

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
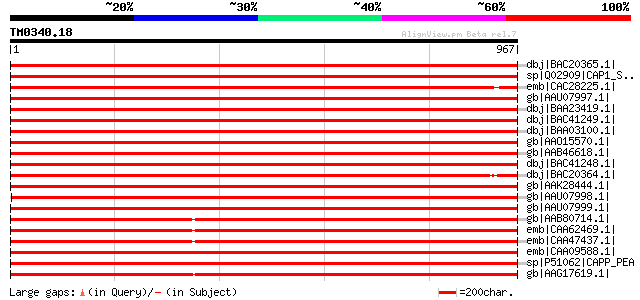
Query= TM0340.18
(967 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC20365.1| phosphoenolpyruvate carboxylase [Lotus cornicula... 1929 0.0
sp|Q02909|CAP1_SOYBN Phosphoenolpyruvate carboxylase, housekeepi... 1830 0.0
emb|CAC28225.1| phosphoenolpyruvate carboxylase [Sesbania rostrata] 1797 0.0
gb|AAU07997.1| phosphoenolpyruvate carboxylase 2; LaPEPC2 [Lupin... 1796 0.0
dbj|BAA23419.1| phosphoenolpyruvate carboxylase [Glycine max] 1774 0.0
dbj|BAC41249.1| phosphoenolpyruvate carboxylase [Glycine max] 1773 0.0
dbj|BAA03100.1| phosphoenolpyruvate carboxylase [Glycine max] gi... 1772 0.0
gb|AAO15570.1| phosphoenolpyruvate carboxylase [Lupinus albus] 1762 0.0
gb|AAB46618.1| phosphoenolpyruvate carboxylase [Medicago sativa]... 1759 0.0
dbj|BAC41248.1| phosphoenolpyruvate carboxylase [Glycine max] 1757 0.0
dbj|BAC20364.1| phosphoenolpyruvate carboxylase [Lotus cornicula... 1749 0.0
gb|AAK28444.1| phosphoenolpyruvate carboxylase [Phaseolus vulgar... 1747 0.0
gb|AAU07998.1| phosphoenolpyruvate carboxylase 3; LaPEPC3 [Lupin... 1744 0.0
gb|AAU07999.1| phosphoenolpyruvate carboxylase 4; LaPEPC4 [Lupin... 1744 0.0
gb|AAB80714.1| phosphoenolpyruvate carboxylase 1 [Gossypium hirs... 1732 0.0
emb|CAA62469.1| phosphoenolpyruvate carboxylase [Solanum tuberosum] 1731 0.0
emb|CAA47437.1| phosphoenolpyruvate carboxylase [Solanum tuberos... 1730 0.0
emb|CAA09588.1| phosphoenolpyruvate-carboxylase [Vicia faba] 1729 0.0
sp|P51062|CAPP_PEA Phosphoenolpyruvate carboxylase (PEPCase) gi|... 1725 0.0
gb|AAG17619.1| phosphoenolpyruvate carboxylase [Flaveria trinervia] 1720 0.0
>dbj|BAC20365.1| phosphoenolpyruvate carboxylase [Lotus corniculatus var. japonicus]
Length = 967
Score = 1929 bits (4997), Expect = 0.0
Identities = 965/967 (99%), Positives = 966/967 (99%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDL+ETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLRETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK
Sbjct: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS
Sbjct: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC
Sbjct: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDR TDVLDAITKHLGIGSYLEWSEE
Sbjct: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRLTDVLDAITKHLGIGSYLEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL
Sbjct: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY
Sbjct: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC
Sbjct: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 961 AGMQNTG 967
>sp|Q02909|CAP1_SOYBN Phosphoenolpyruvate carboxylase, housekeeping isozyme (PEPCase)
gi|320128|pir||S28428 phosphoenolpyruvate carboxylase
(EC 4.1.1.31) - soybean gi|218267|dbj|BAA01560.1|
phosphoenolpyruvate carboxylase [Glycine max]
Length = 967
Score = 1830 bits (4739), Expect = 0.0
Identities = 909/967 (94%), Positives = 941/967 (97%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLR LVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRLLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSIVVAKSF+HMLNLANLAEEVQIAH RR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSIVVAKSFSHMLNLANLAEEVQIAHSRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV +MKKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVVDMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRN LTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNNLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVP FLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPTFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRA+EL SS K++VAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRADELNRSSKKNSVAKHYIEFWKA 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IPP+EPYRV+LGEVRNRLYQTRERSRHLLA+ YSDI EE TFTNVEEFLEPLELCYRSLC
Sbjct: 361 IPPNEPYRVLLGEVRNRLYQTRERSRHLLAHGYSDIPEEETFTNVEEFLEPLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQQWLLSELSGKRPLFGPDLPQTEEI+DVL+TFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIRDVLETFHVIAELPLDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECHVK+PLRVVPLFEKLADLE APAALARLFSV+WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECHVKHPLRVVPLFEKLADLEAAPAALARLFSVDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK+YGVKLTMFHGRGGTVGRGGGPTHLAILSQPP+TI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELIMVAKQYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPETI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMDEMAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFKHVI KD+RN+++LQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY
Sbjct: 781 RFHLPVWLGFGAAFKHVIEKDVRNIHVLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLWSFGEQLRTM+EETK+LLLQVA H+DLLEGDPYLKQRLRLRDSYITTLNVC
Sbjct: 841 DRLLVSEDLWSFGEQLRTMYEETKELLLQVAGHRDLLEGDPYLKQRLRLRDSYITTLNVC 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVKLRPHISKE+I++SKPADEL+TLNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKLRPHISKESIEISKPADELITLNPTSEYAPGLEDTLILTMKGIA 960
Query: 961 AGMQNTG 967
AG+QNTG
Sbjct: 961 AGLQNTG 967
>emb|CAC28225.1| phosphoenolpyruvate carboxylase [Sesbania rostrata]
Length = 961
Score = 1797 bits (4655), Expect = 0.0
Identities = 895/967 (92%), Positives = 930/967 (95%), Gaps = 6/967 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLR LVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRLLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSIVVAKSF+HMLNLANLAEEVQIAHRRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSIVVAKSFSHMLNLANLAEEVQIAHRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLVGEMKKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 121 LKKGDFADESNATTESDIEETLKKLVGEMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGR+RNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRVRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRA+EL SS KDAVAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRADELNRSSKKDAVAKHYIEFWKI 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IPPSEPYRVVLGEVR+RLYQTRERSRHLLA+ YSDI EE TFTN+EEFLEPLELCYRSLC
Sbjct: 361 IPPSEPYRVVLGEVRDRLYQTRERSRHLLAHGYSDIPEEETFTNLEEFLEPLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADG+LLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGTLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQQWLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMAT PSDVL
Sbjct: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATTPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS+EWYRNRING+QEVMIGYSDS
Sbjct: 541 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIEWYRNRINGRQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELIKVAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELIKVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRA+MD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRAMMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI KD+RNL+MLQEMYNQWPFFRVT+DLVEMVFAKGDPGIA+L
Sbjct: 781 RFHLPVWLGFGAAFKKVIEKDVRNLHMLQEMYNQWPFFRVTLDLVEMVFAKGDPGIASLN 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FGEQLR +EETK+LLLQVA H+++LEGDPYLKQRLRLRDSYITTLN
Sbjct: 841 DRLLVSKDLWPFGEQLRNKYEETKKLLLQVAGHREILEGDPYLKQRLRLRDSYITTLNAF 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK++P ISKE+ A ELVTLNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKVKPRISKES------AVELVTLNPTSEYAPGLEDTLILTMKGIA 954
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 955 AGMQNTG 961
>gb|AAU07997.1| phosphoenolpyruvate carboxylase 2; LaPEPC2 [Lupinus albus]
Length = 967
Score = 1796 bits (4651), Expect = 0.0
Identities = 888/967 (91%), Positives = 935/967 (95%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKLVEYDALLLDRFLDILQDLHGED+KETVQE+
Sbjct: 1 MANRNLEKMASIDAQLRQLAPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDMKETVQEI 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSIV AK+F+HMLNLANLAEEVQIAHRRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSIVFAKAFSHMLNLANLAEEVQIAHRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEETFK+LV E+KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 121 LKKGDFADENNATTESDIEETFKRLVRELKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRN LT+LYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNNLTELYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVP+FLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPQFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYS I+DLMFELSMWRCNDELRVRA+E++ SS KDAVAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSMIDDLMFELSMWRCNDELRVRADEISRSSKKDAVAKHYIEFWKI 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IP SEPYRV+LG+VR+RLY+TRERSR+LLA YSDI EE T T+VEEFLEPLELCYRSL
Sbjct: 361 IPSSEPYRVILGDVRDRLYRTRERSRYLLAQGYSDIPEEETLTSVEEFLEPLELCYRSLR 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
CGDRAIADG+LLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT+HLGIGSY EWSEE
Sbjct: 421 VCGDRAIADGTLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITQHLGIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDT HVIAELP DNFGAYIISMATA SDVL
Sbjct: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTLHVIAELPPDNFGAYIISMATAASDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQREC +K+PLRVVPLFEKLADLE+APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECRIKHPLRVVPLFEKLADLESAPAALARLFSIDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI+VAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPP+TI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELIEVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPETI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQRYTAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVF+EPRFVEYFRLATPELEYGRMNIGSRPAKR+P+GGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFQEPRFVEYFRLATPELEYGRMNIGSRPAKRRPTGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAF+ VI KD+RNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY
Sbjct: 781 RFHLPVWLGFGAAFRQVITKDVRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLWSFGEQLRT FEETK+LLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC
Sbjct: 841 DRLLVSEDLWSFGEQLRTKFEETKKLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNY+VKLRPHISKE I++SK ADEL+TLNPTSEYAPGLEDTLILT+KGIA
Sbjct: 901 QAYTLKRIRDPNYDVKLRPHISKECIEISKAADELITLNPTSEYAPGLEDTLILTVKGIA 960
Query: 961 AGMQNTG 967
AG+QNTG
Sbjct: 961 AGLQNTG 967
>dbj|BAA23419.1| phosphoenolpyruvate carboxylase [Glycine max]
Length = 967
Score = 1774 bits (4594), Expect = 0.0
Identities = 878/967 (90%), Positives = 921/967 (94%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSI+VAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSILVAKSFSHMLNLANLAEEVQISRRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV +KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVFGLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP+FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWNGVPRFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL SS KD VAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRSSKKDEVAKHYIEFWKK 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE TFTNVEEFLE LELCYRSLC
Sbjct: 361 VPPNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEEATFTNVEEFLESLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDF+RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGSLLDFMRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 481 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK++G+KLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELINVAKKFGIKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI K++ NLNMLQEMYNQWPFFRVT+DLVEMVFAKGDP IA L
Sbjct: 781 RFHLPVWLGFGAAFKEVIEKNVNNLNMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIAGLK 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FG+QLR +EETK+LLLQVA HK++LEGDPYLKQRLRLR + ITTLN+
Sbjct: 841 DRLLVSKDLWRFGDQLRNKYEETKKLLLQVAGHKEILEGDPYLKQRLRLRHAPITTLNIV 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK+RP ISKE+ + SK ADEL+ LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKVRPRISKESAEASKSADELIKLNPTSEYAPGLEDTLILTMKGIA 960
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 961 AGMQNTG 967
>dbj|BAC41249.1| phosphoenolpyruvate carboxylase [Glycine max]
Length = 967
Score = 1773 bits (4593), Expect = 0.0
Identities = 878/967 (90%), Positives = 921/967 (94%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSI+VAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSILVAKSFSHMLNLANLAEEVQISRRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV +KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVFGLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP+FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWNGVPRFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL SS KD VAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRSSKKDEVAKHYIEFWKK 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE TFTNVEEFLE LELCYRSLC
Sbjct: 361 VPPNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEEATFTNVEEFLESLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDF+RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGSLLDFMRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 481 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK++G+KLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELINVAKKFGIKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI K++ NLNMLQEMYNQWPFFRVT+DLVEMVFAKGDP IA L
Sbjct: 781 RFHLPVWLGFGAAFKEVIEKNVNNLNMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIAGLN 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FG+QLR +EETK+LLLQVA HK++LEGDPYLKQRLRLR + ITTLN+
Sbjct: 841 DRLLVSKDLWLFGDQLRNKYEETKKLLLQVAGHKEILEGDPYLKQRLRLRHAPITTLNIV 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK+RP ISKE+ + SK ADEL+ LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKVRPRISKESAEASKSADELIKLNPTSEYAPGLEDTLILTMKGIA 960
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 961 AGMQNTG 967
>dbj|BAA03100.1| phosphoenolpyruvate carboxylase [Glycine max]
gi|1705587|sp|P51061|CAP2_SOYBN Phosphoenolpyruvate
carboxylase (PEPCase)
Length = 967
Score = 1772 bits (4589), Expect = 0.0
Identities = 877/967 (90%), Positives = 923/967 (94%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA RNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MATRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSI+VAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSILVAKSFSHMLNLANLAEEVQISRRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV ++KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVFDLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP+FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWNGVPRFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL SS KD VAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRSSKKDEVAKHYIEFWKK 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE TFTNVEEFLE LELCYRSLC
Sbjct: 361 VPPNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEEATFTNVEEFLESLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDF+RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGSLLDFMRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 481 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELINVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI ++++NLNMLQEMYNQWPFFRVT+DLVEMVFAKGDP IAAL
Sbjct: 781 RFHLPVWLGFGAAFKKVIEENVKNLNMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIAALN 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FG+QLR +EET++LLLQVA HK++LEGDPYLKQRLRLR + ITTLN+
Sbjct: 841 DRLLVSKDLWPFGDQLRNKYEETRKLLLQVAGHKEILEGDPYLKQRLRLRHAPITTLNIV 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK+RP ISKE+ + SK ADELV LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKVRPRISKESAEASKSADELVKLNPTSEYAPGLEDTLILTMKGIA 960
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 961 AGMQNTG 967
>gb|AAO15570.1| phosphoenolpyruvate carboxylase [Lupinus albus]
Length = 967
Score = 1762 bits (4564), Expect = 0.0
Identities = 871/967 (90%), Positives = 924/967 (95%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKLVEYDALLLDRFLDILQDLHGED+KETVQE+
Sbjct: 1 MANRNLEKMASIDAQLRQLAPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDMKETVQEI 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSIV AK+F+HMLNLANLAEEVQIAHRRR
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSIVFAKAFSHMLNLANLAEEVQIAHRRRNN 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEETFK+LV E+KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 121 LKKGDFADENNATTESDIEETFKRLVRELKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRN LT+LYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNNLTELYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVP+FLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPQFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYS I+DLMFELSMWRCNDELRVRA+E++ SS KDAVAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSMIDDLMFELSMWRCNDELRVRADEISRSSKKDAVAKHYIEFWKI 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IP SEPYRV+LG+VR++LY+ S HLLA YS+I E T+V+ FLE LELCYRSLC
Sbjct: 361 IPSSEPYRVILGDVRDKLYRKSRGSCHLLAQGYSEIPGEEILTSVDGFLELLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
CGDRAIADG+LLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT+HLGIGSY EWSEE
Sbjct: 421 VCGDRAIADGTLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITQHLGIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDT HVIAELP DNFGAYIISMATA SDVL
Sbjct: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTLHVIAELPPDNFGAYIISMATAASDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQREC +K+PLRVVPLFEKLADLE+APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECRIKHPLRVVPLFEKLADLESAPAALARLFSIDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI+VAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPP+TI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELIEVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPETI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQRYTAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVF+EPRFVEYFRLATPELEYGRMNIGSRPAKR+P+GGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFQEPRFVEYFRLATPELEYGRMNIGSRPAKRRPTGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAF+ V+ KD+RNLNMLQEMYNQW FFRVTIDLVEMVFAKGDPGIAALY
Sbjct: 781 RFHLPVWLGFGAAFRQVVPKDVRNLNMLQEMYNQWKFFRVTIDLVEMVFAKGDPGIAALY 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS++LWSFGEQLRT FEETK+LLLQVAAHKDLLEGDPYLKQ+LRLRDSYI+TLNVC
Sbjct: 841 DRLLVSKNLWSFGEQLRTNFEETKKLLLQVAAHKDLLEGDPYLKQKLRLRDSYISTLNVC 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNY+VKLRPHISKE I++SK ADEL+TLNPTSEYAPGLEDT ILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYDVKLRPHISKECIEISKVADELITLNPTSEYAPGLEDTFILTMKGIA 960
Query: 961 AGMQNTG 967
AG+QNTG
Sbjct: 961 AGLQNTG 967
>gb|AAB46618.1| phosphoenolpyruvate carboxylase [Medicago sativa]
gi|1146155|gb|AAB41903.1| phosphoenolpyruvate
carboxylase [Medicago sativa]
gi|399183|sp|Q02735|CAPP_MEDSA Phosphoenolpyruvate
carboxylase (PEPCase) gi|280394|pir||S26235
phosphoenolpyruvate carboxylase (EC 4.1.1.31) - alfalfa
Length = 966
Score = 1759 bits (4557), Expect = 0.0
Identities = 873/967 (90%), Positives = 928/967 (95%), Gaps = 1/967 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MAN+ +EKMASIDAQLRQLVPAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLK++VQEV
Sbjct: 1 MANK-MEKMASIDAQLRQLVPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKDSVQEV 59
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKH+P++LE LGNLITS DAGDSIVVAKSF+HMLNLANLAEEVQIAHRRR K
Sbjct: 60 YELSAEYERKHDPKKLEELGNLITSFDAGDSIVVAKSFSHMLNLANLAEEVQIAHRRRNK 119
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDF DE NATTESDIEET KKLV +MKKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 120 LKKGDFRDESNATTESDIEETLKKLVFDMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 179
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGR+RNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEI+RT PTPQDEMRAGM
Sbjct: 180 LLQKHGRVRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIKRTPPTPQDEMRAGM 239
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 240 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 299
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL +S KD VAKHYIEFWK
Sbjct: 300 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRNSKKDEVAKHYIEFWKK 359
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IP +EPYRVVLGEVR++LY+TRERSR+LLA+ Y +I EE TFTNV+EFLEPLELCYRSLC
Sbjct: 360 IPLNEPYRVVLGEVRDKLYRTRERSRYLLAHGYCEIPEEATFTNVDEFLEPLELCYRSLC 419
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAITKHL IGSY EWSEE
Sbjct: 420 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYQEWSEE 479
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSEL GKRPLFGPDLPQT+EI+DVLDTF VIAELPSDNFGAYIISMATAPSDVL
Sbjct: 480 KRQEWLLSELIGKRPLFGPDLPQTDEIRDVLDTFRVIAELPSDNFGAYIISMATAPSDVL 539
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQREC V+NPLRVVPLFEKL DLE+APAALARLFS++WY NRI+GKQEVMIGYSDS
Sbjct: 540 AVELLQRECKVRNPLRVVPLFEKLDDLESAPAALARLFSIDWYINRIDGKQEVMIGYSDS 599
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQE+LIKVA+++GVKLTMFHGRGGTVGRGGGPTHLAILSQPP+TI
Sbjct: 600 GKDAGRFSAAWQLYKAQEDLIKVAQKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPETI 659
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGM PP SPKPEWRALMD+MAVIA
Sbjct: 660 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMRPPSSPKPEWRALMDQMAVIA 719
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPE+EYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 720 TEEYRSIVFKEPRFVEYFRLATPEMEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 779
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAF+ V+ KD++NL+MLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 780 RFHLPVWLGFGAAFRQVVQKDVKNLHMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALN 839
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FGEQLR+ +EETK+LLLQVAAHK++LEGDPYLKQRLRLRDSYITTLNV
Sbjct: 840 DRLLVSKDLWPFGEQLRSKYEETKKLLLQVAAHKEVLEGDPYLKQRLRLRDSYITTLNVF 899
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNY V++RP ISKE+ + SKPADELVTLNPTSEYAPGLEDTLILTMKGIA
Sbjct: 900 QAYTLKRIRDPNYKVEVRPPISKESAETSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 959
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 960 AGMQNTG 966
>dbj|BAC41248.1| phosphoenolpyruvate carboxylase [Glycine max]
Length = 967
Score = 1757 bits (4551), Expect = 0.0
Identities = 871/967 (90%), Positives = 918/967 (94%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA RNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQ E+LKETVQEV
Sbjct: 1 MATRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQGFTWENLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGNLITSLDAGDSI+VAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNLITSLDAGDSILVAKSFSHMLNLANLAEEVQISRRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV ++KKSPQEVFD LKNQTVDLVLTAHPTQS+RRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVFDLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCL+QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNCLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP+FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWNGVPRFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEEL SS KD VAKHYIEFWK
Sbjct: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELHRSSKKDEVAKHYIEFWKK 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE TFTNVEEFLE LELCYRSLC
Sbjct: 361 VPPNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEEATFTNVEEFLESLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDF+RQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGSLLDFMRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 481 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELINVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGE+HLCFRTLQR+TAATLEHGMHPPISPKPEWRALMD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEQHLCFRTLQRFTAATLEHGMHPPISPKPEWRALMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI ++++NLNMLQEMYNQWPFFRVT+DLVEMVFAKGDP IAAL
Sbjct: 781 RFHLPVWLGFGAAFKKVIEENVKNLNMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIAALN 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS+DLW FG+QLR +EET++LLLQVA HK++LEGDPYLKQRLRLR + ITTLN+
Sbjct: 841 DRLLVSKDLWPFGDQLRNKYEETRKLLLQVAGHKEILEGDPYLKQRLRLRHAPITTLNIV 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNYNVK+RP ISKE+ + K ADELV LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYNVKVRPRISKESAEAXKSADELVKLNPTSEYAPGLEDTLILTMKGIA 960
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 961 AGMQNTG 967
>dbj|BAC20364.1| phosphoenolpyruvate carboxylase [Lotus corniculatus var. japonicus]
Length = 961
Score = 1749 bits (4530), Expect = 0.0
Identities = 874/967 (90%), Positives = 917/967 (94%), Gaps = 6/967 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKH+ ++LE LG +ITSLDAGDSIVVAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 61 YELSAEYERKHDHEKLEELGKVITSLDAGDSIVVAKSFSHMLNLANLAEEVQISRRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV ++KKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVFKLKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKH RIRN L+QLYAKDITPDDKQELDEALQREIQAAFRTDEI+RT PTPQDEMRAGM
Sbjct: 181 LLQKHARIRNSLSQLYAKDITPDDKQELDEALQREIQAAFRTDEIKRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMA+NLYYSQIEDLMFELSMWRCNDELRV A+EL +SNKD VAKHYIEFWK
Sbjct: 301 DVCLLARMMASNLYYSQIEDLMFELSMWRCNDELRVHADELHRNSNKDEVAKHYIEFWKK 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRVVLGEVR+RLY+TRE SR LLA YSDI EE TFTNVEEFLEPLELCYRSLC
Sbjct: 361 VPPNEPYRVVLGEVRDRLYKTREHSRILLAQGYSDIPEEATFTNVEEFLEPLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
CGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAITKHL IGSY EWSEE
Sbjct: 421 DCGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLL ELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELPSDNFGAYIISMA APSDVL
Sbjct: 481 KRQEWLLGELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPSDNFGAYIISMAIAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRE H+KNPLRVVPLFEKL DLE APAALARLFS+EWYRNRINGKQEVMIGYSDS
Sbjct: 541 AVELLQRESHIKNPLRVVPLFEKLDDLEAAPAALARLFSIEWYRNRINGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQE+LIKVAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEDLIKVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGM+PP SPKPEWRALMD++AVIA
Sbjct: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMNPPFSPKPEWRALMDQLAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVF+EPRFVEYFRLATPELEYGRMNIGSRPAKR+P+GGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFQEPRFVEYFRLATPELEYGRMNIGSRPAKRRPTGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAF+ +I KD+RNL+ LQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 781 RFHLPVWLGFGAAFRQLIEKDVRNLHTLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALN 840
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLW FGEQLR +EETK+LLLQVA HK++LEGDPYLKQRLRLRDSYITT+NV
Sbjct: 841 DRLLVSEDLWPFGEQLRNKYEETKKLLLQVAGHKEVLEGDPYLKQRLRLRDSYITTMNVF 900
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDPNY+VK HISKE SKPADELV LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 901 QAYTLKRIRDPNYDVK---HISKEK---SKPADELVRLNPTSEYAPGLEDTLILTMKGIA 954
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 955 AGMQNTG 961
>gb|AAK28444.1| phosphoenolpyruvate carboxylase [Phaseolus vulgaris]
gi|24636262|sp|Q9AU12|CAPP_PHAVU Phosphoenolpyruvate
carboxylase (PEPCase)
Length = 968
Score = 1747 bits (4524), Expect = 0.0
Identities = 866/968 (89%), Positives = 919/968 (94%), Gaps = 1/968 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEKMASIDAQLRQL P+KVSEDDKL+EYDALLLDRFLDILQ+LHGEDLKETVQEV
Sbjct: 1 MANRNLEKMASIDAQLRQLAPSKVSEDDKLIEYDALLLDRFLDILQNLHGEDLKETVQEV 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGN+ITSLDAGDSIVVAKSF+HMLNLANLAEEVQI+ RRR K
Sbjct: 61 YELSAEYEGKHDPKKLEELGNVITSLDAGDSIVVAKSFSHMLNLANLAEEVQISRRRRNK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE NATTESDIEET KKLV E+KKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 121 LKKGDFADENNATTESDIEETLKKLVFELKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKH RIRNCL++LYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHARIRNCLSKLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIW GVP FLRRVDTAL NIGI ERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWNGVPSFLRRVDTALNNIGIKERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAAN+YYSQIEDLMFELSMWRCNDELRV A+E+ SSNKD VAKHYIEFWK
Sbjct: 301 DVCLLARMMAANMYYSQIEDLMFELSMWRCNDELRVHADEVHRSSNKDEVAKHYIEFWKK 360
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+P +EPYRVVLGEVR+RLYQTRERSRHLL+N YSDI EE+TFT+VEEFL+PLELCYRSLC
Sbjct: 361 VPTNEPYRVVLGEVRDRLYQTRERSRHLLSNGYSDIPEENTFTSVEEFLQPLELCYRSLC 420
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDFLRQVSTFGLS+VRLDIRQESDRHTDVLDAITKHL IGSY EWSEE
Sbjct: 421 ACGDRAIADGSLLDFLRQVSTFGLSIVRLDIRQESDRHTDVLDAITKHLEIGSYQEWSEE 480
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLPQTEEI+DVLDTFHVIAELP DNFGAYIISMATAPSDVL
Sbjct: 481 KRQEWLLSELSGKRPLFGPDLPQTEEIRDVLDTFHVIAELPPDNFGAYIISMATAPSDVL 540
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECHVK+PLRVVPLFEKLADLE APAALARLFSV+WY+NRI+GKQEVMIGYSDS
Sbjct: 541 AVELLQRECHVKHPLRVVPLFEKLADLEAAPAALARLFSVDWYKNRIDGKQEVMIGYSDS 600
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEEL+KVAK++G+KLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 601 GKDAGRFSAAWQLYKAQEELVKVAKKFGIKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQ FGE+HLCFRTLQR+TAATLEHGM+PPISPKPEWRA+MD+MAVIA
Sbjct: 661 HGSLRVTVQGEVIEQCFGEQHLCFRTLQRFTAATLEHGMNPPISPKPEWRAMMDQMAVIA 720
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 780
Query: 781 RFHLPVWLGFGAAFKHVIAKDI-RNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL 839
RFHLPVWLGFGAAFK V+ K+ +NL+MLQEMYNQWPFFRVT+DLVEMVFAKGDP I AL
Sbjct: 781 RFHLPVWLGFGAAFKQVLDKNAKKNLSMLQEMYNQWPFFRVTLDLVEMVFAKGDPKIGAL 840
Query: 840 YDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNV 899
DRLLVS+DLW FG+QLR +EETK+LLLQVA HK++LEGDPYLKQRLRLR S ITTLNV
Sbjct: 841 NDRLLVSKDLWPFGDQLRNKYEETKKLLLQVAGHKEILEGDPYLKQRLRLRHSPITTLNV 900
Query: 900 CQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
QAYTLKRIRDPNY VK RP ISKE+ + SK ADEL+ LNPTSEYAPGLEDTLILTMKGI
Sbjct: 901 FQAYTLKRIRDPNYKVKARPRISKESAEASKSADELIKLNPTSEYAPGLEDTLILTMKGI 960
Query: 960 AAGMQNTG 967
AAGMQNTG
Sbjct: 961 AAGMQNTG 968
>gb|AAU07998.1| phosphoenolpyruvate carboxylase 3; LaPEPC3 [Lupinus albus]
Length = 968
Score = 1744 bits (4518), Expect = 0.0
Identities = 860/965 (89%), Positives = 920/965 (95%)
Query: 3 NRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEVYE 62
NRN+EKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLK TVQ+VYE
Sbjct: 4 NRNMEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKNTVQDVYE 63
Query: 63 LSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKLK 122
LS+EYE KH+P++LE +GN ITSLDAGDSIVVAKSF+HMLNLANLAEEVQI+HRRR KLK
Sbjct: 64 LSSEYEGKHDPKKLEEIGNAITSLDAGDSIVVAKSFSHMLNLANLAEEVQISHRRRNKLK 123
Query: 123 KGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSLL 182
KG+FADE NATTESDIEET K+LV ++KKSPQEVFD LKNQTVDLVLTAHPTQS+RRSLL
Sbjct: 124 KGNFADETNATTESDIEETLKRLVFDLKKSPQEVFDALKNQTVDLVLTAHPTQSIRRSLL 183
Query: 183 QKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGMSY 242
QKH RIRNCL+QLYAKDITPDDKQELDE+LQREIQAAFRTDEI+RT PTPQDEMRAGMSY
Sbjct: 184 QKHARIRNCLSQLYAKDITPDDKQELDESLQREIQAAFRTDEIKRTPPTPQDEMRAGMSY 243
Query: 243 FHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTRDV 302
FHETIWKGVPKFLRR+DTALKNIGINERVP+NAP+IQFSSWMGGDRDGNPRVTPEVTRDV
Sbjct: 244 FHETIWKGVPKFLRRIDTALKNIGINERVPHNAPVIQFSSWMGGDRDGNPRVTPEVTRDV 303
Query: 303 CLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKNIP 362
CLLARMMA+N+YYSQIEDLMFELSMWRC+DELR RA+EL SSNKD VAKHYIEFWK +P
Sbjct: 304 CLLARMMASNMYYSQIEDLMFELSMWRCSDELRDRADELHRSSNKDEVAKHYIEFWKKVP 363
Query: 363 PSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLCAC 422
P+EPYRVVLGEVR+RLYQTRERSRHLLA+ YSDI EE TFTNVEEFLEPLE+CYRSLCAC
Sbjct: 364 PNEPYRVVLGEVRDRLYQTRERSRHLLAHGYSDIPEEATFTNVEEFLEPLEVCYRSLCAC 423
Query: 423 GDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEKR 482
GDR+IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAITKHL IGSY EWSEEKR
Sbjct: 424 GDRSIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYQEWSEEKR 483
Query: 483 QQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVLAV 542
Q+WLLSEL GKRPLFGPDLP+TEEI+DVLDTFHV+AELP DNFGAYIISMATAPSDVLAV
Sbjct: 484 QEWLLSELCGKRPLFGPDLPKTEEIRDVLDTFHVLAELPPDNFGAYIISMATAPSDVLAV 543
Query: 543 ELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDSGK 602
ELLQRECH+K+PLRVVPLFEKLADLE APAALARLFSV+WY+NRI+GKQEVMIGYSDSGK
Sbjct: 544 ELLQRECHIKHPLRVVPLFEKLADLEAAPAALARLFSVDWYKNRIDGKQEVMIGYSDSGK 603
Query: 603 DAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIHG 662
DAGRFSAAWQLYKAQE+LIKVA+++GVKLTMFHGRGGTVGRGGGPTHLAILSQPP TIHG
Sbjct: 604 DAGRFSAAWQLYKAQEDLIKVAEKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPGTIHG 663
Query: 663 SLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIATE 722
SLRVTVQGEVIEQSFGE+HLCFRTLQRYTAATLEHGM+PPISPKPEWRALMD+MAV+ATE
Sbjct: 664 SLRVTVQGEVIEQSFGEQHLCFRTLQRYTAATLEHGMNPPISPKPEWRALMDQMAVVATE 723
Query: 723 EYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQTRF 782
EYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+P+GGIETLRAIPWIFAWTQTRF
Sbjct: 724 EYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPTGGIETLRAIPWIFAWTQTRF 783
Query: 783 HLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALYDR 842
HLPVWLGFG AFK VI KD NL MLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL DR
Sbjct: 784 HLPVWLGFGKAFKQVIEKDAGNLLMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALNDR 843
Query: 843 LLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVCQA 902
LLVS+DLW FGEQLR +EETK LLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNV QA
Sbjct: 844 LLVSKDLWPFGEQLRKKYEETKNHLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVFQA 903
Query: 903 YTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIAAG 962
YTLKRIRDPN+NV RPHISK+ ++ SK A ELV+LNPTSEYAPGLED+LIL+MKGIAAG
Sbjct: 904 YTLKRIRDPNFNVPPRPHISKDYLEKSKSATELVSLNPTSEYAPGLEDSLILSMKGIAAG 963
Query: 963 MQNTG 967
MQNTG
Sbjct: 964 MQNTG 968
>gb|AAU07999.1| phosphoenolpyruvate carboxylase 4; LaPEPC4 [Lupinus albus]
Length = 968
Score = 1744 bits (4517), Expect = 0.0
Identities = 859/966 (88%), Positives = 924/966 (94%)
Query: 2 ANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEVY 61
+NRN+EKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLK+TVQEVY
Sbjct: 3 SNRNMEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKDTVQEVY 62
Query: 62 ELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIKL 121
ELS++YE KH+P++LE +GN+ITSLDAGDSIVVAKSF+HMLNLANLAEEVQI+HRRR KL
Sbjct: 63 ELSSQYEGKHDPKKLEEIGNVITSLDAGDSIVVAKSFSHMLNLANLAEEVQISHRRRNKL 122
Query: 122 KKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRSL 181
KKG+FADE NATTESDIEET K+LV ++KKSP EVFD LK+QTVDLVLTAHPTQS+RRSL
Sbjct: 123 KKGNFADETNATTESDIEETLKRLVFDLKKSPHEVFDALKSQTVDLVLTAHPTQSIRRSL 182
Query: 182 LQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGMS 241
LQKH RIR+CL+ LYAKDITPDDK+ELDEALQREIQAAFRTDEI+RT PTPQDEMRAGMS
Sbjct: 183 LQKHARIRDCLSHLYAKDITPDDKKELDEALQREIQAAFRTDEIKRTPPTPQDEMRAGMS 242
Query: 242 YFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTRD 301
YFHETIWKGVPKFLRRVDTALKNIGINE +P+NAP+IQFSSWMGGDRDGNPRVTPEVTRD
Sbjct: 243 YFHETIWKGVPKFLRRVDTALKNIGINECIPHNAPVIQFSSWMGGDRDGNPRVTPEVTRD 302
Query: 302 VCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKNI 361
VCLLARMMAAN+YYSQIEDLMFELSMWRC+DELR RA+EL SSNKD VAKHYIEFWK +
Sbjct: 303 VCLLARMMAANMYYSQIEDLMFELSMWRCSDELRNRADELHRSSNKDEVAKHYIEFWKKV 362
Query: 362 PPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLCA 421
P +EPYRVVLGEVR+RLYQTRERSRHLLA+ YSDI EE TFTNVEEFLEPLE+CYRSLCA
Sbjct: 363 PANEPYRVVLGEVRDRLYQTRERSRHLLAHGYSDIPEEATFTNVEEFLEPLEVCYRSLCA 422
Query: 422 CGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEEK 481
CGDR IADGSLLDFLRQ+STFGLSLVRLDIRQESDRHTDV+DAITKHL IGSYLEWSEEK
Sbjct: 423 CGDRPIADGSLLDFLRQISTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYLEWSEEK 482
Query: 482 RQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVLA 541
RQ+WLLSELSGKRPLFGPDLP+TEEI+DVLDTFHV+AELP DNFGAYIISMATAPSDVLA
Sbjct: 483 RQEWLLSELSGKRPLFGPDLPKTEEIRDVLDTFHVLAELPPDNFGAYIISMATAPSDVLA 542
Query: 542 VELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDSG 601
VELLQRECH+K+PLRVVPLFEKLADLE APAALARLFS++WY NRI+GKQEVMIGYSDSG
Sbjct: 543 VELLQRECHIKHPLRVVPLFEKLADLEVAPAALARLFSIDWYINRIDGKQEVMIGYSDSG 602
Query: 602 KDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIH 661
KDAGRFSAAWQLYKAQEELIKVAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIH
Sbjct: 603 KDAGRFSAAWQLYKAQEELIKVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTIH 662
Query: 662 GSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIAT 721
GSLRVTVQGEVIEQSFGE+HLCFRTLQRYTAATLEHGM+PPISPKPEWRALMD+MAVI+T
Sbjct: 663 GSLRVTVQGEVIEQSFGEQHLCFRTLQRYTAATLEHGMNPPISPKPEWRALMDQMAVIST 722
Query: 722 EEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQTR 781
EEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKR+P+GGIETLRAIPWIFAWTQTR
Sbjct: 723 EEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRRPTGGIETLRAIPWIFAWTQTR 782
Query: 782 FHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALYD 841
FHLPVWLGFGAAFK VI KD+RNL+MLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL D
Sbjct: 783 FHLPVWLGFGAAFKQVIEKDVRNLHMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALND 842
Query: 842 RLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVCQ 901
RLLVS+DLW FGEQLR +EETK LLL+VAAHKDLLEGDPYLKQRLRLR SYITTLNV Q
Sbjct: 843 RLLVSKDLWPFGEQLRKKYEETKNLLLKVAAHKDLLEGDPYLKQRLRLRHSYITTLNVFQ 902
Query: 902 AYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIAA 961
AYTLKRIRDPN+NV+ R HISKE+++ S A ELV+LNPTSEYAPGLED+LILTMKGIAA
Sbjct: 903 AYTLKRIRDPNFNVRPRHHISKESLEKSTSATELVSLNPTSEYAPGLEDSLILTMKGIAA 962
Query: 962 GMQNTG 967
GMQNTG
Sbjct: 963 GMQNTG 968
>gb|AAB80714.1| phosphoenolpyruvate carboxylase 1 [Gossypium hirsutum]
gi|7436543|pir||T09846 phosphoenolpyruvate carboxylase
(EC 4.1.1.31) 1 - upland cotton
Length = 965
Score = 1732 bits (4486), Expect = 0.0
Identities = 847/967 (87%), Positives = 916/967 (94%), Gaps = 2/967 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MA R +EKMASIDAQLR L P KVSEDDKLVEYDA+LLDRFLDILQDLHGED++ETVQE
Sbjct: 1 MAGRKVEKMASIDAQLRLLAPGKVSEDDKLVEYDAVLLDRFLDILQDLHGEDIRETVQEC 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P+ LE LG ++TSLD GDSIVV KSF+HMLNL NLAEEVQIA+RRRIK
Sbjct: 61 YELSAEYEGKHDPKILEELGKVLTSLDPGDSIVVTKSFSHMLNLGNLAEEVQIAYRRRIK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE +ATTESDIEETFK+LVG++ KSP+EVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 121 LKKGDFADESSATTESDIEETFKRLVGQLNKSPEEVFDALKNQTVDLVLTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRR PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRNPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLY+SQIEDLMFELSMWRC+DELR+RA+EL SS KDA KHYIEFWK
Sbjct: 301 DVCLLARMMAANLYFSQIEDLMFELSMWRCSDELRIRADELHRSSKKDA--KHYIEFWKQ 358
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
IPP+EPYR++LG+VR++LY TRER+R LLAN +SDI EE FTNVE+FLEPLELCYRSLC
Sbjct: 359 IPPNEPYRIILGDVRDKLYNTRERARSLLANGFSDIPEEAAFTNVEQFLEPLELCYRSLC 418
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDR IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHL IGSY EW EE
Sbjct: 419 ACGDRPIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLDIGSYREWPEE 478
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
+RQ+WLLSEL GKRPLFGPDLP+TEE+ DVLDTFHVI+ELPSD+FGAYIISMATAPSDVL
Sbjct: 479 RRQEWLLSELRGKRPLFGPDLPKTEEVADVLDTFHVISELPSDSFGAYIISMATAPSDVL 538
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECHVK PLRVVPLFEKLADLE APAA+ARLFS++WYR+RINGKQEVMIGYSDS
Sbjct: 539 AVELLQRECHVKQPLRVVPLFEKLADLEAAPAAVARLFSIDWYRDRINGKQEVMIGYSDS 598
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGR SAAWQLYKAQEEL+KVAK+YGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 599 GKDAGRLSAAWQLYKAQEELVKVAKQYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 658
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGMHPP+SP PEWRALMDEMAV+A
Sbjct: 659 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMHPPVSPNPEWRALMDEMAVVA 718
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
T+EYRS+VF+EPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQT
Sbjct: 719 TKEYRSVVFQEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQT 778
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFKHVI KDI+NL+MLQEM+NQWPFFRVT+DL+EMVFAKGDPGIAALY
Sbjct: 779 RFHLPVWLGFGAAFKHVIQKDIKNLHMLQEMHNQWPFFRVTMDLIEMVFAKGDPGIAALY 838
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
D+LLVS++LW FGE LR +E+TK+L+LQVA H+DLLEGDPYLKQRLRLRD+YITTLNVC
Sbjct: 839 DKLLVSKELWPFGENLRANYEDTKRLVLQVAGHRDLLEGDPYLKQRLRLRDAYITTLNVC 898
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDP+Y+VK+RPH+S+E ++ SK A ELV LNPTSEYAPGLEDTLILTMKGIA
Sbjct: 899 QAYTLKRIRDPDYHVKVRPHLSREYMESSKAAAELVKLNPTSEYAPGLEDTLILTMKGIA 958
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 959 AGMQNTG 965
>emb|CAA62469.1| phosphoenolpyruvate carboxylase [Solanum tuberosum]
Length = 965
Score = 1731 bits (4484), Expect = 0.0
Identities = 855/968 (88%), Positives = 918/968 (94%), Gaps = 4/968 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
M RNLEK+ASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQE
Sbjct: 1 MTTRNLEKLASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEC 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGN++TSLD GDSIV+AK+F+HMLNLANLAEEVQIA+RRR K
Sbjct: 61 YELSAEYEAKHDPKKLEELGNVLTSLDPGDSIVIAKAFSHMLNLANLAEEVQIAYRRRQK 120
Query: 121 LKK-GDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRR 179
LKK GDF DE NATTESDIEETFKKLVG++KKSPQEVFD +KNQTVDLVLTAHPTQSVRR
Sbjct: 121 LKKKGDFGDESNATTESDIEETFKKLVGDLKKSPQEVFDAIKNQTVDLVLTAHPTQSVRR 180
Query: 180 SLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAG 239
SLLQKHGRIR+CL QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAG
Sbjct: 181 SLLQKHGRIRDCLAQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAG 240
Query: 240 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 299
MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT
Sbjct: 241 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 300
Query: 300 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWK 359
RDVCLLARMMAANLYYSQIEDLMFELSMWRCN+ELRVRA++L SS +D KHYIEFWK
Sbjct: 301 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNEELRVRADDLQRSSRRDE--KHYIEFWK 358
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSL 419
+PP+EPYRV+LG+VR++LYQTRER+R LL + YS+I EE T+TN+E+FLEPLELCYRSL
Sbjct: 359 QVPPNEPYRVILGDVRDKLYQTRERARQLLGHGYSEIPEEATYTNIEQFLEPLELCYRSL 418
Query: 420 CACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSE 479
CACGD +IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT+HL IGSY +WSE
Sbjct: 419 CACGDLSIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITQHLEIGSYRDWSE 478
Query: 480 EKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDV 539
E+RQ+WLLSELSGKRPLFGPDLP+TEEI DVLDTFHVIAELP+D FGAYIISMATAPSDV
Sbjct: 479 ERRQEWLLSELSGKRPLFGPDLPKTEEIADVLDTFHVIAELPADCFGAYIISMATAPSDV 538
Query: 540 LAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSD 599
LAVELLQREC V+ PLRVVPLFEKLADL+ APAA+ARLFS+EWYRNRINGKQEVMIGYSD
Sbjct: 539 LAVELLQRECRVRQPLRVVPLFEKLADLDAAPAAVARLFSIEWYRNRINGKQEVMIGYSD 598
Query: 600 SGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT 659
SGKDAGR SAAWQLYKAQEELI+VAKE+ VKLTMFHGRGGTVGRGGGP HLAILSQPP+T
Sbjct: 599 SGKDAGRLSAAWQLYKAQEELIQVAKEFDVKLTMFHGRGGTVGRGGGPAHLAILSQPPET 658
Query: 660 IHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVI 719
IHGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGMHPP+SPKPEWRALMDE+AV+
Sbjct: 659 IHGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMHPPVSPKPEWRALMDEIAVV 718
Query: 720 ATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQ 779
ATE+YRSIVFKEPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQ
Sbjct: 719 ATEKYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQ 778
Query: 780 TRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL 839
TRFHLPVWLGFGAAFK+ I KDI+NL MLQEMYN WPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 779 TRFHLPVWLGFGAAFKYAIEKDIKNLRMLQEMYNAWPFFRVTIDLVEMVFAKGDPGIAAL 838
Query: 840 YDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNV 899
+D+LLVSEDLWSFGE LR+ +EETK LLLQ+A HKDLLEGDPYLKQRLRLRDSYITTLNV
Sbjct: 839 FDKLLVSEDLWSFGELLRSKYEETKSLLLQIAGHKDLLEGDPYLKQRLRLRDSYITTLNV 898
Query: 900 CQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
CQAYTLKRIRDP+Y+V RPHISKE ++ +KPA ELV LNPTSEYAPGLEDTLILTMKGI
Sbjct: 899 CQAYTLKRIRDPDYSVTPRPHISKEYME-AKPATELVNLNPTSEYAPGLEDTLILTMKGI 957
Query: 960 AAGMQNTG 967
AAGMQNTG
Sbjct: 958 AAGMQNTG 965
>emb|CAA47437.1| phosphoenolpyruvate carboxylase [Solanum tuberosum]
gi|1168761|sp|P29196|CAPP_SOLTU Phosphoenolpyruvate
carboxylase (PEPCase)
Length = 965
Score = 1730 bits (4481), Expect = 0.0
Identities = 854/968 (88%), Positives = 918/968 (94%), Gaps = 4/968 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
M RNL+K+ASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQE
Sbjct: 1 MTTRNLDKLASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEC 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGN++TSLD GDSIV+AK+F+HMLNLANLAEEVQIA+RRR K
Sbjct: 61 YELSAEYEAKHDPKKLEELGNVLTSLDPGDSIVIAKAFSHMLNLANLAEEVQIAYRRRQK 120
Query: 121 LKK-GDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRR 179
LKK GDF DE NATTESDIEETFKKLVG++KKSPQEVFD +KNQTVDLVLTAHPTQSVRR
Sbjct: 121 LKKKGDFGDESNATTESDIEETFKKLVGDLKKSPQEVFDAIKNQTVDLVLTAHPTQSVRR 180
Query: 180 SLLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAG 239
SLLQKHGRIR+CL QLYAKDITPDDKQELDEALQREIQAAFRTDEIRRT PTPQDEMRAG
Sbjct: 181 SLLQKHGRIRDCLAQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTPPTPQDEMRAG 240
Query: 240 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 299
MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT
Sbjct: 241 MSYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVT 300
Query: 300 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWK 359
RDVCLLARMMAANLYYSQIEDLMFELSMWRCN+ELRVRA++L SS +D KHYIEFWK
Sbjct: 301 RDVCLLARMMAANLYYSQIEDLMFELSMWRCNEELRVRADDLQRSSRRDE--KHYIEFWK 358
Query: 360 NIPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSL 419
+PP+EPYRV+LG+VR++LYQTRER+R LL + YS+I EE T+TN+E+FLEPLELCYRSL
Sbjct: 359 QVPPNEPYRVILGDVRDKLYQTRERARQLLGHGYSEIPEEATYTNIEQFLEPLELCYRSL 418
Query: 420 CACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSE 479
CACGD +IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT+HL IGSY +WSE
Sbjct: 419 CACGDLSIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITQHLEIGSYRDWSE 478
Query: 480 EKRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDV 539
E+RQ+WLLSELSGKRPLFGPDLP+TEEI DVLDTFHVIAELP+D FGAYIISMATAPSDV
Sbjct: 479 ERRQEWLLSELSGKRPLFGPDLPKTEEIADVLDTFHVIAELPADCFGAYIISMATAPSDV 538
Query: 540 LAVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSD 599
LAVELLQREC V+ PLRVVPLFEKLADL+ APAA+ARLFS+EWYRNRINGKQEVMIGYSD
Sbjct: 539 LAVELLQRECRVRQPLRVVPLFEKLADLDAAPAAVARLFSIEWYRNRINGKQEVMIGYSD 598
Query: 600 SGKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDT 659
SGKDAGR SAAWQLYKAQEELI+VAKE+ VKLTMFHGRGGTVGRGGGP HLAILSQPP+T
Sbjct: 599 SGKDAGRLSAAWQLYKAQEELIQVAKEFDVKLTMFHGRGGTVGRGGGPAHLAILSQPPET 658
Query: 660 IHGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVI 719
IHGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGMHPP+SPKPEWRALMDE+AV+
Sbjct: 659 IHGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMHPPVSPKPEWRALMDEIAVV 718
Query: 720 ATEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQ 779
ATE+YRSIVFKEPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQ
Sbjct: 719 ATEKYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQ 778
Query: 780 TRFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAAL 839
TRFHLPVWLGFGAAFK+ I KDI+NL MLQEMYN WPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 779 TRFHLPVWLGFGAAFKYAIEKDIKNLRMLQEMYNAWPFFRVTIDLVEMVFAKGDPGIAAL 838
Query: 840 YDRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNV 899
+D+LLVSEDLWSFGE LR+ +EETK LLLQ+A HKDLLEGDPYLKQRLRLRDSYITTLNV
Sbjct: 839 FDKLLVSEDLWSFGELLRSKYEETKSLLLQIAGHKDLLEGDPYLKQRLRLRDSYITTLNV 898
Query: 900 CQAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
CQAYTLKRIRDP+Y+V RPHISKE ++ +KPA ELV LNPTSEYAPGLEDTLILTMKGI
Sbjct: 899 CQAYTLKRIRDPDYSVTPRPHISKEYME-AKPATELVNLNPTSEYAPGLEDTLILTMKGI 957
Query: 960 AAGMQNTG 967
AAGMQNTG
Sbjct: 958 AAGMQNTG 965
>emb|CAA09588.1| phosphoenolpyruvate-carboxylase [Vicia faba]
Length = 966
Score = 1729 bits (4477), Expect = 0.0
Identities = 855/967 (88%), Positives = 914/967 (94%), Gaps = 1/967 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MAN+ +EKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLK++VQEV
Sbjct: 1 MANK-MEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKDSVQEV 59
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKH+P++LE LG LIT LDAGDSIVVAKSF+HMLNLANLAEEVQIAHRRR K
Sbjct: 60 YELSAEYERKHDPKKLEELGKLITGLDAGDSIVVAKSFSHMLNLANLAEEVQIAHRRRNK 119
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDF DE NATTES+IEET K+LV MKKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 120 LKKGDFRDESNATTESNIEETLKRLVFNMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 179
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKH RIRNCL+ LYAKDITPDDKQELDE+LQREIQAAFRTDEI+RT PTPQDEMRAGM
Sbjct: 180 LLQKHARIRNCLSHLYAKDITPDDKQELDESLQREIQAAFRTDEIKRTPPTPQDEMRAGM 239
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKN+GINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 240 SYFHETIWKGVPKFLRRVDTALKNVGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 299
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELR RAEEL +S KD VAKHYIEFWK
Sbjct: 300 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRDRAEELHRNSKKDEVAKHYIEFWKK 359
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+P +EPYRVVLG++R++LY+TRERSR+LLA+ YSDI EE TFTNV+EFLEPLELCYRSLC
Sbjct: 360 VPLNEPYRVVLGDIRDKLYRTRERSRYLLAHGYSDIPEEATFTNVDEFLEPLELCYRSLC 419
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAITKHL IGSY EWSEE
Sbjct: 420 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYQEWSEE 479
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSEL GKRPLFGPDLP+T+EI+DVLDTFHVIAELPSDNFGAYIISMATAPSDVL
Sbjct: 480 KRQEWLLSELIGKRPLFGPDLPKTDEIRDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 539
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQREC +KNPLRVVPLFEKL DLE APAALARLFS++WYRNRI+GKQEVMIGYSDS
Sbjct: 540 AVELLQRECKIKNPLRVVPLFEKLDDLEAAPAALARLFSIDWYRNRIDGKQEVMIGYSDS 599
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VA+++ VKLTMFHGRGGTVGRGGGPTHLAILSQPP+TI
Sbjct: 600 GKDAGRFSAAWQLYKAQEELINVAQKFSVKLTMFHGRGGTVGRGGGPTHLAILSQPPETI 659
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HG LRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGM PP SPKPEWR LMD++A+IA
Sbjct: 660 HGILRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMRPPSSPKPEWRILMDQLAIIA 719
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPE+EYGRMNIGSRPAKR+PSGGIETLRAIPWIFAWTQT
Sbjct: 720 TEEYRSIVFKEPRFVEYFRLATPEMEYGRMNIGSRPAKRRPSGGIETLRAIPWIFAWTQT 779
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK VI KD++NL+MLQ+MYNQWPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 780 RFHLPVWLGFGAAFKQVIEKDVKNLHMLQDMYNQWPFFRVTIDLVEMVFAKGDPGIAALN 839
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVSEDLW FGEQLR +EETK+LLLQVA HK++LEGDPYLKQRLRLRDSYITTLNV
Sbjct: 840 DRLLVSEDLWPFGEQLRNKYEETKELLLQVATHKEVLEGDPYLKQRLRLRDSYITTLNVF 899
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGIA 960
QAYTLKRIRDP + RP +SK++ + +KPADELVTLNPTSEYAPGLEDTLILTMKGIA
Sbjct: 900 QAYTLKRIRDPKSSANGRPPLSKDSPEATKPADELVTLNPTSEYAPGLEDTLILTMKGIA 959
Query: 961 AGMQNTG 967
AGMQNTG
Sbjct: 960 AGMQNTG 966
>sp|P51062|CAPP_PEA Phosphoenolpyruvate carboxylase (PEPCase) gi|984039|dbj|BAA10902.1|
phosphoenolpyruvate carboxylase [Pisum sativum]
Length = 967
Score = 1725 bits (4468), Expect = 0.0
Identities = 857/968 (88%), Positives = 913/968 (93%), Gaps = 2/968 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MAN+ +EKMASIDAQLRQL PAKVSEDDKL+EYDALLLDRFLDILQDLHGEDLK++VQEV
Sbjct: 1 MANK-MEKMASIDAQLRQLAPAKVSEDDKLIEYDALLLDRFLDILQDLHGEDLKDSVQEV 59
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYERKH+P++LE LG LIT LDAGDSIVVAKSF+HMLNLANLAEEVQIAHRRR K
Sbjct: 60 YELSAEYERKHDPKKLEELGKLITGLDAGDSIVVAKSFSHMLNLANLAEEVQIAHRRRNK 119
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDF DE NATTESDIEET KKLV MKKSPQEVFD LKNQTVDLVLTAHPTQSVRRS
Sbjct: 120 LKKGDFRDESNATTESDIEETLKKLVFNMKKSPQEVFDALKNQTVDLVLTAHPTQSVRRS 179
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKH R+RNCL+QLYAKDITPDDKQELDE+LQREIQAAFRTDEI+RT PTPQDEMRAGM
Sbjct: 180 LLQKHARVRNCLSQLYAKDITPDDKQELDESLQREIQAAFRTDEIKRTPPTPQDEMRAGM 239
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 240 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 299
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELR RAEEL +S KD VAKHYIEFWK
Sbjct: 300 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRDRAEELHRNSKKDEVAKHYIEFWKK 359
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+P +EPYRV+LG VR++LY+TRERSR+LLA+ YSDI EE TFTN +EFLEPLELCYRSLC
Sbjct: 360 VPLNEPYRVILGHVRDKLYRTRERSRYLLAHGYSDIPEEDTFTNFDEFLEPLELCYRSLC 419
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
CGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDV+DAITKHL IGSY EWSEE
Sbjct: 420 FCGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVMDAITKHLEIGSYQEWSEE 479
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSEL GKRPLFGPDLP T+EI+DVLDTFHVIAELPSDNFGAYIISMATAPSDVL
Sbjct: 480 KRQEWLLSELVGKRPLFGPDLPTTDEIRDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 539
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQREC +KNPLRVVPLFEKLADLE APAALARLFS++WYRNRI+GKQEVMIGYSDS
Sbjct: 540 AVELLQRECKIKNPLRVVPLFEKLADLEAAPAALARLFSIDWYRNRIDGKQEVMIGYSDS 599
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGRFSAAWQLYKAQEELI VA+++ VKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI
Sbjct: 600 GKDAGRFSAAWQLYKAQEELINVAQKFSVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 659
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQR+TAATLEHGM PP SPKPEWRALMD+MA+IA
Sbjct: 660 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRFTAATLEHGMRPPSSPKPEWRALMDQMAIIA 719
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TEEYRSIVFKEPRFVEYFRLATPE+EYGRMNIGSRPAKR+PSGGIETLRAIPWIF WTQT
Sbjct: 720 TEEYRSIVFKEPRFVEYFRLATPEMEYGRMNIGSRPAKRRPSGGIETLRAIPWIFPWTQT 779
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFG+AFK VI KD++NL+MLQ+MYNQWPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 780 RFHLPVWLGFGSAFKQVIEKDVKNLHMLQDMYNQWPFFRVTIDLVEMVFAKGDPGIAALN 839
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
DRLLVS++LW FGEQLR +EETK+LLLQVA HK++LEGDPYLKQRLRLRDSYITTLNV
Sbjct: 840 DRLLVSQNLWPFGEQLRNKYEETKKLLLQVATHKEVLEGDPYLKQRLRLRDSYITTLNVF 899
Query: 901 QAYTLKRIRDPNYNVKL-RPHISKEAIDVSKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
QAYTLKRIRDP +V R +S+E+ + +KPADELVTLNPTSEYAPGLEDTLILTMKGI
Sbjct: 900 QAYTLKRIRDPKSSVNASRLPLSRESPEATKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
Query: 960 AAGMQNTG 967
AAGMQNTG
Sbjct: 960 AAGMQNTG 967
>gb|AAG17619.1| phosphoenolpyruvate carboxylase [Flaveria trinervia]
Length = 967
Score = 1720 bits (4455), Expect = 0.0
Identities = 849/968 (87%), Positives = 912/968 (93%), Gaps = 2/968 (0%)
Query: 1 MANRNLEKMASIDAQLRQLVPAKVSEDDKLVEYDALLLDRFLDILQDLHGEDLKETVQEV 60
MANRNLEK+ASIDAQLR LVP KVSEDDKL+EYDALLLD+FLDILQDLHGE LKETVQE
Sbjct: 1 MANRNLEKLASIDAQLRLLVPGKVSEDDKLIEYDALLLDKFLDILQDLHGEGLKETVQEC 60
Query: 61 YELSAEYERKHEPQRLEVLGNLITSLDAGDSIVVAKSFAHMLNLANLAEEVQIAHRRRIK 120
YELSAEYE KH+P++LE LGN++TSLD GDSIV+AK+F+HMLNLANLAEEVQIA+RRRIK
Sbjct: 61 YELSAEYEGKHDPKKLEELGNVLTSLDPGDSIVIAKAFSHMLNLANLAEEVQIAYRRRIK 120
Query: 121 LKKGDFADEGNATTESDIEETFKKLVGEMKKSPQEVFDELKNQTVDLVLTAHPTQSVRRS 180
LKKGDFADE +ATTESDIEETFKKLV ++KKSP+EVFD LKNQTVDLV TAHPTQSVRRS
Sbjct: 121 LKKGDFADEASATTESDIEETFKKLVHKLKKSPEEVFDALKNQTVDLVFTAHPTQSVRRS 180
Query: 181 LLQKHGRIRNCLTQLYAKDITPDDKQELDEALQREIQAAFRTDEIRRTAPTPQDEMRAGM 240
LLQKHGRIR+CL QLYAKDITPDDKQELDEAL REIQAAFRTDEIRRT PTPQDEMRAGM
Sbjct: 181 LLQKHGRIRDCLAQLYAKDITPDDKQELDEALHREIQAAFRTDEIRRTPPTPQDEMRAGM 240
Query: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR
Sbjct: 241 SYFHETIWKGVPKFLRRVDTALKNIGINERVPYNAPLIQFSSWMGGDRDGNPRVTPEVTR 300
Query: 301 DVCLLARMMAANLYYSQIEDLMFELSMWRCNDELRVRAEELTSSSNKDAVAKHYIEFWKN 360
DVCLLARMMAAN+Y+SQIEDLMFE+SMWRC+DELRVRAEEL SS+K V KHYIEFWK
Sbjct: 301 DVCLLARMMAANMYFSQIEDLMFEMSMWRCSDELRVRAEELHRSSSKRDV-KHYIEFWKQ 359
Query: 361 IPPSEPYRVVLGEVRNRLYQTRERSRHLLANEYSDILEEHTFTNVEEFLEPLELCYRSLC 420
+PP+EPYRV+LG+VR++LY TRER+RHLLA++ SDI EE +TNVE+FLEPLELCYRSLC
Sbjct: 360 VPPTEPYRVILGDVRDKLYNTRERARHLLAHDVSDIPEESVYTNVEQFLEPLELCYRSLC 419
Query: 421 ACGDRAIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITKHLGIGSYLEWSEE 480
ACGDR IADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAIT+HL IGSY EWSEE
Sbjct: 420 ACGDRVIADGSLLDFLRQVSTFGLSLVRLDIRQESDRHTDVLDAITQHLEIGSYREWSEE 479
Query: 481 KRQQWLLSELSGKRPLFGPDLPQTEEIKDVLDTFHVIAELPSDNFGAYIISMATAPSDVL 540
KRQ+WLLSELSGKRPLFGPDLP+TEEI DVLDTFHV+AELP+D FGAYIISMAT+PSDVL
Sbjct: 480 KRQEWLLSELSGKRPLFGPDLPKTEEIADVLDTFHVLAELPADCFGAYIISMATSPSDVL 539
Query: 541 AVELLQRECHVKNPLRVVPLFEKLADLETAPAALARLFSVEWYRNRINGKQEVMIGYSDS 600
AVELLQRECHVK PLRVVPLFEKLADLE APAA+ARLFS+EWY+NRI+GKQEVMIGYSDS
Sbjct: 540 AVELLQRECHVKQPLRVVPLFEKLADLEAAPAAVARLFSIEWYKNRIDGKQEVMIGYSDS 599
Query: 601 GKDAGRFSAAWQLYKAQEELIKVAKEYGVKLTMFHGRGGTVGRGGGPTHLAILSQPPDTI 660
GKDAGR SAAWQLYKAQEELI VAK++GVKLTMFHGRGGTVGRGGGPTHLAILSQPP+TI
Sbjct: 600 GKDAGRLSAAWQLYKAQEELINVAKKFGVKLTMFHGRGGTVGRGGGPTHLAILSQPPETI 659
Query: 661 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRYTAATLEHGMHPPISPKPEWRALMDEMAVIA 720
HGSLRVTVQGEVIEQSFGEEHLCFRTLQR+ AATLEHGM+PPISP+PEWRALMDEMAV A
Sbjct: 660 HGSLRVTVQGEVIEQSFGEEHLCFRTLQRFCAATLEHGMNPPISPRPEWRALMDEMAVYA 719
Query: 721 TEEYRSIVFKEPRFVEYFRLATPELEYGRMNIGSRPAKRKPSGGIETLRAIPWIFAWTQT 780
TE+YR IVFKEPRFVEYFRLATPELEYGRMNIGSRP+KRKPSGGIE+LRAIPWIFAWTQT
Sbjct: 720 TEQYREIVFKEPRFVEYFRLATPELEYGRMNIGSRPSKRKPSGGIESLRAIPWIFAWTQT 779
Query: 781 RFHLPVWLGFGAAFKHVIAKDIRNLNMLQEMYNQWPFFRVTIDLVEMVFAKGDPGIAALY 840
RFHLPVWLGFGAAFK+ I KDI+NL+MLQEMY WPFFRVTIDLVEMVFAKGDPGIAAL
Sbjct: 780 RFHLPVWLGFGAAFKYAIEKDIKNLHMLQEMYKTWPFFRVTIDLVEMVFAKGDPGIAALN 839
Query: 841 DRLLVSEDLWSFGEQLRTMFEETKQLLLQVAAHKDLLEGDPYLKQRLRLRDSYITTLNVC 900
D+LLVSEDLWSFGE LR +EETK LLL++A HKDLLEGDPYL+QRLRLRDSYITTLNVC
Sbjct: 840 DKLLVSEDLWSFGESLRANYEETKNLLLKIAGHKDLLEGDPYLRQRLRLRDSYITTLNVC 899
Query: 901 QAYTLKRIRDPNYNVKLRPHISKEAID-VSKPADELVTLNPTSEYAPGLEDTLILTMKGI 959
QAYTLKRIRDPNY+V RPHISKE + SKPADE + LNP SEYAPGLEDTLILTMKGI
Sbjct: 900 QAYTLKRIRDPNYHVTFRPHISKEYSEPSSKPADEYIKLNPKSEYAPGLEDTLILTMKGI 959
Query: 960 AAGMQNTG 967
AAGMQNTG
Sbjct: 960 AAGMQNTG 967
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,658,270,312
Number of Sequences: 2540612
Number of extensions: 72050760
Number of successful extensions: 185786
Number of sequences better than 10.0: 676
Number of HSP's better than 10.0 without gapping: 616
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 183116
Number of HSP's gapped (non-prelim): 873
length of query: 967
length of database: 863,360,394
effective HSP length: 138
effective length of query: 829
effective length of database: 512,755,938
effective search space: 425074672602
effective search space used: 425074672602
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0340.18


