

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0348.15
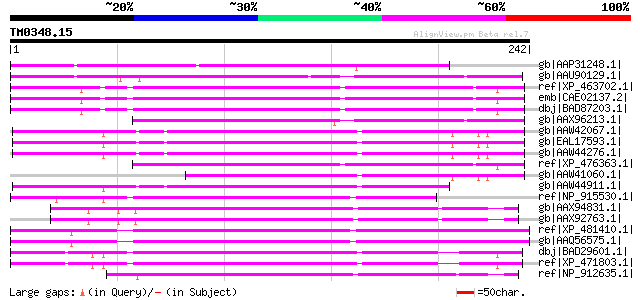
(242 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP31248.1| transposase [Fusarium oxysporum f. sp. melonis] 129 5e-29
gb|AAU90129.1| unknown protein [Oryza sativa (japonica cultivar-... 100 4e-20
ref|XP_463702.1| putative far-red impaired response protein [Ory... 98 2e-19
emb|CAE02137.2| OSJNBa0074L08.5 [Oryza sativa (japonica cultivar... 98 2e-19
dbj|BAD87203.1| far-red impaired response-like [Oryza sativa (ja... 98 2e-19
gb|AAX96213.1| transposon protein, putative, unclassified [Oryza... 96 7e-19
gb|AAW42067.1| conserved hypothetical protein [Cryptococcus neof... 93 6e-18
gb|EAL17593.1| hypothetical protein CNBM0450 [Cryptococcus neofo... 93 6e-18
gb|AAW44276.1| conserved hypothetical protein [Cryptococcus neof... 93 6e-18
ref|XP_476363.1| far-red impaired response-like protein [Oryza s... 93 7e-18
gb|AAW41060.1| conserved hypothetical protein [Cryptococcus neof... 92 1e-17
gb|AAW44911.1| conserved hypothetical protein [Cryptococcus neof... 91 3e-17
ref|NP_915530.1| P0529E05.9 [Oryza sativa (japonica cultivar-gro... 86 7e-16
gb|AAX94831.1| transposon protein, putative, unclassified [Oryza... 86 9e-16
gb|AAX92763.1| transposon protein, putative, unclassified [Oryza... 86 9e-16
ref|XP_481410.1| far-red impaired response protein -like [Oryza ... 85 2e-15
gb|AAQ56575.1| putative transposase [Oryza sativa (japonica cult... 85 2e-15
dbj|BAD29601.1| putative far-red impaired response protein [Oryz... 84 3e-15
ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultiva... 84 4e-15
ref|NP_912635.1| Putative far-red impaired response protein [Ory... 83 8e-15
>gb|AAP31248.1| transposase [Fusarium oxysporum f. sp. melonis]
Length = 836
Score = 129 bits (325), Expect = 5e-29
Identities = 72/206 (34%), Positives = 125/206 (59%), Gaps = 3/206 (1%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKELDCGPLTEMQY 60
L+ +E++ V+ + N + P+ + + L+ + L+T +YN I K + + +
Sbjct: 129 LSRQEEITVNQLTNAGIAPKEIGSFLRITS-NTLATQQDIYNCIAKGRRDLSKGQSNIHA 187
Query: 61 LLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLL 120
L +L E + + +E S V LF +HP +++ T+P V+I+D TYKTN+F++PLL
Sbjct: 188 LADQLNEEGFWNRICLDESSRVTAVLF-AHPKSLEYLKTYPEVLILDSTYKTNRFKMPLL 246
Query: 121 EMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLAD-PARLPGVIVTDRELALLSA 179
++VG+ + T+ IAF +++ E D+ WAL+ ++S+ D LP VI+TDR LA ++A
Sbjct: 247 DIVGVDACQRTFCIAFAFLSGEEEGDFTWALQALRSVYEDHNIGLPSVILTDRCLACMNA 306
Query: 180 VRNIFPGATHLLCLFHINKNVEAKCK 205
V + FPG+ LCL+HINK V++ C+
Sbjct: 307 VSSCFPGSALFLCLWHINKAVQSYCR 332
>gb|AAU90129.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 904
Score = 100 bits (249), Expect = 4e-20
Identities = 63/245 (25%), Positives = 121/245 (48%), Gaps = 15/245 (6%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYNRIKKVKEL---DCGPLTE 57
+ ++EK+ + ++N VP R +++ L G L + + ++ + G
Sbjct: 459 MIDQEKLLIRTLNKINVPTRMIMSVLSYVR-GGLFAVPYTKKAMSNYRDFVRRESGKNDM 517
Query: 58 MQ---YLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNK 114
MQ + KK++E +F+ +++ V+ LFWS N+ K + F +V D TYKTN+
Sbjct: 518 MQCLDFFEKKISEDPLFYFRFRTDENNVVKSLFWSDRNSRKFYEMFGDIVSFDTTYKTNR 577
Query: 115 FQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDREL 174
+ +P VG+TS G + ++ ++ T + L+CM ++P I+TD++L
Sbjct: 578 YDLPFAPFVGITSHGDNCLFGYAFL-QDETMVVQYVLDCM------GGKVPATIITDQDL 630
Query: 175 ALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFE 234
A+ +A+ +FP H C+FH+ N K ++ D + + + V ++T +FE
Sbjct: 631 AMKAAIVIVFPDTVHRNCMFHMLSNARDKTGRTFNSED-EEVYKDFHDIVTKSQTEAEFE 689
Query: 235 EEWRD 239
W+D
Sbjct: 690 YLWKD 694
>ref|XP_463702.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)]
Length = 1114
Score = 98.2 bits (243), Expect = 2e-19
Identities = 66/250 (26%), Positives = 121/250 (48%), Gaps = 17/250 (6%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPG---------NLSTITQVYNRIKKVKELD 51
+T+EEK + ++ +P R+M++ L G ++S + N + + D
Sbjct: 473 MTDEEKKLIRTLKQCNIPTRNMISILSFLRGGIAALPYTRKDVSNVGTAIN--SETRNTD 530
Query: 52 CGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYK 111
+ M+YL +K A+ ++K +++ + +FW+ + +L+ + V D TYK
Sbjct: 531 MNQV--MEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYK 588
Query: 112 TNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTD 171
TN++ +P +VG+T G A ++ E T + W E L A + P I+TD
Sbjct: 589 TNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETF--LTAMGGKHPETIITD 646
Query: 172 RELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVY-AETT 230
++LA+ +A+R +FP + H CLFHI K + K L ++N++V+ + T
Sbjct: 647 QDLAMRAAIRQVFPNSKHRNCLFHILKKCRERSGNTFSDKRRKDL-YAEFNDIVHNSLTR 705
Query: 231 TQFEEEWRDM 240
+FE W M
Sbjct: 706 AEFESLWLQM 715
>emb|CAE02137.2| OSJNBa0074L08.5 [Oryza sativa (japonica cultivar-group)]
gi|50926620|ref|XP_473257.1| OSJNBa0074L08.5 [Oryza
sativa (japonica cultivar-group)]
Length = 1061
Score = 98.2 bits (243), Expect = 2e-19
Identities = 66/250 (26%), Positives = 121/250 (48%), Gaps = 17/250 (6%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPG---------NLSTITQVYNRIKKVKELD 51
+T+EEK + ++ +P R+M++ L G ++S + N + + D
Sbjct: 420 MTDEEKKLIRTLKQCNIPTRNMISILSFLRGGIAALPYTGKDVSNVGTAIN--SETRNTD 477
Query: 52 CGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYK 111
+ M+YL +K A+ ++K +++ + +FW+ + +L+ + V D TYK
Sbjct: 478 MNQV--MEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYK 535
Query: 112 TNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTD 171
TN++ +P +VG+T G A ++ E T + W E L A + P I+TD
Sbjct: 536 TNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETF--LTAMGGKHPETIITD 593
Query: 172 RELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVY-AETT 230
++LA+ +A+R +FP + H CLFHI K + K L ++N++V+ + T
Sbjct: 594 QDLAMRAAIRQVFPNSKHRNCLFHILKKCRERSGNTFSDKRRKDL-YAEFNDIVHNSLTR 652
Query: 231 TQFEEEWRDM 240
+FE W M
Sbjct: 653 AEFESLWLQM 662
>dbj|BAD87203.1| far-red impaired response-like [Oryza sativa (japonica
cultivar-group)]
Length = 1130
Score = 98.2 bits (243), Expect = 2e-19
Identities = 66/250 (26%), Positives = 121/250 (48%), Gaps = 17/250 (6%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPG---------NLSTITQVYNRIKKVKELD 51
+T+EEK + ++ +P R+M++ L G ++S + N + + D
Sbjct: 489 MTDEEKKLIRTLKQCNIPTRNMISILSFLRGGIAALPYTRKDVSNVGTAIN--SETRNTD 546
Query: 52 CGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYK 111
+ M+YL +K A+ ++K +++ + +FW+ + +L+ + V D TYK
Sbjct: 547 MNQV--MEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYK 604
Query: 112 TNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTD 171
TN++ +P +VG+T G A ++ E T + W E L A + P I+TD
Sbjct: 605 TNRYNMPFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETF--LTAMGGKHPETIITD 662
Query: 172 RELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVY-AETT 230
++LA+ +A+R +FP + H CLFHI K + K L ++N++V+ + T
Sbjct: 663 QDLAMRAAIRQVFPNSKHRNCLFHILKKCRERSGNTFSDKRRKDL-YAEFNDIVHNSLTR 721
Query: 231 TQFEEEWRDM 240
+FE W M
Sbjct: 722 AEFESLWLQM 731
>gb|AAX96213.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1318
Score = 96.3 bits (238), Expect = 7e-19
Identities = 52/187 (27%), Positives = 93/187 (48%), Gaps = 11/187 (5%)
Query: 58 MQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQI 117
+ + KK +E +F+ +++ V+ LFWS N K + F +V D YKTN++ +
Sbjct: 739 LDFFEKKRSEDPLFYFRFRTDENNVVKSLFWSDGNNRKFYEMFDDIVSFDTMYKTNRYDL 798
Query: 118 PLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWA----LECMKSLLADPARLPGVIVTDRE 173
P VG+TS G + ++ E + + W L+CM ++P I+TD++
Sbjct: 799 PFAPFVGITSHGDNCLFGYAFLQDETSETFQWLFKTFLDCM------GGKVPATIITDQD 852
Query: 174 LALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQF 233
LA+ +A+ +FP H CLFH+ N K ++ D + + + V ++T +F
Sbjct: 853 LAMKAAIAIVFPDTVHRNCLFHMLSNARDKTGRTFNSED-EEVYKDFHDIVTKSQTEAEF 911
Query: 234 EEEWRDM 240
E W+D+
Sbjct: 912 EYLWKDL 918
>gb|AAW42067.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|50260537|gb|EAL23192.1|
hypothetical protein CNBA5360 [Cryptococcus neoformans
var. neoformans B-3501A] gi|50259285|gb|EAL21958.1|
hypothetical protein CNBC0980 [Cryptococcus neoformans
var. neoformans B-3501A] gi|50256296|gb|EAL19021.1|
hypothetical protein CNBH1230 [Cryptococcus neoformans
var. neoformans B-3501A] gi|57229113|gb|AAW45547.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21] gi|58271396|ref|XP_572854.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21] gi|58264436|ref|XP_569374.1|
conserved hypothetical protein [Cryptococcus neoformans
var. neoformans JEC21]
Length = 932
Score = 93.2 bits (230), Expect = 6e-18
Identities = 66/245 (26%), Positives = 124/245 (49%), Gaps = 10/245 (4%)
Query: 2 TEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYN-RIKKVKELDCGPLTEMQY 60
T E K ++++ ++ P +++T+ + P + T ++ ++ +L G
Sbjct: 223 TPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRRADLRAGSSASAA- 281
Query: 61 LLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLL 120
+++L + +H + ++ L ++ A L + FP V+ DCTYKTN +++P+L
Sbjct: 282 CIRQLVSNDELH-RPFVNTRDELIGLVYTKRTARALIHRFPEVLFFDCTYKTNLYRMPML 340
Query: 121 EMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAV 180
+VG TSTG+TY+ M RE T Y AL L+ P V++TDR+ AL++A+
Sbjct: 341 HIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVVITDRDPALINAL 398
Query: 181 RNIFPGATHLLCLFHINKNVEAKCK-LWVDTTDFKALE---MQKW-NEVVYAETTTQFEE 235
++ P A C +H+ +NV++ + +VD + + W V+A++ Q EE
Sbjct: 399 MSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWVRYCVHAKSDEQLEE 458
Query: 236 EWRDM 240
+R M
Sbjct: 459 GYRKM 463
>gb|EAL17593.1| hypothetical protein CNBM0450 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57226908|gb|AAW43368.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58267038|ref|XP_570675.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 580
Score = 93.2 bits (230), Expect = 6e-18
Identities = 66/245 (26%), Positives = 124/245 (49%), Gaps = 10/245 (4%)
Query: 2 TEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYN-RIKKVKELDCGPLTEMQY 60
T E K ++++ ++ P +++T+ + P + T ++ ++ +L G
Sbjct: 223 TPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRRADLRAGSSASAA- 281
Query: 61 LLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLL 120
+++L + +H + ++ L ++ A L + FP V+ DCTYKTN +++P+L
Sbjct: 282 CIRQLVSNDELH-RPFVNTRDELIGLVYTKRTARALIHRFPEVLFFDCTYKTNLYRMPML 340
Query: 121 EMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAV 180
+VG TSTG+TY+ M RE T Y AL L+ P V++TDR+ AL++A+
Sbjct: 341 HIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVVITDRDPALINAL 398
Query: 181 RNIFPGATHLLCLFHINKNVEAKCK-LWVDTTDFKALE---MQKW-NEVVYAETTTQFEE 235
++ P A C +H+ +NV++ + +VD + + W V+A++ Q EE
Sbjct: 399 MSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWVRYCVHAKSDEQLEE 458
Query: 236 EWRDM 240
+R M
Sbjct: 459 GYRKM 463
>gb|AAW44276.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58268854|ref|XP_571583.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 895
Score = 93.2 bits (230), Expect = 6e-18
Identities = 66/245 (26%), Positives = 124/245 (49%), Gaps = 10/245 (4%)
Query: 2 TEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYN-RIKKVKELDCGPLTEMQY 60
T E K ++++ ++ P +++T+ + P + T ++ ++ +L G
Sbjct: 223 TPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRRADLRAGSSASAA- 281
Query: 61 LLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLL 120
+++L + +H + ++ L ++ A L + FP V+ DCTYKTN +++P+L
Sbjct: 282 CIRQLVSNDELH-RPFVNTRDELIGLVYTKRTARALIHRFPEVLFFDCTYKTNLYRMPML 340
Query: 121 EMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAV 180
+VG TSTG+TY+ M RE T Y AL L+ P V++TDR+ AL++A+
Sbjct: 341 HIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVVITDRDPALINAL 398
Query: 181 RNIFPGATHLLCLFHINKNVEAKCK-LWVDTTDFKALE---MQKW-NEVVYAETTTQFEE 235
++ P A C +H+ +NV++ + +VD + + W V+A++ Q EE
Sbjct: 399 MSVLPKAYRFSCFWHLQENVKSNIRPCFVDEENIVMAVSNFCKAWVRYCVHAKSDEQLEE 458
Query: 236 EWRDM 240
+R M
Sbjct: 459 GYRKM 463
>ref|XP_476363.1| far-red impaired response-like protein [Oryza sativa (japonica
cultivar-group)] gi|22324476|dbj|BAC10390.1| far-red
impaired response-like protein [Oryza sativa (japonica
cultivar-group)] gi|50508936|dbj|BAD31841.1| far-red
impaired response-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 772
Score = 92.8 bits (229), Expect = 7e-18
Identities = 54/184 (29%), Positives = 94/184 (50%), Gaps = 4/184 (2%)
Query: 58 MQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQI 117
M+YL +K A+ ++K +++ + +FW+ + +L+ + V D TYKTN++ +
Sbjct: 220 MEYLRQKEAKDPGYYYKLTLDENNKVKSMFWTDGRSTQLYEQYGEFVSFDTTYKTNRYNM 279
Query: 118 PLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALL 177
P +VG+T G A ++ E T + W E L A + P I+TD++LA+
Sbjct: 280 PFAPIVGVTGHGNICIFACAFLGDETTETFKWVFETF--LTAMGRKHPETIITDQDLAMR 337
Query: 178 SAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVY-AETTTQFEEE 236
+A+R +FP + H CLFHI K + K L ++N++V+ + T +FE
Sbjct: 338 AAIRQVFPNSKHRNCLFHILKKYRERSGNTFSDKRRKDL-YAEFNDIVHNSLTRAEFESL 396
Query: 237 WRDM 240
W M
Sbjct: 397 WLQM 400
>gb|AAW41060.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58258933|ref|XP_566879.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 678
Score = 92.0 bits (227), Expect = 1e-17
Identities = 55/163 (33%), Positives = 90/163 (54%), Gaps = 7/163 (4%)
Query: 83 IMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRE 142
++ L ++ A L + FP V+ DCTYKTN +++P+L +VG TSTG+TY+ M RE
Sbjct: 49 LIGLVYTKRTARALIHRFPEVLFFDCTYKTNLYRMPMLHIVGSTSTGMTYTAGVILMLRE 108
Query: 143 RTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEA 202
T Y AL L+ P V++TDR+ AL++A+ ++ P A C +H+ +NV++
Sbjct: 109 TTNWYTQALNSFFELVGKPD--VKVVITDRDPALINALMSVLPKAYRFSCFWHLQENVKS 166
Query: 203 KCK-LWVDTTDFKALE---MQKW-NEVVYAETTTQFEEEWRDM 240
+ +VD + + W V+A++ Q EE +R M
Sbjct: 167 NIRPCFVDEENIVMAVSNFCKAWVRYCVHAKSDEQLEEGYRKM 209
>gb|AAW44911.1| conserved hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21] gi|58270124|ref|XP_572218.1| conserved
hypothetical protein [Cryptococcus neoformans var.
neoformans JEC21]
Length = 920
Score = 90.9 bits (224), Expect = 3e-17
Identities = 56/205 (27%), Positives = 107/205 (51%), Gaps = 5/205 (2%)
Query: 2 TEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTITQVYN-RIKKVKELDCGPLTEMQY 60
T E K ++++ ++ P +++T+ + P + T ++ ++ +L G
Sbjct: 223 TPEAKELINTLIDSRTPFPSIISTMSQRFPTVMLTSEDLHRLAFRRRADLRAGSSASAA- 281
Query: 61 LLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLL 120
+++L + +H + ++ L ++ A L + FP V+ DCTYKTN +++P+L
Sbjct: 282 CIRQLVSNDELH-RPFVNTRDELIGLVYTKRTARALIHRFPEVLFFDCTYKTNLYRMPML 340
Query: 121 EMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAV 180
+VG TSTG+TY+ M RE T Y AL L+ P V++TDR+ AL++A+
Sbjct: 341 HIVGSTSTGMTYTAGVILMLRETTNWYTQALNSFFELVGKPD--VKVVITDRDPALINAL 398
Query: 181 RNIFPGATHLLCLFHINKNVEAKCK 205
++ P A C +H+ +NV++ +
Sbjct: 399 MSVLPKAYRFSCFWHLQENVKSNIR 423
>ref|NP_915530.1| P0529E05.9 [Oryza sativa (japonica cultivar-group)]
Length = 773
Score = 86.3 bits (212), Expect = 7e-16
Identities = 53/207 (25%), Positives = 102/207 (48%), Gaps = 12/207 (5%)
Query: 1 LTEEEKVQVDSMNNNWVPPR--HMLATLKENNPGNLSTITQVYN------RIKKVKELDC 52
+TE +K Q+D +N++ V + H + + + +L+ + Y R+K ++E D
Sbjct: 265 MTEAQKAQIDILNDSGVRSKEGHEVMSRQAGGRQSLTFTRKDYKNYLRSKRMKSIQEGDT 324
Query: 53 GPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKT 112
G + +QYL K E + ++ ++ +FW+ ++ F+ F V+ D TY+T
Sbjct: 325 GAI--LQYLQDKQMENPSFFYAIQVDEDEMMTNIFWADARSVLDFDYFGDVICFDTTYRT 382
Query: 113 NKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDR 172
N + P VG+ T + E T + W E K ++ + P I+TD+
Sbjct: 383 NNYGRPFALFVGVNHHKQTVVFGAALLYDETTSTFEWLFETFKRAMS--GKEPRTILTDQ 440
Query: 173 ELALLSAVRNIFPGATHLLCLFHINKN 199
A+++A+ +FP +TH LC++H+ +N
Sbjct: 441 CAAIINAIGTVFPNSTHRLCVWHMYQN 467
>gb|AAX94831.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1023
Score = 85.9 bits (211), Expect = 9e-16
Identities = 56/225 (24%), Positives = 107/225 (46%), Gaps = 17/225 (7%)
Query: 20 RHMLATLKENNPGNLS----TITQVYNRIKKVKE--LDCGPLTE-MQYLLKKLAEANYVH 72
+H + + E + G T +YN K+K+ + G ++Y+ + + +
Sbjct: 714 QHQVMDIMERDHGGYEGTGFTSRDMYNFFVKLKKKRIKGGDADHVLKYMQARQKDDMEFY 773
Query: 73 FKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTY 132
+ + +G + LFWS P + ++ F VV+ D TY+ NK+ +P + VG+ G T
Sbjct: 774 YDYEIDQAGCLKRLFWSDPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTV 833
Query: 133 SIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLC 192
A ++ ER Y W L + + + P ++TD + A+ A+ ++FP + H LC
Sbjct: 834 IFACAVVSDERVETYEWVLRQFLTCMCQ--KHPKSVITDGDNAMRRAILHVFPNSDHRLC 891
Query: 193 LFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
+HI +N+ A+ +DF+ L +++E +FE +W
Sbjct: 892 TWHIEQNM-ARNLSPAMLSDFRTLVHSEFDE-------DEFERKW 928
>gb|AAX92763.1| transposon protein, putative, unclassified [Oryza sativa (japonica
cultivar-group)]
Length = 1185
Score = 85.9 bits (211), Expect = 9e-16
Identities = 56/225 (24%), Positives = 107/225 (46%), Gaps = 17/225 (7%)
Query: 20 RHMLATLKENNPGNLS----TITQVYNRIKKVKE--LDCGPLTE-MQYLLKKLAEANYVH 72
+H + + E + G T +YN K+K+ + G ++Y+ + + +
Sbjct: 714 QHQVMDIMERDHGGYEGTGFTSRDMYNFFVKLKKKRIKGGDADHVLKYMQARQKDDMEFY 773
Query: 73 FKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYKTNKFQIPLLEMVGLTSTGLTY 132
+ + +G + LFWS P + ++ F VV+ D TY+ NK+ +P + VG+ G T
Sbjct: 774 YDYEIDQAGCLKRLFWSDPQSRIDYDAFGDVVVFDSTYRVNKYNLPFIPFVGVNHHGSTV 833
Query: 133 SIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTDRELALLSAVRNIFPGATHLLC 192
A ++ ER Y W L + + + P ++TD + A+ A+ ++FP + H LC
Sbjct: 834 IFACAVVSDERVETYEWVLRQFLTCMCQ--KHPKSVITDGDNAMRRAILHVFPNSDHRLC 891
Query: 193 LFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAETTTQFEEEW 237
+HI +N+ A+ +DF+ L +++E +FE +W
Sbjct: 892 TWHIEQNM-ARNLSPAMLSDFRTLVHSEFDE-------DEFERKW 928
>ref|XP_481410.1| far-red impaired response protein -like [Oryza sativa (japonica
cultivar-group)] gi|35215248|dbj|BAC92598.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)] gi|35215057|dbj|BAC92415.1|
putative far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 1132
Score = 85.1 bits (209), Expect = 2e-15
Identities = 57/254 (22%), Positives = 106/254 (41%), Gaps = 21/254 (8%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLK------------ENNPGNLSTITQVYNRIKKVK 48
+T +EK + ++ +P R M+ L + + N+ T R +K
Sbjct: 523 MTIKEKRLIRTLKECNIPTRSMIVILSFLRGGLPALPYTKKDVSNVGTAINSETRNNDMK 582
Query: 49 ELDCGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDC 108
++ + YL KK E + +K +++ + +FW+ + +L+ + + D
Sbjct: 583 QV-------LAYLRKKEIEDPGISYKFKLDENNKVTSMFWTDGRSTQLYEEYGDCISFDT 635
Query: 109 TYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVI 168
TY+TN++ +P VG+ G T ++ E + W E + + + P I
Sbjct: 636 TYRTNRYNMPFAPFVGVIGHGTTCLFGCAFLGDETAETFKWVFETFATAMG--GKHPKTI 693
Query: 169 VTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAE 228
+TD++ A+ SA+ +F H CLFHI KN K K L + + +
Sbjct: 694 ITDQDNAMRSAIAQVFQNTKHRNCLFHIKKNCREKTGSMFSQKSNKNLYDEYDDILSNCL 753
Query: 229 TTTQFEEEWRDMCD 242
T +FE W M +
Sbjct: 754 TEAEFESLWPQMIE 767
>gb|AAQ56575.1| putative transposase [Oryza sativa (japonica cultivar-group)]
Length = 1037
Score = 85.1 bits (209), Expect = 2e-15
Identities = 57/254 (22%), Positives = 106/254 (41%), Gaps = 21/254 (8%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLK------------ENNPGNLSTITQVYNRIKKVK 48
+T +EK + ++ +P R M+ L + + N+ T R +K
Sbjct: 428 MTIKEKRLIRTLKECNIPTRSMIVILSFLRGGLPALPYTKKDVSNVGTAINSETRNNDMK 487
Query: 49 ELDCGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDC 108
++ + YL KK E + +K +++ + +FW+ + +L+ + + D
Sbjct: 488 QV-------LAYLRKKEIEDPGISYKFKLDENNKVTSMFWTDGRSTQLYEEYGDCISFDT 540
Query: 109 TYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVI 168
TY+TN++ +P VG+ G T ++ E + W E + + + P I
Sbjct: 541 TYRTNRYNMPFAPFVGVIGHGTTCLFGCAFLGDETAETFKWVFETFATAMG--GKHPKTI 598
Query: 169 VTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVYAE 228
+TD++ A+ SA+ +F H CLFHI KN K K L + + +
Sbjct: 599 ITDQDNAMRSAIAQVFQNTKHRNCLFHIKKNCREKTGSMFSQKSNKNLYDEYDDILSNCL 658
Query: 229 TTTQFEEEWRDMCD 242
T +FE W M +
Sbjct: 659 TEAEFESLWPQMIE 672
>dbj|BAD29601.1| putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|50251625|dbj|BAD29488.1| putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 688
Score = 84.3 bits (207), Expect = 3e-15
Identities = 58/249 (23%), Positives = 111/249 (44%), Gaps = 24/249 (9%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTI----TQVYN-----RIKKVKELD 51
+++ +K Q + + + P ++ + ENN L + +YN +K +K D
Sbjct: 133 MSDSKKAQAVELRMSGLRPFQVMEVM-ENNHDELDEVGFVMKDLYNFFTRYEMKNIKGHD 191
Query: 52 CGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYK 111
+ ++YL +K E FK ++ G + +FW+ + + F VVI D TY+
Sbjct: 192 AEDV--LKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYR 249
Query: 112 TNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTD 171
NK+ +P + +G+ T ++ E Y W LE + P ++TD
Sbjct: 250 VNKYNLPFIPFIGVNHHRSTTIFGCGILSNESVNSYCWLLETFLEAMRQVH--PKSLITD 307
Query: 172 RELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVY-AETT 230
+LA+ A+ + PGA H LC +HI +N+ + + ++ + +++Y +
Sbjct: 308 GDLAMAKAISKVMPGAYHRLCTWHIEENM---------SRHLRKPKLDELRKLIYESMDE 358
Query: 231 TQFEEEWRD 239
+FE W D
Sbjct: 359 EEFERRWAD 367
>ref|XP_471803.1| OSJNBa0050F15.2 [Oryza sativa (japonica cultivar-group)]
gi|38345091|emb|CAD40514.2| OSJNBa0050F15.2 [Oryza
sativa (japonica cultivar-group)]
Length = 688
Score = 83.6 bits (205), Expect = 4e-15
Identities = 58/249 (23%), Positives = 111/249 (44%), Gaps = 24/249 (9%)
Query: 1 LTEEEKVQVDSMNNNWVPPRHMLATLKENNPGNLSTI----TQVYN-----RIKKVKELD 51
+++ +K Q + + + P ++ + ENN L + +YN +K +K D
Sbjct: 133 MSDSKKAQAVELRMSGLRPFQVMEVM-ENNHDELDEVGFVMKDLYNFFTRYDMKNIKGHD 191
Query: 52 CGPLTEMQYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVVIMDCTYK 111
+ ++YL +K E FK ++ G + +FW+ + + F VVI D TY+
Sbjct: 192 AEDV--LKYLTRKQEEDAEFFFKYTTDEEGRLRNVFWADAESRLDYAAFGGVVIFDSTYR 249
Query: 112 TNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARLPGVIVTD 171
NK+ +P + +G+ T ++ E Y W LE + P ++TD
Sbjct: 250 VNKYNLPFIPFIGVNHHRSTTIFGCGILSNESVNSYCWLLETFLEAMRQVH--PKSLITD 307
Query: 172 RELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEVVY-AETT 230
+LA+ A+ + PGA H LC +HI +N+ + + ++ + +++Y +
Sbjct: 308 GDLAMAKAISKVMPGAYHRLCTWHIEENM---------SRHLRKPKLDELRKLIYESMDE 358
Query: 231 TQFEEEWRD 239
+FE W D
Sbjct: 359 EEFERRWAD 367
>ref|NP_912635.1| Putative far-red impaired response protein [Oryza sativa (japonica
cultivar-group)] gi|20219041|gb|AAM15785.1| Putative
far-red impaired response protein [Oryza sativa
(japonica cultivar-group)]
Length = 611
Score = 82.8 bits (203), Expect = 8e-15
Identities = 49/193 (25%), Positives = 92/193 (47%), Gaps = 11/193 (5%)
Query: 46 KVKELDCGPLTEM-QYLLKKLAEANYVHFKRHEEDSGVIMELFWSHPNAIKLFNTFPHVV 104
K K +D G + +Y+ + + +++ +++G + LFW+ P + ++ F VV
Sbjct: 185 KKKRIDGGDADRVIKYMQARQKDDMDFYYEYETDEAGCLKRLFWADPQSRIDYDAFGDVV 244
Query: 105 IMDCTYKTNKFQIPLLEMVGLTSTGLTYSIAFCYMTRERTPDYVWALECMKSLLADPARL 164
+ D TY+ NK+ +P + VG+ G T ++ ER Y W L+ S + +
Sbjct: 245 VFDSTYRVNKYNLPFIPFVGVNHHGSTVIFGCAVVSDERVGTYEWVLKQFLSCMCQ--KH 302
Query: 165 PGVIVTDRELALLSAVRNIFPGATHLLCLFHINKNVEAKCKLWVDTTDFKALEMQKWNEV 224
P ++TD + A+ A+ +FP + H LC +HI +N+ + +DF+ L E
Sbjct: 303 PKSVITDGDNAMRRAILLVFPNSDHRLCTWHIEQNMARNLSPTM-LSDFRVLVHAPLEE- 360
Query: 225 VYAETTTQFEEEW 237
+FE +W
Sbjct: 361 ------DEFERKW 367
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.135 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 428,390,891
Number of Sequences: 2540612
Number of extensions: 16379071
Number of successful extensions: 37210
Number of sequences better than 10.0: 400
Number of HSP's better than 10.0 without gapping: 295
Number of HSP's successfully gapped in prelim test: 105
Number of HSP's that attempted gapping in prelim test: 36729
Number of HSP's gapped (non-prelim): 420
length of query: 242
length of database: 863,360,394
effective HSP length: 124
effective length of query: 118
effective length of database: 548,324,506
effective search space: 64702291708
effective search space used: 64702291708
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Lotus: description of TM0348.15


