

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0309.4
(116 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
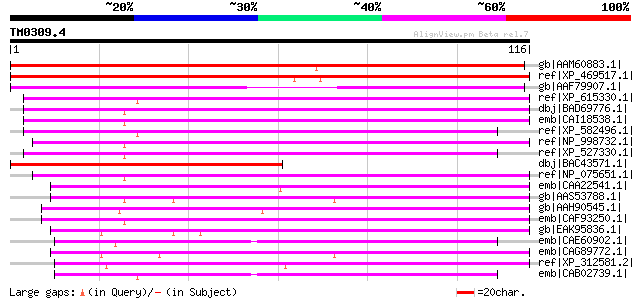
gb|AAM60883.1| putative RNA polymerase I subunit [Arabidopsis th... 140 7e-33
ref|XP_469517.1| putative RNA polymerase I subunit [Oryza sativa... 134 4e-31
gb|AAF79907.1| Contains a weak similarity to transcription elong... 109 1e-23
ref|XP_615330.1| PREDICTED: similar to zinc ribbon domain contai... 84 1e-15
dbj|BAD69776.1| zinc ribbon domain containing, 1 [Macaca mulatta] 82 3e-15
emb|CAI18538.1| zinc ribbon domain containing, 1 [Homo sapiens] ... 81 5e-15
ref|XP_582496.1| PREDICTED: similar to zinc ribbon domain contai... 81 5e-15
ref|NP_998732.1| zinc ribbon domain containing, 1 [Rattus norveg... 79 3e-14
ref|XP_527330.1| PREDICTED: similar to zinc ribbon domain contai... 79 3e-14
dbj|BAC43571.1| unknown protein [Arabidopsis thaliana] 78 6e-14
ref|NP_075651.1| nuclear RNA polymerase I small specific subunit... 77 1e-13
emb|CAA22541.1| SPCC1259.03 [Schizosaccharomyces pombe] gi|19075... 76 2e-13
gb|AAS53788.1| AFR417Wp [Ashbya gossypii ATCC 10895] gi|45198935... 73 2e-12
gb|AAH90545.1| Zgc:110825 [Danio rerio] gi|61806687|ref|NP_00101... 72 2e-12
emb|CAF93250.1| unnamed protein product [Tetraodon nigroviridis] 72 4e-12
gb|EAK95836.1| hypothetical protein CaO19.2287 [Candida albicans... 70 1e-11
emb|CAE60902.1| Hypothetical protein CBG04618 [Caenorhabditis br... 70 1e-11
emb|CAG89772.1| unnamed protein product [Debaryomyces hansenii C... 70 2e-11
ref|XP_312581.2| ENSANGP00000015033 [Anopheles gambiae str. PEST... 69 3e-11
emb|CAB02739.1| Hypothetical protein C15H11.8 [Caenorhabditis el... 68 5e-11
>gb|AAM60883.1| putative RNA polymerase I subunit [Arabidopsis thaliana]
gi|9279599|dbj|BAB01057.1| unnamed protein product
[Arabidopsis thaliana] gi|18404735|ref|NP_566786.1|
transcription factor S-II (TFIIS) domain-containing
protein [Arabidopsis thaliana]
Length = 119
Score = 140 bits (353), Expect = 7e-33
Identities = 67/118 (56%), Positives = 81/118 (67%), Gaps = 3/118 (2%)
Query: 1 MAYSRERDFLFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELG 60
M SRE +FLFC+ CGTML + S YAECP CK N KDI KEI+YT+SAE+IRRELG
Sbjct: 1 MEKSRESEFLFCNLCGTMLVLKSTKYAECPHCKTTRNAKDIIDKEIAYTVSAEDIRRELG 60
Query: 61 IETIEEQ---KVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQE 115
I E+ + +L K+ K CEKC + E + TRQ RSADEGQTT+Y C C H+F E
Sbjct: 61 ISLFGEKTQAEAELPKIKKACEKCQHPELVYTTRQTRSADEGQTTYYTCPNCAHRFTE 118
>ref|XP_469517.1| putative RNA polymerase I subunit [Oryza sativa]
gi|13324793|gb|AAK18841.1| putative RNA polymerase I
subunit [Oryza sativa]
Length = 126
Score = 134 bits (338), Expect = 4e-31
Identities = 66/125 (52%), Positives = 84/125 (66%), Gaps = 9/125 (7%)
Query: 1 MAYSRERDFLFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELG 60
MA+ + RDFLFC CGT+L SV A CPLC + KDIEGKE YT++AE+IRREL
Sbjct: 1 MAFWQARDFLFCGVCGTLLKFDSVRSASCPLCGFKRKAKDIEGKETRYTVTAEDIRRELK 60
Query: 61 IE-------TIEEQK--VQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGH 111
++ T++E+ V+ + VNK CEKC N E +YT+Q+RSADEGQT FY C C H
Sbjct: 61 LDPYVILETTLKEEDVIVERATVNKECEKCKNPELQYYTKQLRSADEGQTVFYKCANCRH 120
Query: 112 QFQEN 116
+F EN
Sbjct: 121 EFNEN 125
>gb|AAF79907.1| Contains a weak similarity to transcription elongation factor S-II
from Drosophila melanogaster gi|135660 and contains a
transcription factor S-II domain PF|01096. [Arabidopsis
thaliana] gi|25518086|pir||B86334 T20H2.16 protein -
Arabidopsis thaliana
Length = 122
Score = 109 bits (273), Expect = 1e-23
Identities = 53/115 (46%), Positives = 66/115 (57%), Gaps = 20/115 (17%)
Query: 1 MAYSRERDFLFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELG 60
M SRE D LFC+ CGTML + S YAECPLC+ N K+I K ++YT++ E
Sbjct: 27 MEKSRESDILFCNLCGTMLVLKSTKYAECPLCETTRNGKEIIDKNLAYTVTTE------- 79
Query: 61 IETIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQE 115
+ K CEKC + E + TRQ RSADEGQTT+Y C CGH+F E
Sbjct: 80 -------------IKKACEKCQHPELVYTTRQTRSADEGQTTYYTCPNCGHRFTE 121
>ref|XP_615330.1| PREDICTED: similar to zinc ribbon domain containing, 1 [Bos taurus]
Length = 123
Score = 83.6 bits (205), Expect = 1e-15
Identities = 42/114 (36%), Positives = 64/114 (55%), Gaps = 1/114 (0%)
Query: 4 SRERDFLFCHYCGTMLTVPSVNYA-ECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIE 62
S + D FC CG++L +P V A C C N++D EGK + ++ ++ + +
Sbjct: 9 SFQSDLDFCPDCGSVLPLPGVQDAVACTRCGFSINVRDFEGKVVKTSVVFNKLGTAMPLS 68
Query: 63 TIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
E + Q V++ C +CG+ ++TRQMRSADEGQT FY CT C Q +E+
Sbjct: 69 MEEGPEFQGPVVDRRCSRCGHEGMAYHTRQMRSADEGQTVFYTCTNCKFQEKED 122
>dbj|BAD69776.1| zinc ribbon domain containing, 1 [Macaca mulatta]
Length = 126
Score = 82.0 bits (201), Expect = 3e-15
Identities = 40/114 (35%), Positives = 63/114 (55%), Gaps = 1/114 (0%)
Query: 4 SRERDFLFCHYCGTMLTVPSV-NYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIE 62
S + D FC CG++L +P + C C N++D EGK + ++ ++ + +
Sbjct: 12 SFQSDLDFCSDCGSVLPLPGAQDTVTCTRCGFNINVRDFEGKVVKTSVVFHQLGTAMPMS 71
Query: 63 TIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
E + Q V++ C +CG+ ++TRQMRSADEGQT FY CT C Q +E+
Sbjct: 72 VEEGPECQGPVVDRRCPRCGHEGMAYHTRQMRSADEGQTVFYTCTNCKFQEKED 125
>emb|CAI18538.1| zinc ribbon domain containing, 1 [Homo sapiens]
gi|66347826|emb|CAI95579.1| OTTHUMP00000068350 [Homo
sapiens] gi|55961255|emb|CAI17577.1| zinc ribbon domain
containing, 1 [Homo sapiens] gi|55961202|emb|CAI18176.1|
zinc ribbon domain containing, 1 [Homo sapiens]
gi|7212805|gb|AAF40469.1| transcription-associated zinc
ribbon protein [Homo sapiens] gi|29792038|gb|AAH50608.1|
Zinc ribbon domain containing, 1 [Homo sapiens]
gi|15012007|gb|AAH10898.1| Zinc ribbon domain
containing, 1 [Homo sapiens] gi|12275850|gb|AAG50160.1|
nuclear RNA polymerase I small specific subunit Rpa12
[Homo sapiens] gi|12275847|gb|AAG50159.1| nuclear RNA
polymerase I small specific subunit Rpa12 [Homo sapiens]
gi|48146335|emb|CAG33390.1| ZNRD1 [Homo sapiens]
gi|25777707|ref|NP_740753.1| zinc ribbon domain
containing, 1 [Homo sapiens] gi|7657709|ref|NP_055411.1|
zinc ribbon domain containing, 1 [Homo sapiens]
Length = 126
Score = 81.3 bits (199), Expect = 5e-15
Identities = 40/114 (35%), Positives = 63/114 (55%), Gaps = 1/114 (0%)
Query: 4 SRERDFLFCHYCGTMLTVPSV-NYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIE 62
S + D FC CG++L +P + C C N++D EGK + ++ ++ + +
Sbjct: 12 SFQSDLDFCSDCGSVLPLPGAQDTVTCIRCGFNINVRDFEGKVVKTSVVFHQLGTAMPMS 71
Query: 63 TIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
E + Q V++ C +CG+ ++TRQMRSADEGQT FY CT C Q +E+
Sbjct: 72 VEEGPECQGPVVDRRCPRCGHEGMAYHTRQMRSADEGQTVFYTCTNCKFQEKED 125
>ref|XP_582496.1| PREDICTED: similar to zinc ribbon domain containing, 1, partial
[Bos taurus]
Length = 117
Score = 81.3 bits (199), Expect = 5e-15
Identities = 40/107 (37%), Positives = 60/107 (55%), Gaps = 1/107 (0%)
Query: 4 SRERDFLFCHYCGTMLTVPSVNYA-ECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIE 62
S + D FC CG++L +P V A C C N++D EGK + ++ ++ + +
Sbjct: 9 SFQSDLDFCPDCGSVLPLPGVQDAVACTRCGFSINVRDFEGKVVKTSVVFNKLGTAMPLS 68
Query: 63 TIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRC 109
E + Q V++ C +CG+ ++TRQMRSADEGQT FY CT C
Sbjct: 69 MEEGPEFQGPVVDRRCSRCGHEGMAYHTRQMRSADEGQTVFYTCTNC 115
>ref|NP_998732.1| zinc ribbon domain containing, 1 [Rattus norvegicus]
gi|46237691|emb|CAE84062.1| zinc ribbon domain
containing, 1 [Rattus norvegicus]
Length = 123
Score = 79.0 bits (193), Expect = 3e-14
Identities = 39/112 (34%), Positives = 62/112 (54%), Gaps = 1/112 (0%)
Query: 6 ERDFLFCHYCGTMLTVPSV-NYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETI 64
+ D FC CG++L +P V + CP C +++D GK + ++ ++ + +
Sbjct: 11 QSDLDFCPDCGSVLPLPGVQDTVICPRCGFSIDVRDFGGKVVKTSVVFNKLGTVIPMSVD 70
Query: 65 EEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
E + Q V++ C +CG+ +YTRQMRSADEGQT FY C C Q +E+
Sbjct: 71 EGPESQGPVVDRRCSRCGHEGMAYYTRQMRSADEGQTVFYTCINCKFQEKED 122
>ref|XP_527330.1| PREDICTED: similar to zinc ribbon domain containing, 1;
transcription-associated zinc ribbon protein; RNA
polymerase I small specific subunit Rpa12 [Pan
troglodytes]
Length = 663
Score = 79.0 bits (193), Expect = 3e-14
Identities = 38/107 (35%), Positives = 59/107 (54%), Gaps = 1/107 (0%)
Query: 4 SRERDFLFCHYCGTMLTVPSV-NYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIE 62
S + D FC CG++L +P + C C N++D EGK + ++ ++ + +
Sbjct: 12 SFQSDLDFCSDCGSVLPLPGAQDTVTCIRCGFNINVRDFEGKVVKTSVVFHQLGTAMPMS 71
Query: 63 TIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRC 109
E + Q V++ C +CG+ ++TRQMRSADEGQT FY CT C
Sbjct: 72 VEEGPECQGPVVDRRCPRCGHEGMAYHTRQMRSADEGQTVFYTCTNC 118
>dbj|BAC43571.1| unknown protein [Arabidopsis thaliana]
Length = 63
Score = 77.8 bits (190), Expect = 6e-14
Identities = 35/61 (57%), Positives = 44/61 (71%)
Query: 1 MAYSRERDFLFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELG 60
M SRE D LFC+ CGTML + S YAECPLC+ N K+I K ++YT++ E+IRRELG
Sbjct: 1 MEKSRESDILFCNLCGTMLVLKSTKYAECPLCETTRNGKEIIDKNLAYTVTTEDIRRELG 60
Query: 61 I 61
I
Sbjct: 61 I 61
>ref|NP_075651.1| nuclear RNA polymerase I small specific subunit [Mus musculus]
gi|12275854|gb|AAG50162.1| nuclear RNA polymerase I
small specific subunit Rpa12 [Mus musculus]
gi|12275852|gb|AAG50161.1| nuclear RNA polymerase I
small specific subunit Rpa12 [Mus musculus]
gi|27695655|gb|AAH43016.1| Znrd1 protein [Mus musculus]
gi|12838841|dbj|BAB24350.1| unnamed protein product [Mus
musculus] gi|12834550|dbj|BAB22954.1| unnamed protein
product [Mus musculus]
Length = 123
Score = 77.0 bits (188), Expect = 1e-13
Identities = 36/112 (32%), Positives = 63/112 (56%), Gaps = 1/112 (0%)
Query: 6 ERDFLFCHYCGTMLTVPSV-NYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETI 64
+ D FC CG++L +P + + C C +++D EGK + ++ ++ + +
Sbjct: 11 QSDLDFCPDCGSVLPLPGIQDTVICSRCGFSIDVRDCEGKVVKTSVVFNKLGATIPLSVD 70
Query: 65 EEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
E ++Q +++ C +CG+ ++TRQMRSADEGQT FY C C Q +E+
Sbjct: 71 EGPELQGPVIDRRCPRCGHEGMAYHTRQMRSADEGQTVFYTCINCKFQEKED 122
>emb|CAA22541.1| SPCC1259.03 [Schizosaccharomyces pombe]
gi|19075559|ref|NP_588059.1| hypothetical protein
SPCC1259.03 [Schizosaccharomyces pombe 972h-]
gi|9297043|sp|O94703|RPA9_SCHPO DNA-directed RNA
polymerase I 13.1 kDa polypeptide
gi|6518899|dbj|BAA87928.1| RPA12 [Schizosaccharomyces
pombe] gi|6519808|dbj|BAA87930.1| SpRPA12
[Schizosaccharomyces pombe] gi|8886803|gb|AAF80580.1|
RNA polymerase I specific subunit Rpa12
[Schizosaccharomyces pombe]
Length = 119
Score = 76.3 bits (186), Expect = 2e-13
Identities = 37/111 (33%), Positives = 54/111 (48%), Gaps = 4/111 (3%)
Query: 10 LFCHYCGTMLTVPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRREL----GIETIE 65
+FC CG +L + + C C++ + + SA L I +E
Sbjct: 8 IFCSECGNLLESTTAQWTTCDQCQSVYPSEQFANLVVETKSSASAFPSALKLKHSIVQVE 67
Query: 66 EQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
QK + + + + C KCGN TF+T Q+RSADEG T FY C RC ++F N
Sbjct: 68 SQKEEAATIEEKCPKCGNDHMTFHTLQLRSADEGSTVFYECPRCAYKFSTN 118
>gb|AAS53788.1| AFR417Wp [Ashbya gossypii ATCC 10895] gi|45198935|ref|NP_985964.1|
AFR417Wp [Eremothecium gossypii]
Length = 125
Score = 72.8 bits (177), Expect = 2e-12
Identities = 39/117 (33%), Positives = 64/117 (54%), Gaps = 10/117 (8%)
Query: 10 LFCHYCGTMLTVPSV---NYAECPLCKNR------SNIKDIEGKEISYTISAEEIRRELG 60
+FC YCG +L PS ++ C LC + SN+K + SA +R +
Sbjct: 8 IFCVYCGNLLDNPSAVAGDHVACALCDAQYDKATFSNLKVVTATADDAFPSALRAKRSVV 67
Query: 61 IETIEEQKVQL-SKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
T+ + +++ + + + C +CG+ E ++T Q+RSADEG T FY CT CG++F+ N
Sbjct: 68 KTTLRKGELEDGATIREKCPQCGHDEMQYHTLQLRSADEGATVFYTCTSCGYRFRTN 124
>gb|AAH90545.1| Zgc:110825 [Danio rerio] gi|61806687|ref|NP_001013572.1|
hypothetical protein LOC541428 [Danio rerio]
Length = 118
Score = 72.4 bits (176), Expect = 2e-12
Identities = 34/111 (30%), Positives = 62/111 (55%), Gaps = 2/111 (1%)
Query: 8 DFLFCHYCGTMLTVPS-VNYAECPLCKNRSNIKDIEGKEISYTISAEEI-RRELGIETIE 65
D FC CG +L +PS +N CP C + +++D + I ++ + + + + + E
Sbjct: 7 DVDFCPECGNILPLPSRLNTITCPRCSFKISVQDFTSQVIKSSVMFNPLDQSNVAVGSAE 66
Query: 66 EQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
+ +++ +++ C +C ++TRQMRSADEGQT F+ C C Q +E+
Sbjct: 67 DAELKGPVIDRKCSRCNKEGMVYHTRQMRSADEGQTVFFTCIHCRFQEKED 117
>emb|CAF93250.1| unnamed protein product [Tetraodon nigroviridis]
Length = 147
Score = 71.6 bits (174), Expect = 4e-12
Identities = 33/110 (30%), Positives = 55/110 (50%), Gaps = 1/110 (0%)
Query: 8 DFLFCHYCGTMLTVPSV-NYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIEE 66
D FC CG +L P + + CP C + + +G++I T+ + + E+
Sbjct: 19 DLNFCPECGNILPPPGLQDTVRCPRCSFSIPVAEFDGQQIRSTVVLNPAEKSAAVVEEED 78
Query: 67 QKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
+Q +++ C +C ++TRQMRSADEGQT F+ C C + E+
Sbjct: 79 SDLQGPVIDRRCVRCNKEGMVYHTRQMRSADEGQTVFFTCVHCRRRTLES 128
>gb|EAK95836.1| hypothetical protein CaO19.2287 [Candida albicans SC5314]
gi|46436409|gb|EAK95772.1| hypothetical protein
CaO19.9827 [Candida albicans SC5314]
Length = 123
Score = 70.1 bits (170), Expect = 1e-11
Identities = 35/115 (30%), Positives = 64/115 (55%), Gaps = 8/115 (6%)
Query: 10 LFCHYCGTML-TVPSVNYAECPLCKNR------SNIKDI-EGKEISYTISAEEIRRELGI 61
+FC+YCG +L + S + +C +C +N+K + + + ++ + R +
Sbjct: 8 IFCNYCGNLLDSHSSSSEIKCTVCSAAYPKSKFANLKVVTKSSDDAFPSKLKSARSVVKT 67
Query: 62 ETIEEQKVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
+++ + + + + C KCGN E ++T Q+RSADEG T FY CT CG++F+ N
Sbjct: 68 SLKKDELDEGATIKEKCPKCGNEEMQYHTLQLRSADEGATVFYTCTNCGYRFRTN 122
>emb|CAE60902.1| Hypothetical protein CBG04618 [Caenorhabditis briggsae]
Length = 119
Score = 70.1 bits (170), Expect = 1e-11
Identities = 37/101 (36%), Positives = 55/101 (53%), Gaps = 3/101 (2%)
Query: 11 FCHYCGTMLTVP--SVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIEEQK 68
FC YCG +L +P + + C +C R N+K+ + +S E R + IE +
Sbjct: 12 FCGYCGAILELPPQAPSTVTCKVCSTRWNVKERVDQVVSRVEKIYE-RTVADTDGIENDE 70
Query: 69 VQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRC 109
+ V+ C KCG+ +A++ T Q RSADEGQT FY C +C
Sbjct: 71 SADAVVDHICTKCGHTKASYSTMQTRSADEGQTVFYTCLKC 111
>emb|CAG89772.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50425541|ref|XP_461366.1| unnamed protein product
[Debaryomyces hansenii]
Length = 123
Score = 69.7 bits (169), Expect = 2e-11
Identities = 39/115 (33%), Positives = 63/115 (53%), Gaps = 8/115 (6%)
Query: 10 LFCHYCGTML-TVPSVNYAECPLC------KNRSNIKDIEGKEISYTISAEEIRRELGIE 62
+FC CG +L TV S + C LC N +N+K I SA +++R +
Sbjct: 8 IFCTDCGNLLDTVGSKSTLNCKLCHKSYPTSNFANLKVITQSSEDAFPSALKMKRSVVKT 67
Query: 63 TIEEQKVQL-SKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
+++ +++ + + + C KCG E ++ Q+RSADEG T FY CT CG++F+ N
Sbjct: 68 SLKNDELEEGATIKEKCPKCGTEEMQYHVLQLRSADEGATVFYTCTGCGYRFRTN 122
>ref|XP_312581.2| ENSANGP00000015033 [Anopheles gambiae str. PEST]
gi|55242409|gb|EAA07851.3| ENSANGP00000015033 [Anopheles
gambiae str. PEST]
Length = 114
Score = 68.9 bits (167), Expect = 3e-11
Identities = 36/109 (33%), Positives = 57/109 (52%), Gaps = 3/109 (2%)
Query: 11 FCHYCGTMLT-VPSVNYAECPLCKNRSNIKDIEGKEISYTISAEEIRRELG--IETIEEQ 67
FC CG++L + + N C C++ + E YTI + + E +
Sbjct: 5 FCPDCGSILPPLKNSNRVSCYGCQSEFDAAAFGTMETEYTIHFNSYANKKSDQADRAEGE 64
Query: 68 KVQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRCGHQFQEN 116
+ + VN+ C KCGN + ++ T Q+RSADEGQT F+ CT+C ++ EN
Sbjct: 65 EAEGPIVNRQCPKCGNDQMSYATLQLRSADEGQTVFFTCTKCKYKMSEN 113
>emb|CAB02739.1| Hypothetical protein C15H11.8 [Caenorhabditis elegans]
gi|17558010|ref|NP_506572.1| RNA polymerase (13.2 kD)
(5O904) [Caenorhabditis elegans] gi|7496068|pir||T19321
hypothetical protein C15H11.8 - Caenorhabditis elegans
Length = 119
Score = 68.2 bits (165), Expect = 5e-11
Identities = 37/101 (36%), Positives = 54/101 (52%), Gaps = 3/101 (2%)
Query: 11 FCHYCGTMLTVPSVNYA--ECPLCKNRSNIKDIEGKEISYTISAEEIRRELGIETIEEQK 68
FC YCG +L +P+ A C +C R +K+ + +S E R + IE +
Sbjct: 12 FCGYCGAILELPAQAPATVSCKVCSTRWAVKERVDQVVSRVEKIYE-RTVADTDGIENDE 70
Query: 69 VQLSKVNKTCEKCGNGEATFYTRQMRSADEGQTTFYACTRC 109
+ V+ C KCG+ +A++ T Q RSADEGQT FY C +C
Sbjct: 71 SADAVVDHICTKCGHSKASYSTMQTRSADEGQTVFYTCLKC 111
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 181,986,427
Number of Sequences: 2540612
Number of extensions: 6144329
Number of successful extensions: 24758
Number of sequences better than 10.0: 360
Number of HSP's better than 10.0 without gapping: 310
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 24320
Number of HSP's gapped (non-prelim): 451
length of query: 116
length of database: 863,360,394
effective HSP length: 92
effective length of query: 24
effective length of database: 629,624,090
effective search space: 15110978160
effective search space used: 15110978160
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0309.4


