

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0282.6
(303 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
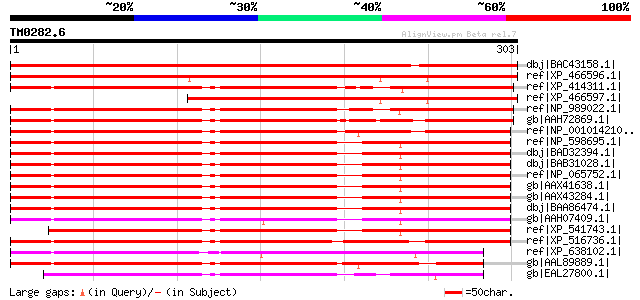
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC43158.1| unknown protein [Arabidopsis thaliana] gi|929390... 491 e-137
ref|XP_466596.1| 2 coiled coil domains of eukaryotic origin (31.... 440 e-122
ref|XP_414311.1| PREDICTED: similar to Hypothetical protein MGC7... 273 3e-72
ref|XP_466597.1| 2 coiled coil domains of eukaryotic origin (31.... 271 2e-71
ref|NP_989022.1| hypothetical protein LOC394618 [Xenopus tropica... 268 2e-70
gb|AAH72869.1| MGC80278 protein [Xenopus laevis] 266 4e-70
ref|NP_001014210.1| hypothetical protein LOC362394 [Rattus norve... 265 2e-69
ref|NP_598695.1| hypothetical protein LOC57905 [Mus musculus] gi... 264 3e-69
dbj|BAD32394.1| mKIAA1160 protein [Mus musculus] 264 3e-69
dbj|BAB31028.1| unnamed protein product [Mus musculus] 263 6e-69
ref|NP_065752.1| hypothetical protein LOC57461 [Homo sapiens] gi... 261 1e-68
gb|AAX41638.1| KIAA1160 protein [synthetic construct] 261 1e-68
gb|AAX43284.1| KIAA1160 protein [synthetic construct] 261 1e-68
dbj|BAA86474.1| KIAA1160 protein [Homo sapiens] 261 1e-68
gb|AAH07409.1| KIAA1160 protein [Homo sapiens] 251 1e-65
ref|XP_541743.1| PREDICTED: similar to RIKEN cDNA 5830446M03 [Ca... 244 2e-63
ref|XP_516736.1| PREDICTED: similar to KIAA1160 protein [Pan tro... 239 6e-62
ref|XP_638102.1| hypothetical protein DDB0186544 [Dictyostelium ... 232 9e-60
gb|AAL89889.1| RE31520p [Drosophila melanogaster] 230 4e-59
gb|EAL27800.1| GA21951-PA [Drosophila pseudoobscura] 218 2e-55
>dbj|BAC43158.1| unknown protein [Arabidopsis thaliana] gi|9293904|dbj|BAB01807.1|
unnamed protein product [Arabidopsis thaliana]
gi|19699138|gb|AAL90935.1| AT3g18790/MVE11_17
[Arabidopsis thaliana] gi|18086381|gb|AAL57650.1|
AT3g18790/MVE11_17 [Arabidopsis thaliana]
gi|15230193|ref|NP_188509.1| expressed protein
[Arabidopsis thaliana]
Length = 300
Score = 491 bits (1264), Expect = e-137
Identities = 241/304 (79%), Positives = 270/304 (88%), Gaps = 5/304 (1%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKAQSMLNRFIT K EKKKPKERRP+LA+ECRDL+EADKWRQQI+REIG KVAE
Sbjct: 1 MARNEEKAQSMLNRFITQKESEKKKPKERRPYLASECRDLAEADKWRQQILREIGSKVAE 60
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQNEGLGEHRLRDLNDEINKL+RE+ HWERRIVELGG NY++HSAKMTDL+GNI+DVPNP
Sbjct: 61 IQNEGLGEHRLRDLNDEINKLLRERYHWERRIVELGGHNYSKHSAKMTDLEGNIIDVPNP 120
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
SGRGPGYRYFGAAKKLPGVRELF+KPPELRKR+TRYDIYKRIDA+YYGYRDDEDGILE+L
Sbjct: 121 SGRGPGYRYFGAAKKLPGVRELFEKPPELRKRKTRYDIYKRIDASYYGYRDDEDGILEKL 180
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKS-GEVAEVSAAVREILREEEEDVVEEERM 239
E +E MR+ + EEW RLD +R+EARKG V +AA RE+L EEEEDVVEEERM
Sbjct: 181 ERKSEGGMRKRSVEEWRRLDEVRKEARKGASEVVSVGAAAAAAREVLFEEEEDVVEEERM 240
Query: 240 RGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
+E+ EE EREFVVHVPLPDEKEIEKMVLEKKKM+LL+KY SEDL+++Q+EAK ML
Sbjct: 241 E----REKEEEKEREFVVHVPLPDEKEIEKMVLEKKKMDLLSKYASEDLVEQQTEAKSML 296
Query: 300 NIHR 303
NIHR
Sbjct: 297 NIHR 300
>ref|XP_466596.1| 2 coiled coil domains of eukaryotic origin (31.3 kD)-like protein
[Oryza sativa (japonica cultivar-group)]
gi|47848307|dbj|BAD22171.1| 2 coiled coil domains of
eukaryotic origin (31.3 kD)-like protein [Oryza sativa
(japonica cultivar-group)] gi|47497303|dbj|BAD19345.1| 2
coiled coil domains of eukaryotic origin (31.3 kD)-like
protein [Oryza sativa (japonica cultivar-group)]
Length = 311
Score = 440 bits (1131), Expect = e-122
Identities = 221/311 (71%), Positives = 258/311 (82%), Gaps = 8/311 (2%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKAQSMLNRFITMK EEK+KP+ERRP+LA+ECRDL++A++WR +I+REIG KVAE
Sbjct: 1 MARNEEKAQSMLNRFITMKQEEKRKPRERRPYLASECRDLADAERWRSEILREIGAKVAE 60
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAK--MTDLDGNIVDVP 118
IQNEGLGEHRLRDLNDEINKL+RE+ HWERRIVELGG +++R S MTDLDGNIV +P
Sbjct: 61 IQNEGLGEHRLRDLNDEINKLLRERGHWERRIVELGGRDHSRSSNAPLMTDLDGNIVAIP 120
Query: 119 NPSGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILE 178
NPSGRGPGYRYFGAAKKLPGVRELFDKPPE+RKRRTRY+I+KRI+A YYGY DDEDG+LE
Sbjct: 121 NPSGRGPGYRYFGAAKKLPGVRELFDKPPEVRKRRTRYEIHKRINAGYYGYYDDEDGMLE 180
Query: 179 RLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSA----AVREILREEEEDVV 234
RLE AE MR E EW+R++R+R+EA KGV SGEVA A RE+L EE E+ V
Sbjct: 181 RLEAVAEKRMRNEVITEWHRVERVRREAMKGVVSGEVASAGGRGGEAAREVLFEEVEEEV 240
Query: 235 EEERMRGREMKERL--EENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQ 292
EEER E +ER EE +EF+ HVPLPDEKEIE+MVLE+KK ELL+KY S+ L EQ
Sbjct: 241 EEERRLEEEKREREKGEEAGKEFIAHVPLPDEKEIERMVLERKKKELLSKYTSDALQVEQ 300
Query: 293 SEAKDMLNIHR 303
EAK+MLN+ R
Sbjct: 301 EEAKEMLNVRR 311
>ref|XP_414311.1| PREDICTED: similar to Hypothetical protein MGC76019 [Gallus gallus]
Length = 280
Score = 273 bits (699), Expect = 3e-72
Identities = 154/303 (50%), Positives = 203/303 (66%), Gaps = 25/303 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECNELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y R +M D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIRELGGPDYARIGPRMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K IDA YYGYRD++DGILE L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKSIDAEYYGYRDEDDGILEPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDV--VEEER 238
E E + EA E+W ++ +EAR + GE E EEEE++ V++E
Sbjct: 174 EQEHEKKVIAEAVEKW----KMEREAR--LARGEEEE----------EEEENIYAVDDES 217
Query: 239 MRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDM 298
++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE LM + EAK +
Sbjct: 218 DEEGGKEKEGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLMAQSEEAKRL 277
Query: 299 LNI 301
L +
Sbjct: 278 LGL 280
>ref|XP_466597.1| 2 coiled coil domains of eukaryotic origin (31.3 kD)-like protein
[Oryza sativa (japonica cultivar-group)]
gi|47848308|dbj|BAD22172.1| 2 coiled coil domains of
eukaryotic origin (31.3 kD)-like protein [Oryza sativa
(japonica cultivar-group)] gi|47497304|dbj|BAD19346.1| 2
coiled coil domains of eukaryotic origin (31.3 kD)-like
protein [Oryza sativa (japonica cultivar-group)]
Length = 203
Score = 271 bits (693), Expect = 2e-71
Identities = 138/203 (67%), Positives = 160/203 (77%), Gaps = 6/203 (2%)
Query: 107 MTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAY 166
MTDLDGNIV +PNPSGRGPGYRYFGAAKKLPGVRELFDKPPE+RKRRTRY+I+KRI+A Y
Sbjct: 1 MTDLDGNIVAIPNPSGRGPGYRYFGAAKKLPGVRELFDKPPEVRKRRTRYEIHKRINAGY 60
Query: 167 YGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSA----AV 222
YGY DDEDG+LERLE AE MR E EW+R++R+R+EA KGV SGEVA A
Sbjct: 61 YGYYDDEDGMLERLEAVAEKRMRNEVITEWHRVERVRREAMKGVVSGEVASAGGRGGEAA 120
Query: 223 REILREEEEDVVEEERMRGREMKERL--EENEREFVVHVPLPDEKEIEKMVLEKKKMELL 280
RE+L EE E+ VEEER E +ER EE +EF+ HVPLPDEKEIE+MVLE+KK ELL
Sbjct: 121 REVLFEEVEEEVEEERRLEEEKREREKGEEAGKEFIAHVPLPDEKEIERMVLERKKKELL 180
Query: 281 NKYVSEDLMDEQSEAKDMLNIHR 303
+KY S+ L EQ EAK+MLN+ R
Sbjct: 181 SKYTSDALQVEQEEAKEMLNVRR 203
>ref|NP_989022.1| hypothetical protein LOC394618 [Xenopus tropicalis]
gi|38174132|gb|AAH61420.1| Hypothetical protein MGC76019
[Xenopus tropicalis]
Length = 282
Score = 268 bits (684), Expect = 2e-70
Identities = 154/305 (50%), Positives = 195/305 (63%), Gaps = 27/305 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECNELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKLIREK HWE RI ELGGP+Y R KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLIREKGHWEVRIKELGGPDYGRIGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K IDA YYGYRD++DG+L L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKSIDAEYYGYRDEDDGVLVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEE----DVVEE 236
E E EA E+W RQE + +G+ E EEEE V +
Sbjct: 174 EQEEEKKAIGEALEKW------RQEKEARLANGDRNE----------EEEEVNIYAVAAD 217
Query: 237 ERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAK 296
E + EE +++F+ HVP+P +KEIE+ ++ +KKMELL KY SE L+ + +AK
Sbjct: 218 ESGDDSDSGAEGEEGQQKFIAHVPVPTQKEIEEALIRRKKMELLQKYASETLLAQSEDAK 277
Query: 297 DMLNI 301
+L I
Sbjct: 278 RLLGI 282
>gb|AAH72869.1| MGC80278 protein [Xenopus laevis]
Length = 284
Score = 266 bits (681), Expect = 4e-70
Identities = 150/302 (49%), Positives = 201/302 (65%), Gaps = 19/302 (6%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + E+ K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEDGKV-KERRPFLASECSELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE ++RD+NDEINKLIREK HWE RI ELGGP+Y R KM D +G V P
Sbjct: 60 IQNAGLGEFKIRDVNDEINKLIREKGHWEVRIKELGGPDYGRIGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K IDA YYGYRD++DG+L L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRTELMKSIDAEYYGYRDEDDGVLVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEE-EDVVEEERM 239
E E EA E+W RL++ + G K+ E EV+ + + +E ED ++E
Sbjct: 174 EQEQEKKAIAEALEKW-RLEK-EERLANGDKNEEEEEVN--IYAVAADESGEDSDDDEGA 229
Query: 240 RGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
G EE +++F+ HVP+P +KE+E+ ++ +KKMELL KY SE L+ + +AK +L
Sbjct: 230 EG-------EEGQQKFIAHVPVPSQKEMEEALVRRKKMELLQKYASETLLAQSEDAKRLL 282
Query: 300 NI 301
+
Sbjct: 283 GV 284
>ref|NP_001014210.1| hypothetical protein LOC362394 [Rattus norvegicus]
gi|50926969|gb|AAH79119.1| Hypothetical LOC362394
[Rattus norvegicus]
Length = 284
Score = 265 bits (676), Expect = 2e-69
Identities = 146/301 (48%), Positives = 195/301 (64%), Gaps = 21/301 (6%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEAR--KGVKSGEVAEVSAAVREILREEEEDVVEEER 238
E E +R E E+W + +EAR +G K E E + + EE ++ +E+
Sbjct: 174 EQEYEKRLRAELVEKW----KAEREARLARGEKEEEEEEEEINIYAVTEEESDEEGSQEK 229
Query: 239 MRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDM 298
E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EAK +
Sbjct: 230 AG--------EDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEAKRL 281
Query: 299 L 299
L
Sbjct: 282 L 282
>ref|NP_598695.1| hypothetical protein LOC57905 [Mus musculus]
gi|18043689|gb|AAH20103.1| RIKEN cDNA 5830446M03 [Mus
musculus] gi|22902359|gb|AAH37695.1| RIKEN cDNA
5830446M03 [Mus musculus]
Length = 285
Score = 264 bits (674), Expect = 3e-69
Identities = 147/304 (48%), Positives = 194/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGNQEKAGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
K +L
Sbjct: 280 KRLL 283
>dbj|BAD32394.1| mKIAA1160 protein [Mus musculus]
Length = 300
Score = 264 bits (674), Expect = 3e-69
Identities = 147/304 (48%), Positives = 194/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 16 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 74
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 75 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 130
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 131 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 188
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 189 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 234
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 235 EESDEEGNQEKAGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 294
Query: 296 KDML 299
K +L
Sbjct: 295 KRLL 298
>dbj|BAB31028.1| unnamed protein product [Mus musculus]
Length = 285
Score = 263 bits (671), Expect = 6e-69
Identities = 146/304 (48%), Positives = 194/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGV+ELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVKELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ E+ +++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGNQEKAGEDGQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
K +L
Sbjct: 280 KRLL 283
>ref|NP_065752.1| hypothetical protein LOC57461 [Homo sapiens]
gi|60817802|gb|AAX36438.1| KIAA1160 protein [synthetic
construct] gi|18043796|gb|AAH19849.1| KIAA1160 protein
[Homo sapiens] gi|13325246|gb|AAH04442.1| KIAA1160
protein [Homo sapiens]
Length = 285
Score = 261 bits (668), Expect = 1e-68
Identities = 145/304 (47%), Positives = 195/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
+ +L
Sbjct: 280 RRLL 283
>gb|AAX41638.1| KIAA1160 protein [synthetic construct]
Length = 285
Score = 261 bits (668), Expect = 1e-68
Identities = 145/304 (47%), Positives = 195/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
+ +L
Sbjct: 280 RRLL 283
>gb|AAX43284.1| KIAA1160 protein [synthetic construct]
Length = 286
Score = 261 bits (668), Expect = 1e-68
Identities = 145/304 (47%), Positives = 195/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 174 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 219
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 220 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 279
Query: 296 KDML 299
+ +L
Sbjct: 280 RRLL 283
>dbj|BAA86474.1| KIAA1160 protein [Homo sapiens]
Length = 351
Score = 261 bits (668), Expect = 1e-68
Identities = 145/304 (47%), Positives = 195/304 (63%), Gaps = 26/304 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 21 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 79
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 80 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 135
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 136 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 193
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEED-----VVE 235
E E +R E E+W K+ A ++ +E EEEE+ V E
Sbjct: 194 EQEYEKKLRAELVEKW--------------KAEREARLARGEKEEEEEEEEEINIYAVTE 239
Query: 236 EERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEA 295
EE ++ ++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA
Sbjct: 240 EESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEA 299
Query: 296 KDML 299
+ +L
Sbjct: 300 RRLL 303
>gb|AAH07409.1| KIAA1160 protein [Homo sapiens]
Length = 307
Score = 251 bits (642), Expect = 1e-65
Identities = 145/326 (44%), Positives = 197/326 (59%), Gaps = 48/326 (14%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELR----------------------KRRTRYDI 158
R GY+YFGAAK LPGVRELF+K ++R R+TR ++
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKERQVRWLMPVIPALWEAEAGGSQALPPPRKTRAEL 173
Query: 159 YKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEV 218
K ID YYGY D++DG++ LE E +R E E+W K+ A +
Sbjct: 174 MKAIDFEYYGYLDEDDGVIVPLEQEYEKKLRAELVEKW--------------KAEREARL 219
Query: 219 SAAVREILREEEED-----VVEEERMRGREMKERLEENEREFVVHVPLPDEKEIEKMVLE 273
+ +E EEEE+ V EEE ++ ++++++F+ HVP+P ++EIE+ ++
Sbjct: 220 ARGEKEEEEEEEEEINIYAVTEEESDEEGSQEKGGDDSQQKFIAHVPVPSQQEIEEALVR 279
Query: 274 KKKMELLNKYVSEDLMDEQSEAKDML 299
+KKMELL KY SE L + EA+ +L
Sbjct: 280 RKKMELLQKYASETLQAQSEEARRLL 305
>ref|XP_541743.1| PREDICTED: similar to RIKEN cDNA 5830446M03 [Canis familiaris]
Length = 558
Score = 244 bits (623), Expect = 2e-63
Identities = 133/281 (47%), Positives = 178/281 (63%), Gaps = 25/281 (8%)
Query: 24 KKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINKLIR 83
+K ERRPFLA+EC +L +A+KWR+QI+ EI +KVA+IQN GLGE R+RDLNDEINKL+R
Sbjct: 296 RKGHERRPFLASECTELPKAEKWRRQIIGEISKKVAQIQNAGLGEFRIRDLNDEINKLLR 355
Query: 84 EKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVRELF 143
EK HWE RI ELGGP+Y + KM D +G V P R GY+YFGAAK LPGVRELF
Sbjct: 356 EKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----PGNR--GYKYFGAAKDLPGVRELF 409
Query: 144 DKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLDRIR 203
+K P R+TR ++ K ID YYGY D++DG++ LE E R E+W
Sbjct: 410 EKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPLEQEYEKKFRAGLVEKW------- 462
Query: 204 QEARKGVKSGEVAEVSAAVREILREEEED-----VVEEERMRGREMKERLEENEREFVVH 258
K+ A ++ +E EEEE+ V EEE ++ E+ +++F+ H
Sbjct: 463 -------KAEREARLARGEKEEEEEEEEEINIYAVTEEESDEEGSQEKGGEDGQQKFIAH 515
Query: 259 VPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
VP+P ++EIE+ ++ +KKMELL KY SE L + EAK +L
Sbjct: 516 VPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEAKRLL 556
>ref|XP_516736.1| PREDICTED: similar to KIAA1160 protein [Pan troglodytes]
Length = 805
Score = 239 bits (611), Expect = 6e-62
Identities = 134/299 (44%), Positives = 188/299 (62%), Gaps = 22/299 (7%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L RF + EE K KERRPFLA+EC +L +A+KWR+QI+ EI +KVA+
Sbjct: 1 MARNAEKAMTALARFRQAQLEEGKV-KERRPFLASECTELPKAEKWRRQIIGEISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE RI ELGGP+Y + KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKGHWEVRIKELGGPDYGKVGPKMLDHEGKEV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF+K P R+TR ++ K ID YYGY D++DG++ L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEKEPLPPPRKTRAELMKAIDFEYYGYLDEDDGVIVPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMR 240
E E + ++++ + K + ++ L +E+ +E+
Sbjct: 174 EQEYEKKLIHSQG------SKVKKNIKIFHKDSLSSPCPSSTLVSLPSSDEEGSQEKGG- 226
Query: 241 GREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKYVSEDLMDEQSEAKDML 299
++++++F+ HVP+P ++EIE+ ++ +KKMELL KY SE L + EA+ +L
Sbjct: 227 --------DDSQQKFIAHVPVPSQQEIEEALVRRKKMELLQKYASETLQAQSEEARRLL 277
>ref|XP_638102.1| hypothetical protein DDB0186544 [Dictyostelium discoideum]
gi|60466546|gb|EAL64598.1| hypothetical protein
DDB0186544 [Dictyostelium discoideum]
Length = 299
Score = 232 bits (592), Expect = 9e-60
Identities = 126/303 (41%), Positives = 183/303 (59%), Gaps = 27/303 (8%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARNEEKA+SMLNR++ +K E K+ +ERRP+L+ EC L +A+KWR+QI++EI R + E
Sbjct: 1 MARNEEKAKSMLNRYLQLKGTESKQ-EERRPYLSNECDSLVDAEKWRRQILKEITRGITE 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN L E+++RDLND INKL+REK HWERRI++LGGPN+ + K+ D DG P
Sbjct: 60 IQNSALDEYKIRDLNDHINKLVREKGHWERRILQLGGPNHRALAPKLFDADGK-----EP 114
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPEL---RKRRTRYDIYKRIDAAYYGYRDDEDGIL 177
G G YRY+G AK LPGV ELF+KP + +++TR D+YK ID+ YYGYRDDEDG L
Sbjct: 115 LGTGT-YRYYGEAKNLPGVAELFEKPEQTLHGTEKKTRQDLYKYIDSDYYGYRDDEDGKL 173
Query: 178 ERLEPSAEAAMRREAAEEWYRLDRIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEE 237
E +E E + E W + R + + + + S A+ + ++E ++++
Sbjct: 174 EEIEREYEKYAIEQNVENWKKQQRDKFQINTNRFNNNNIKNSIAINKNTENKDEINIDDD 233
Query: 238 RMRG-----------------REMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELL 280
++E E+ + +F +V LP +IE ++LEK+K EL
Sbjct: 234 EENDDNNNNNNNNNTENTENIENIQEEEEKEDIKFKSYVSLPSTSQIETILLEKRKEELR 293
Query: 281 NKY 283
+Y
Sbjct: 294 KRY 296
>gb|AAL89889.1| RE31520p [Drosophila melanogaster]
Length = 272
Score = 230 bits (586), Expect = 4e-59
Identities = 128/285 (44%), Positives = 179/285 (61%), Gaps = 20/285 (7%)
Query: 1 MARNEEKAQSMLNRFITMKAEEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAE 60
MARN EKA + L R+ K E + KERRP+LA+EC DL +K+R +I+R+I +KVA+
Sbjct: 1 MARNAEKAMTTLARWRAAKEVESGE-KERRPYLASECHDLPRCEKFRLEIIRDISKKVAQ 59
Query: 61 IQNEGLGEHRLRDLNDEINKLIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNP 120
IQN GLGE R+RDLNDEINKL+REK HWE +I LGGP+Y R+ KM D +G V P
Sbjct: 60 IQNAGLGEFRIRDLNDEINKLLREKRHWENQISSLGGPHYRRYGPKMFDAEGREV----P 115
Query: 121 SGRGPGYRYFGAAKKLPGVRELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERL 180
R GY+YFGAAK LPGVRELF++ P R++R ++ K IDA YYGYRDDEDG+L L
Sbjct: 116 GNR--GYKYFGAAKDLPGVRELFEQDPPPPPRKSRAELMKDIDAEYYGYRDDEDGVLIPL 173
Query: 181 EPSAEAAMRREAAEEWYRLDRIRQEAR--KGVKSGEVAEVSAAVREILREEEEDVVEEER 238
E E A ++A +EW +++ ++ R + E+ +S RE + + + E+
Sbjct: 174 EERIERAAIQKAVQEW--REKMARDGRIIDDEEEDEIYPLSGPAREAIEDAKARAAVEDP 231
Query: 239 MRGREMKERLEENEREFVVHVPLPDEKEIEKMVLEKKKMELLNKY 283
K F HVP+P ++++++ +L ++K ELL KY
Sbjct: 232 HGLLASK---------FTAHVPVPTQQDVQEALLRQRKRELLEKY 267
>gb|EAL27800.1| GA21951-PA [Drosophila pseudoobscura]
Length = 266
Score = 218 bits (554), Expect = 2e-55
Identities = 120/273 (43%), Positives = 163/273 (58%), Gaps = 33/273 (12%)
Query: 21 EEKKKPKERRPFLATECRDLSEADKWRQQIMREIGRKVAEIQNEGLGEHRLRDLNDEINK 80
E + KERRP+LA+EC DL +K+R +I+REI +KVA+IQN GLGE R+RDLNDEINK
Sbjct: 12 EVESGEKERRPYLASECGDLPRCEKFRLEIIREISKKVAQIQNAGLGEFRIRDLNDEINK 71
Query: 81 LIREKSHWERRIVELGGPNYTRHSAKMTDLDGNIVDVPNPSGRGPGYRYFGAAKKLPGVR 140
L+REK HWE +I LGGP+Y R+ KM D +G V P R GY+YFGAAK LPGVR
Sbjct: 72 LLREKRHWENQISALGGPHYRRYGPKMFDAEGREV----PGNR--GYKYFGAAKDLPGVR 125
Query: 141 ELFDKPPELRKRRTRYDIYKRIDAAYYGYRDDEDGILERLEPSAEAAMRREAAEEWYRLD 200
ELF++ P R++R ++ K +DA YYGYRDDEDG+L LE E ++A +EW
Sbjct: 126 ELFEQDPPPPPRKSRAELMKDVDAEYYGYRDDEDGVLIPLEERIERVALQKAVQEW---- 181
Query: 201 RIRQEARKGVKSGEVAEVSAAVREILREEEEDVVEEERMRGREMKERLEENER------- 253
K + G + + +EEED + RE E +
Sbjct: 182 -----REKMARDGRIVDDD--------DEEEDEIYPLSGPAREAAEDAKARAAVDDPHGL 228
Query: 254 ---EFVVHVPLPDEKEIEKMVLEKKKMELLNKY 283
+F HVP+P ++++++ +L K+K ELL KY
Sbjct: 229 LAPKFTAHVPVPTQQDVQEALLRKRKQELLEKY 261
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 521,268,076
Number of Sequences: 2540612
Number of extensions: 24377163
Number of successful extensions: 131104
Number of sequences better than 10.0: 1890
Number of HSP's better than 10.0 without gapping: 241
Number of HSP's successfully gapped in prelim test: 1749
Number of HSP's that attempted gapping in prelim test: 114115
Number of HSP's gapped (non-prelim): 8239
length of query: 303
length of database: 863,360,394
effective HSP length: 127
effective length of query: 176
effective length of database: 540,702,670
effective search space: 95163669920
effective search space used: 95163669920
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0282.6


