

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
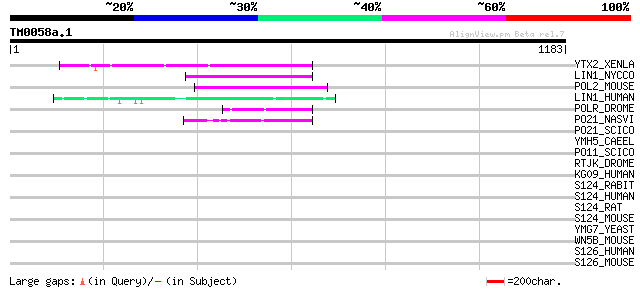
Query= TM0058a.1
(1183 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 142 4e-33
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 96 4e-19
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 93 4e-18
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 87 3e-16
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 52 7e-06
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 45 9e-04
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 45 0.002
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 42 0.008
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 42 0.013
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 38 0.19
KG09_HUMAN (Q9H0A0) UPF0202 protein KIAA1709 37 0.32
S124_RABIT (Q28677) Solute carrier family 12 member 4 (Electrone... 37 0.41
S124_HUMAN (Q9UP95) Solute carrier family 12 member 4 (Electrone... 37 0.41
S124_RAT (Q63632) Solute carrier family 12 member 4 (Electroneut... 36 0.54
S124_MOUSE (Q9JIS8) Solute carrier family 12 member 4 (Electrone... 36 0.54
YMG7_YEAST (Q04651) Hypothetical 40.7 kDa protein in DAK1-ORC1 i... 34 2.7
WN5B_MOUSE (P22726) Wnt-5b protein precursor 33 6.0
S126_HUMAN (Q9UHW9) Solute carrier family 12 member 6 (Electrone... 33 6.0
S126_MOUSE (Q924N4) Solute carrier family 12 member 6 (Electrone... 32 7.8
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 142 bits (359), Expect = 4e-33
Identities = 146/561 (26%), Positives = 250/561 (44%), Gaps = 33/561 (5%)
Query: 106 IINIYSPCDIDGKRAMWDALCAWRGTCDVGE-WCLAGDFNAVRYAEERRGSSGVLSSHRR 164
++N+Y+P + +++L A+ T D E + GDFN A +R SS
Sbjct: 106 LMNVYAPTTGPERARFFESLSAYMETIDSDEALIIGGDFNYTLDARDRNVPKKRDSSES- 164
Query: 165 EMREFNVFIANMDLVDI-----PLTGRRFTWRRANDQ--AQSRIDRFLVSTSWCDVWPNC 217
+RE IA+ LVD+ P T FT+ R D +QSRIDR +S+
Sbjct: 165 VLREL---IAHFSLVDVWREQNPETVA-FTYVRVRDGHVSQSRIDRIYISSHLMSRAQ-- 218
Query: 218 SHLVLNRDVSDHCPLLMRQVVADWGPKP--FRVLNCWLDDPRLFPFVEQSWKGMKIQGWG 275
S + SDH + +R +A PK + N L+D V +W+G +
Sbjct: 219 SSTIRLAPFSDHNCVSLRMSIAPSLPKAAYWHFNNSLLEDEGFAKSVRDTWRGWRAFQDE 278
Query: 276 AYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKIN--LLDVKEEEVGLQPEELVEMR 333
L + + L++ +E + +R ++ +N +LD+++ G + + L +
Sbjct: 279 FATLNQWWDVGKVHLKLLCQEYTKSVSGQRNAEIEALNGEVLDLEQRLSGSEDQAL---Q 335
Query: 334 DLLIEFWRVLRYQESFLCQKAC----MKWLVEGDLNSGFFHSTINWKRRADSIVGLMV-D 388
+E LR E + A M+ L + D S FF++ K I L D
Sbjct: 336 CEYLERKEALRNMEQRQARGAFVRSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAED 395
Query: 389 GMWEDEPVRVKAAIHDFFQKKFK----SVAWLRPKLDGVQFSTITDRDNTELVGPFDEVE 444
G ++P ++ F+Q F S DG+ +++R L P E
Sbjct: 396 GTPLEDPEAIRDRARSFYQNLFSPDPISPDACEELWDGLP--VVSERRKERLETPITLDE 453
Query: 445 IKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLI 504
+ QA+ KSPG DG F Q FWD L D + + E + +G P + + L+
Sbjct: 454 LSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLL 513
Query: 505 PKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVV 564
PK +L+ ++RP+SL+ YK+V K ++RL+ V+ +VI Q V R + D+V +
Sbjct: 514 PKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFL 573
Query: 565 LNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATV 624
+ +++H A+ F +D EKA++ V+ +L + +F +++G++ SA
Sbjct: 574 IRDLLHFARRTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAEC 633
Query: 625 SVLVNGSPSGEFHMEKGLRQG 645
V +N S + +G+RQG
Sbjct: 634 LVKINWSLTAPLAFGRGVRQG 654
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 96.3 bits (238), Expect = 4e-19
Identities = 66/278 (23%), Positives = 132/278 (46%), Gaps = 8/278 (2%)
Query: 376 KRRADSIVGLMVDGMWE--DEPVRVKAAIHDFFQK----KFKSVAWLRPKLDGVQFSTIT 429
K+R S++ + +G E +P ++ ++++++K K++++ + L+ ++
Sbjct: 382 KKRVKSLISSIRNGNDEITTDPSEIQKILNEYYKKLYSHKYENLKEIDQYLEACHLPRLS 441
Query: 430 DRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMR 489
++ L P EI + KSPGPDGF F Q F + L ++ +
Sbjct: 442 QKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKE 501
Query: 490 GA*PRGSNASFIVLIPKV-KIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDER 548
G P + I LIPK K P ++RPISL+ K++ K+ R++ I K+I
Sbjct: 502 GILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHD 561
Query: 549 QFTFV-GNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNF 607
Q F+ G++ + +N + H K+K + + +D EKA+++++ F+ + ++
Sbjct: 562 QVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGI 621
Query: 608 CAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
++ I + T ++++NG F + G RQG
Sbjct: 622 EGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQG 659
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 93.2 bits (230), Expect = 4e-18
Identities = 70/290 (24%), Positives = 131/290 (45%), Gaps = 6/290 (2%)
Query: 394 EPVRVKAAIHDFFQK----KFKSVAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAV 449
+P ++ I F+++ K +++ + LD Q + L P EI+ +
Sbjct: 429 DPEEIQNTIRSFYKRLYSTKLENLDEMDKFLDRYQVPKLNQDQVDHLNSPISPKEIEAVI 488
Query: 450 WECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPK-VK 508
KSPGPDGF+ F Q F + L + K + + G P + I LIPK K
Sbjct: 489 NSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFYEATITLIPKPQK 548
Query: 509 IPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFV-GNRNMLDSVVVLNE 567
P + +FRPISL+ K++ K+ A R++ I +I Q F+ G + + +N
Sbjct: 549 DPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINV 608
Query: 568 VMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVL 627
+ + K+K + + +D EKA++ ++ F+ ++ R ++ I + ++
Sbjct: 609 IHYINKLKDKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIK 668
Query: 628 VNGSPSGEFHMEKGLRQGILLRRFFS*LLLKA*MGYSRMQLELGNLKVSR 677
VNG ++ G RQG L + ++L+ R Q E+ +++ +
Sbjct: 669 VNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQKEIKGIQIGK 718
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 87.0 bits (214), Expect = 3e-16
Identities = 131/647 (20%), Positives = 252/647 (38%), Gaps = 83/647 (12%)
Query: 93 LMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCAWRGTCDVGEWCLAGDFNAVRYAEER 152
+M + Q+ I+NIY+P + R + L + D + GDFN +R
Sbjct: 97 IMVKGSIQQEELTILNIYAP-NTGAPRFIKQVLSDLQRDLD-SHTIIMGDFNTPLSTLDR 154
Query: 153 RGSSGVLSSHRREMREFNVFIANMDLVDIPLT----GRRFTWRRANDQAQSRIDRFLVST 208
+ ++++E N + DL+DI T +T+ A S+ D L S
Sbjct: 155 STRQKI----NKDIQELNSALHQADLIDIYRTLHPKSTEYTFFSAPHHTYSKTDHILGSK 210
Query: 209 SWCDVWPNCSHL-VLNRDVSDHCPL--------LMRQVVADWGPKPFRVLNCWLDDP--- 256
+ + C ++ +SDH + L + W + + W+ +
Sbjct: 211 T---LLSKCKRTEIITNCLSDHSAIKLELRIKKLTQNHSTTWKLNNLLLNDYWVHNEMKA 267
Query: 257 RLFPFVEQS----------WKGMKIQGWGAYVL---------KEKLKGLRGSLRI*NKEV 297
+ F E + W K G ++ + K+ L L+ K+
Sbjct: 268 EIKKFFETNENKDTTYQNLWDTAKAVCRGKFIALNAHKRKQERSKIDTLISQLKELEKQE 327
Query: 298 FGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMK 357
+ K R + + KI ++KE E +++ E R E + + L +K
Sbjct: 328 QTNSKASRRQEIIKIRA-ELKEIETQKTLQKINESRSWFFEKINKIDRPLARLIKK---- 382
Query: 358 WLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDEPVRVKAAIHDFFQ----KKFKS 412
KR + I + D G +P ++ I ++++ K ++
Sbjct: 383 ------------------KREKNQIDTIKNDRGDITTDPTEIQTTIREYYKHLYANKLEN 424
Query: 413 VAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFW 472
+ + LD + + L P EI+ + KSPGP+GF F QR+
Sbjct: 425 LEEMDKFLDTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYK 484
Query: 473 DVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKV-KIPQLLNDFRPISLIGCIYKVVPK 531
+ L ++K + G P + I+LIPK + +FRPISL+ K++ K
Sbjct: 485 EELVPFLLKLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNK 544
Query: 532 LFAIRLRLVIGKVIDERQFTFV----GNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYE 587
+ A +++ I K+I Q F+ G N+ S+ N + H + K + +D E
Sbjct: 545 ILANQIQQHIKKLIHHDQVGFIPAMQGWFNIRKSI---NIIQHINRTKDTNHMIISIDAE 601
Query: 588 KAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGIL 647
KA++ ++ F+ + ++ ++ I + T ++++NG ++ G RQG
Sbjct: 602 KAFDKIQQPFMLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCP 661
Query: 648 LRRFFS*LLLKA*MGYSRMQLELGNLKVSRLGQEAILWCLYCSLLMI 694
L ++L+ R + E +K +LG+E + L+ +++
Sbjct: 662 LSPLLPNIVLEVLARAIRQEKE---IKGIQLGKEEVKLSLFADDMIV 705
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 52.4 bits (124), Expect = 7e-06
Identities = 44/191 (23%), Positives = 81/191 (42%), Gaps = 5/191 (2%)
Query: 455 TKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLN 514
+ SPGPDG + + +V SG +++ + G P + V IPK +
Sbjct: 357 SSSPGPDGITPKSAR---EVPSGIMLRIMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQ 413
Query: 515 DFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKV 574
DFRPIS+ + + + + A RL I D RQ F+ D+ +++ V+ +
Sbjct: 414 DFRPISVPSVLVRQLNAILATRLNSSIN--WDPRQRGFLPTDGCADNATIVDLVLRHSHK 471
Query: 575 KKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSG 634
R +D KA++S+ + + ++ ++ S+ +G S
Sbjct: 472 HFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSE 531
Query: 635 EFHMEKGLRQG 645
EF +G++QG
Sbjct: 532 EFVPARGVKQG 542
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 45.4 bits (106), Expect = 9e-04
Identities = 62/279 (22%), Positives = 113/279 (40%), Gaps = 23/279 (8%)
Query: 371 STINWKRRADSIVGLMVDGMWEDEPVRVKA--AIHDFFQKKFKSVAWLRPKLDGVQFSTI 428
S +WK+ +++DG + P ++ A + + LR ++ G
Sbjct: 257 SNNSWKKNMSKAAHIVLDGDTDACPAGLEGTEASGAIMRAGCPTTRHLRSRMQG------ 310
Query: 429 TDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWM 488
+ L P EIK+ V C T + GPDG W+ + +
Sbjct: 311 ---EIKNLWRPISNDEIKE-VEACKRTAA-GPDGMT----TTAWNSIDECIKSLFNMIMY 361
Query: 489 RGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGK--VID 546
G PR S VLIPK FRP+S+ + ++ A R IG+ ++D
Sbjct: 362 HGQCPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANR----IGEHGLLD 417
Query: 547 ERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMN 606
RQ F+ + ++ +L+ ++ EA++K + +D +KA++SVE + + R
Sbjct: 418 TRQRAFIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKK 477
Query: 607 FCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
+ +IM ++ + V + +G+RQG
Sbjct: 478 LPLEMRNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQG 516
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 44.7 bits (104), Expect = 0.002
Identities = 44/156 (28%), Positives = 65/156 (41%), Gaps = 9/156 (5%)
Query: 439 PFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVRE-FWMRGA*PRGSN 497
P +EIK A K GPDG R W+ L + + F G P
Sbjct: 158 PVSLIEIKSA--RASNEKGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRVPSPIK 211
Query: 498 ASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRN 557
S V PK++ FRP+S+ I + K+ A R V DERQ ++
Sbjct: 212 GSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILA--RRFVSCYTYDERQTAYLPIDG 269
Query: 558 MLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSV 593
+ +V +L ++ EAK ++ +D KA+NSV
Sbjct: 270 VCINVSMLTAIIAEAKRLRKELHIAILDLVKAFNSV 305
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 42.4 bits (98), Expect = 0.008
Identities = 33/146 (22%), Positives = 60/146 (40%), Gaps = 1/146 (0%)
Query: 501 IVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLD 560
I+ IPK P +++RPISL +++ ++ R+R ++ Q F+ R+
Sbjct: 670 IIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPS 729
Query: 561 SVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLN 620
S+V + H ++ D+ KA++ V L + W L+
Sbjct: 730 SLVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLH 789
Query: 621 SATVSVLVNG-SPSGEFHMEKGLRQG 645
T SV +N S + + G+ QG
Sbjct: 790 LRTFSVKINKFVSSNAYPISSGVPQG 815
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 41.6 bits (96), Expect = 0.013
Identities = 55/278 (19%), Positives = 113/278 (39%), Gaps = 28/278 (10%)
Query: 327 EELVEMRDLLIEFWRVLRYQESFLCQKACMKW--LVEGDLNS---GFFHSTINWKRRADS 381
+ L RD IE+ R+L C+ W V + N G +R+
Sbjct: 303 DRLQAFRDCKIEYKRML-------CEAKLRCWQEFVASESNENPWGRVFKLCRGRRKPVD 355
Query: 382 IVGLMVDGMWEDE-PVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTITDRDNTELVGPF 440
+ + VDG++ D V A ++ FF + + +L + D + E
Sbjct: 356 VCSVKVDGVYTDTWEGSVNAMMNVFFPASIDDASEI-DRLKAIARPLPPDLEMDE----- 409
Query: 441 DEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASF 500
+ +V C KSPGPDG ++ W + + ++ + P+ +
Sbjct: 410 ----VSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIAS 465
Query: 501 IVLIPKV--KIPQLLNDFRPISLIGCIYKVVPKLFAIRL-RLVIGKVIDERQFTFVGNRN 557
+V++ K+ +I +RPI L+ + KV+ + RL + ++ + QF F ++
Sbjct: 466 LVILLKLLDRIRSDPGSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKS 525
Query: 558 MLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEW 595
D+ + + +++K I +D++ A++++ W
Sbjct: 526 TEDAWRCVQRHVECSEMKY--VIGLNIDFQGAFDNLGW 561
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 37.7 bits (86), Expect = 0.19
Identities = 49/208 (23%), Positives = 88/208 (41%), Gaps = 4/208 (1%)
Query: 444 EIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVL 503
E+K + + K+PG D + R I+ D + G P+ ++ I++
Sbjct: 442 EVKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSASIIM 501
Query: 504 IPKV-KIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLV--IGKVIDERQFTFVGNRNMLD 560
I K K P ++ +RP SL+ + K++ +L RL + K I + QF F +
Sbjct: 502 IHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQFGFRLQHGTPE 561
Query: 561 SVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLN 620
+ + EA K ++ +D ++A++ V L Y R+ F + + L
Sbjct: 562 QLHRVVNFALEAMENKEYAVGAFLDIQQAFDRVWHPGLLYKAKRL-FPPQLYLVVKSFLE 620
Query: 621 SATVSVLVNGSPSGEFHMEKGLRQGILL 648
T V V+G S + G+ QG +L
Sbjct: 621 ERTFHVSVDGYKSSIKPIAAGVPQGSVL 648
>KG09_HUMAN (Q9H0A0) UPF0202 protein KIAA1709
Length = 1025
Score = 37.0 bits (84), Expect = 0.32
Identities = 22/60 (36%), Positives = 36/60 (59%), Gaps = 4/60 (6%)
Query: 536 RLRLVIGKVIDERQ---FTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNS 592
R+R++I + ERQ F VG+R D VV+L+ ++ +A VK RPS+ + E ++S
Sbjct: 9 RIRILIENGVAERQRSLFVVVGDRGK-DQVVILHHMLSKATVKARPSVLWCYKKELGFSS 67
>S124_RABIT (Q28677) Solute carrier family 12 member 4 (Electroneutral
potassium-chloride cotransporter 1) (Erythroid K-Cl
cotransporter 1) (rbKCC1)
Length = 1085
Score = 36.6 bits (83), Expect = 0.41
Identities = 34/116 (29%), Positives = 48/116 (41%), Gaps = 7/116 (6%)
Query: 910 GEMFRRRVLSGR*IG-LMKLFL----SRFGEVMQLHSGWMIGPGGVLLGSSFI-ACIICP 963
GE RRR +G LM ++L + FG ++ L WM+G GVL + C C
Sbjct: 105 GEGTRRRAAKAPSMGTLMGVYLPCLQNIFGVILFLRLTWMVGTAGVLQALLIVLICCCCT 164
Query: 964 L*SVLLLRAVVTGIKTNGGGIYLGDGRWVCVSKAGWGRCCILYLVPCFGRVRWTLG 1019
L + + + A+ T GG Y R + G C YL F + LG
Sbjct: 165 LLTAISMSAIATNGVVPAGGSYFMISRSLGPEFGGAVGLC-FYLGTTFAAAMYILG 219
>S124_HUMAN (Q9UP95) Solute carrier family 12 member 4 (Electroneutral
potassium-chloride cotransporter 1) (Erythroid K-Cl
cotransporter 1) (hKCC1)
Length = 1085
Score = 36.6 bits (83), Expect = 0.41
Identities = 34/116 (29%), Positives = 48/116 (41%), Gaps = 7/116 (6%)
Query: 910 GEMFRRRVLSGR*IG-LMKLFL----SRFGEVMQLHSGWMIGPGGVLLGSSFI-ACIICP 963
GE RRR +G LM ++L + FG ++ L WM+G GVL + C C
Sbjct: 105 GEGTRRRAAEAPSMGTLMGVYLPCLQNIFGVILFLRLTWMVGTAGVLQALLIVLICCCCT 164
Query: 964 L*SVLLLRAVVTGIKTNGGGIYLGDGRWVCVSKAGWGRCCILYLVPCFGRVRWTLG 1019
L + + + A+ T GG Y R + G C YL F + LG
Sbjct: 165 LLTAISMSAIATNGVVPAGGSYFMISRSLGPEFGGAVGLC-FYLGTTFAAAMYILG 219
>S124_RAT (Q63632) Solute carrier family 12 member 4 (Electroneutral
potassium-chloride cotransporter 1) (Erythroid K-Cl
cotransporter 1) (rKCC1) (Furosemide-sensitive K-Cl
cotransporter)
Length = 1085
Score = 36.2 bits (82), Expect = 0.54
Identities = 34/116 (29%), Positives = 48/116 (41%), Gaps = 7/116 (6%)
Query: 910 GEMFRRRVLSGR*IG-LMKLFL----SRFGEVMQLHSGWMIGPGGVLLGSSFI-ACIICP 963
GE RRR +G LM ++L + FG ++ L WM+G GVL + C C
Sbjct: 105 GEGGRRRAAKAPSMGTLMGVYLPCLQNIFGVILFLRLTWMVGTAGVLQALLIVLICCCCT 164
Query: 964 L*SVLLLRAVVTGIKTNGGGIYLGDGRWVCVSKAGWGRCCILYLVPCFGRVRWTLG 1019
L + + + A+ T GG Y R + G C YL F + LG
Sbjct: 165 LLTAISMSAIATNGVVPAGGSYFMISRSLGPEFGGAVGLC-FYLGTTFAAAMYILG 219
>S124_MOUSE (Q9JIS8) Solute carrier family 12 member 4 (Electroneutral
potassium-chloride cotransporter 1) (Erythroid K-Cl
cotransporter 1) (mKCC1)
Length = 1085
Score = 36.2 bits (82), Expect = 0.54
Identities = 34/116 (29%), Positives = 48/116 (41%), Gaps = 7/116 (6%)
Query: 910 GEMFRRRVLSGR*IG-LMKLFL----SRFGEVMQLHSGWMIGPGGVLLGSSFI-ACIICP 963
GE RRR +G LM ++L + FG ++ L WM+G GVL + C C
Sbjct: 105 GEGGRRRAAKAPSMGTLMGVYLPCLQNIFGVILFLRLTWMVGTAGVLQALLIVLICCCCT 164
Query: 964 L*SVLLLRAVVTGIKTNGGGIYLGDGRWVCVSKAGWGRCCILYLVPCFGRVRWTLG 1019
L + + + A+ T GG Y R + G C YL F + LG
Sbjct: 165 LLTAISMSAIATNGVVPAGGSYFMISRSLGPEFGGAVGLC-FYLGTTFAAAMYILG 219
>YMG7_YEAST (Q04651) Hypothetical 40.7 kDa protein in DAK1-ORC1
intergenic region
Length = 352
Score = 33.9 bits (76), Expect = 2.7
Identities = 28/120 (23%), Positives = 51/120 (42%), Gaps = 19/120 (15%)
Query: 514 NDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAK 573
N P++ VVP LF +G +D Q++ R + V AK
Sbjct: 226 NQDEPLTTYVYYTSVVPTLFK-----KLGAEVDTNQYSVNDYRYLYKDVA--------AK 272
Query: 574 VKKRPSIFFKVDYEK---AYNSVEWGFLEYMMGRMNFCAKWI---GWIMGCLNSATVSVL 627
K P IFFK ++E + V F+++++ + C+ + WI L+ A ++++
Sbjct: 273 GDKMPGIFFKYNFEPLSIVVSDVRLSFIQFLVRLVAICSFLVYCASWIFTLLDMALITIM 332
>WN5B_MOUSE (P22726) Wnt-5b protein precursor
Length = 359
Score = 32.7 bits (73), Expect = 6.0
Identities = 18/43 (41%), Positives = 22/43 (50%), Gaps = 9/43 (20%)
Query: 1140 CSLTLEGM-----ICSGAGYWRFGSLALGRC----GWCVMVLC 1173
C+ T EGM +C G GY RF S+ + RC WC V C
Sbjct: 304 CNKTSEGMDGCELMCCGRGYDRFKSVQVERCHCRFHWCCFVRC 346
>S126_HUMAN (Q9UHW9) Solute carrier family 12 member 6 (Electroneutral
potassium-chloride cotransporter 3) (K-Cl cotransporter
3)
Length = 1150
Score = 32.7 bits (73), Expect = 6.0
Identities = 23/88 (26%), Positives = 36/88 (40%), Gaps = 2/88 (2%)
Query: 933 FGEVMQLHSGWMIGPGGVLLGSSFI-ACIICPL*SVLLLRAVVTGIKTNGGGIYLGDGRW 991
FG ++ L W++G GVL + + C C + + + + A+ T GG Y R
Sbjct: 200 FGVILFLRLTWVVGTAGVLQAFAIVLICCCCTMLTAISMSAIATNGVVPAGGSYFMISRA 259
Query: 992 VCVSKAGWGRCCILYLVPCFGRVRWTLG 1019
+ G C YL F + LG
Sbjct: 260 LGPEFGGAVGLC-FYLGTTFAAAMYILG 286
>S126_MOUSE (Q924N4) Solute carrier family 12 member 6 (Electroneutral
potassium-chloride cotransporter 3) (K-Cl cotransporter
3)
Length = 1150
Score = 32.3 bits (72), Expect = 7.8
Identities = 22/88 (25%), Positives = 36/88 (40%), Gaps = 2/88 (2%)
Query: 933 FGEVMQLHSGWMIGPGGVLLGSSFI-ACIICPL*SVLLLRAVVTGIKTNGGGIYLGDGRW 991
FG ++ L W++G G+L + + C C + + + + A+ T GG Y R
Sbjct: 200 FGVILFLRLTWVVGTAGILQAFAIVLICCCCTMLTAISMSAIATNGVVPAGGSYFMISRA 259
Query: 992 VCVSKAGWGRCCILYLVPCFGRVRWTLG 1019
+ G C YL F + LG
Sbjct: 260 LGPEFGGAVGLC-FYLGTTFAAAMYILG 286
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.339 0.151 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 135,948,296
Number of Sequences: 164201
Number of extensions: 5789712
Number of successful extensions: 15845
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 15824
Number of HSP's gapped (non-prelim): 25
length of query: 1183
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1062
effective length of database: 40,105,733
effective search space: 42592288446
effective search space used: 42592288446
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0058a.1


