSAPLSASLAWAPWVCGSPRRGFG 150
+ L + + +L+ S +RG G
Sbjct: 180 YHTINLHNCEVKKALSKQEMQTASAQRGRG 209
>RNP2_MOUSE (Q8VH51) RNA-binding region containing protein 2
(Coactivator of activating protein-1 and estrogen
receptors) (Coactivator of AP-1 and ERs) (Transcription
coactivator CAPER)
Length = 530
Score = 40.4 bits (93), Expect = 0.008
Identities = 27/121 (22%), Positives = 51/121 (41%)
Query: 11 FAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDGIEES 70
F +S+ IG T R +G + A + + G +++ + +
Sbjct: 201 FVDVSSVRLAIGLTGQRVLGVPIIVQASQAEKNRAAAMANNLQKGSAGPMRLYVGSLHFN 260
Query: 71 FDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAP 130
LR I FG++ ++ + ++GR +GF + + A KA++ LNGF L P
Sbjct: 261 ITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQLNGFELAGRP 320
Query: 131 L 131
+
Sbjct: 321 M 321
>RNP2_HUMAN (Q14498) RNA-binding region containing protein 2
(Hepatocellular carcinoma protein 1) (Splicing factor
HCC1)
Length = 530
Score = 40.4 bits (93), Expect = 0.008
Identities = 27/121 (22%), Positives = 51/121 (41%)
Query: 11 FAPISAAARQIGSTTARGVGDVAGEEEGGELDGEADGGSRSCSAGRRSCFSVFIDGIEES 70
F +S+ IG T R +G + A + + G +++ + +
Sbjct: 201 FVDVSSVPLAIGLTGQRVLGVPIIVQASQAEKNRAAAMANNLQKGSAGPMRLYVGSLHFN 260
Query: 71 FDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRLGSAP 130
LR I FG++ ++ + ++GR +GF + + A KA++ LNGF L P
Sbjct: 261 ITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQLNGFELAGRP 320
Query: 131 L 131
+
Sbjct: 321 M 321
>PAB4_HUMAN (Q13310) Polyadenylate-binding protein 4
(Poly(A)-binding protein 4) (PABP 4) (Inducible
poly(A)-binding protein) (iPABP) (Activated-platelet
protein-1) (APP-1)
Length = 644
Score = 40.0 bits (92), Expect = 0.011
Identities = 24/76 (31%), Positives = 41/76 (53%), Gaps = 2/76 (2%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
+++I ++++ D LR S FG + + V + GR FGF + ++A KA+
Sbjct: 295 NLYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLE--DGRSKGFGFVCFSSPEEATKAVTE 352
Query: 121 LNGFRLGSAPLSASLA 136
+NG +GS PL +LA
Sbjct: 353 MNGRIVGSKPLYVALA 368
Score = 38.5 bits (88), Expect = 0.031
Identities = 21/63 (33%), Positives = 36/63 (56%), Gaps = 1/63 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
+V+I E D L+++ S+FGK +++ V R +G+ FGF Y + A KA++
Sbjct: 192 NVYIKNFGEEVDDESLKELFSQFGKTLSVKVMRD-PNGKSKGFGFVSYEKHEDANKAVEE 250
Query: 121 LNG 123
+NG
Sbjct: 251 MNG 253
Score = 31.2 bits (69), Expect = 5.0
Identities = 19/66 (28%), Positives = 33/66 (49%), Gaps = 2/66 (3%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
+VFI +++S D L D S FG +++ V + + F + ++ A KAI+
Sbjct: 100 NVFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSK--GYAFVHFETQEAADKAIEK 157
Query: 121 LNGFRL 126
+NG L
Sbjct: 158 MNGMLL 163
>IF34_CAEEL (Q19706) Probable eukaryotic translation initiation
factor 3 subunit 4 (eIF-3 delta) (eIF3 p44) (eIF-3
RNA-binding subunit) (eIF3g)
Length = 256
Score = 40.0 bits (92), Expect = 0.011
Identities = 19/51 (37%), Positives = 29/51 (56%)
Query: 76 LRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQGLNGFRL 126
LRD+ K G+V+ +F+ R + +G F F + RD A +AI LN R+
Sbjct: 192 LRDLFGKIGRVIRIFIARDKVTGLPKGFAFVTFESRDDAARAIAELNDIRM 242
>CST2_HUMAN (P33240) Cleavage stimulation factor, 64 kDa subunit
(CSTF 64 kDa subunit) (CF-1 64 kDa subunit)
Length = 577
Score = 40.0 bits (92), Expect = 0.011
Identities = 24/67 (35%), Positives = 37/67 (54%)
Query: 57 RSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALK 116
RS SVF+ I L+DI S+ G VV+ + R++G+ +GF Y ++ AL
Sbjct: 13 RSLRSVFVGNIPYEATEEQLKDIFSEVGPVVSFRLVYDRETGKPKGYGFCEYQDQETALS 72
Query: 117 AIQGLNG 123
A++ LNG
Sbjct: 73 AMRNLNG 79
>ROD_MOUSE (Q60668) Heterogeneous nuclear ribonucleoprotein D0
(hnRNP D0) (AU-rich element RNA-binding protein 1)
Length = 355
Score = 39.7 bits (91), Expect = 0.014
Identities = 32/111 (28%), Positives = 52/111 (46%), Gaps = 10/111 (9%)
Query: 5 ARASRSFAPISAAARQIGSTTARGVGDVAGEEEGGELDG----EADGGSRS------CSA 54
A A++ A + + G + A G + E EG ++D E +G S S +A
Sbjct: 32 AAAAQGPAAAAGSGSGGGGSAAGGTEGGSAEAEGAKIDASKNEEDEGHSNSSPRHTEAAA 91
Query: 55 GRRSCFSVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGF 105
+R + +FI G+ L+D SKFG+VV+ ++ +GR FGF
Sbjct: 92 AQREEWKMFIGGLSWDTTKKDLKDYFSKFGEVVDCTLKLDPITGRSRGFGF 142
>PAB3_ARATH (O64380) Polyadenylate-binding protein 3
(Poly(A)-binding protein 3) (PABP 3)
Length = 660
Score = 39.7 bits (91), Expect = 0.014
Identities = 22/76 (28%), Positives = 43/76 (55%), Gaps = 1/76 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++++ +++S D L+++ S++G V + V + G FGF Y ++AL+A+
Sbjct: 333 NLYLKNLDDSVDDEKLKEMFSEYGNVTSSKVMLNPQ-GMSRGFGFVAYSNPEEALRALSE 391
Query: 121 LNGFRLGSAPLSASLA 136
+NG +G PL +LA
Sbjct: 392 MNGKMIGRKPLYIALA 407
Score = 33.5 bits (75), Expect = 1.0
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
+V++ + + LR KFG + + V R + SG FGF + + A A++
Sbjct: 230 NVYVKNLPKEIGEDELRKTFGKFGVISSAVVMRDQ-SGNSRCFGFVNFECTEAAASAVEK 288
Query: 121 LNGFRLG 127
+NG LG
Sbjct: 289 MNGISLG 295
Score = 30.8 bits (68), Expect = 6.5
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Query: 61 SVFIDGIEESFDYLHLRDICSKFGKVVNLFVQRQRKSGRFFRFGFFRYGLRDQALKAIQG 120
++FI ++ S D L + S FG +++ V +GR +GF ++ + A AI
Sbjct: 137 NIFIKNLDASIDNKALFETFSSFGTILSCKVAMD-VTGRSKGYGFVQFEKEESAQAAIDK 195
Query: 121 LNG 123
LNG
Sbjct: 196 LNG 198
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.142 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,860,261
Number of Sequences: 164201
Number of extensions: 2177213
Number of successful extensions: 6491
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 90
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 6280
Number of HSP's gapped (non-prelim): 224
length of query: 407
length of database: 59,974,054
effective HSP length: 113
effective length of query: 294
effective length of database: 41,419,341
effective search space: 12177286254
effective search space used: 12177286254
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0058a.9


