

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
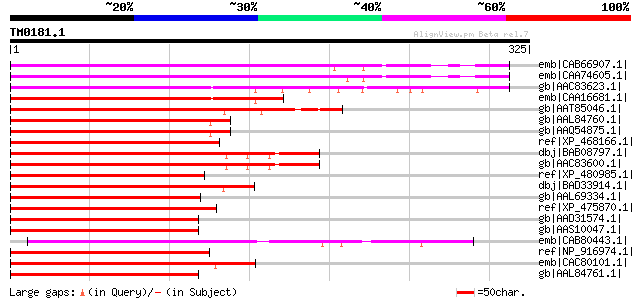
Query= TM0181.1
(325 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB66907.1| AtMYB84 [Arabidopsis thaliana] gi|15229192|ref|N... 280 4e-74
emb|CAA74605.1| R2R3-MYB transcription factor [Arabidopsis thali... 268 1e-70
gb|AAC83623.1| putative transcription factor [Arabidopsis thalia... 261 2e-68
emb|CAA16681.1| myb - related protein [Arabidopsis thaliana] gi|... 260 4e-68
gb|AAT85046.1| P-type R2R3 Myb proteind [Oryza sativa (japonica ... 256 6e-67
gb|AAL84760.1| typical P-type R2R3 Myb protein [Sorghum bicolor] 248 2e-64
gb|AAQ54875.1| P-type R2R3 Myb protein [Sorghum bicolor] 248 2e-64
ref|XP_468166.1| R2R3-MYB transcription factor-like [Oryza sativ... 246 9e-64
dbj|BAB08797.1| Myb-related transcription factor-like protein [A... 245 1e-63
gb|AAC83600.1| putative transcription factor [Arabidopsis thalia... 245 1e-63
ref|XP_480985.1| putative P-type R2R3 Myb protein [Oryza sativa ... 235 2e-60
dbj|BAD33914.1| R2R3-MYB transcription factor-like protein [Oryz... 232 1e-59
gb|AAL69334.1| blind [Lycopersicon esculentum] 229 6e-59
ref|XP_475870.1| putative myb transctiption factor [Oryza sativa... 223 6e-57
gb|AAD31574.1| light-regulated myb protein, putative [Arabidopsi... 221 2e-56
gb|AAS10047.1| MYB transcription factor [Arabidopsis thaliana] 221 2e-56
emb|CAB80443.1| myb transcription factor-like protein [Arabidops... 221 2e-56
ref|NP_916974.1| putative Myb-related transcription factor-l [Or... 219 7e-56
emb|CAC80101.1| R2R3-MYB transcription factor [Arabidopsis thali... 218 1e-55
gb|AAL84761.1| typical P-type R2R3 Myb protein [Sorghum bicolor] 203 5e-51
>emb|CAB66907.1| AtMYB84 [Arabidopsis thaliana] gi|15229192|ref|NP_190538.1| myb
family transcription factor [Arabidopsis thaliana]
gi|41619286|gb|AAS10067.1| MYB transcription factor
[Arabidopsis thaliana] gi|11273965|pir||T46035 AtMYB84 -
Arabidopsis thaliana
Length = 310
Score = 280 bits (716), Expect = 4e-74
Identities = 162/325 (49%), Positives = 197/325 (59%), Gaps = 33/325 (10%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK+YI++ GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDAKLKSYIENSGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEE+NIIC+LY++IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL+ K
Sbjct: 61 YLRPNIKHGGFSEEEENIICSLYLTIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLINK 120
Query: 121 QRKEQQAQARRVSTMKQEIKRE-TDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQD 179
QRKE Q M +KR+ Q + + + F WP + A Q
Sbjct: 121 QRKELQEACMEQQEMMVMMKRQHQQQQIQTSFMMRQDQTMFTWPLHHHNVQVPALFMNQT 180
Query: 180 YDFHNQTSLKSLLINKQGGVRFS-------YDHQQPYSNAITATANSQ---DPCDIYPAS 229
F +Q +K +LI + + HQ +NA + SQ DP + S
Sbjct: 181 NSFCDQEDVKPVLIKNMVKIEDQELEKTNPHHHQDSMTNAFDHLSFSQLLLDPNHNHLGS 240
Query: 230 TMNMINPVAMVSNANIGGTTCQQSNVFQGFENFTSDFSELVCVNPQTIDGFYGMESLEMS 289
+ + +AN SN Q F NF + +T++ F G
Sbjct: 241 GEGF--SMNSILSANTNSPLLNTSNDNQWFGNFQA----------ETVNLFSG------- 281
Query: 290 NVSSTNTPSAEST-SWGDMSSLVYS 313
+ST+T + +ST SW D+SSLVYS
Sbjct: 282 --ASTSTSADQSTISWEDISSLVYS 304
>emb|CAA74605.1| R2R3-MYB transcription factor [Arabidopsis thaliana]
Length = 309
Score = 268 bits (686), Expect = 1e-70
Identities = 159/324 (49%), Positives = 192/324 (59%), Gaps = 32/324 (9%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK+YI++ GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDAKLKSYIENSGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEE+NIIC+LY++IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL+ K
Sbjct: 61 YLRPNIKHGGFSEEEENIICSLYLTIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLINK 120
Query: 121 QRKEQQAQARRVSTMKQEIKRE-TDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQD 179
QRKE Q M +KR+ Q + + + F WP + A I+
Sbjct: 121 QRKELQEACMEQQEMMVMMKRQHQQQQIQTSFMMRQDQTMFTWPLHHHNVQVPALFRIKP 180
Query: 180 YDFHNQTSLKSLLINKQGGVRFSYDHQQPYS------NAITATANSQ---DPCDIYPAST 230
F + L + +Q S NA + SQ DP + S
Sbjct: 181 TRFATKKMLSQCSSRTWSRSKIKNWRKQTSSSSRFNDNAFDHLSFSQLLLDPNHNHLGSG 240
Query: 231 MNMINPVAMVSNANIGGTTCQQSNVFQGFENFTSDFSELVCVNPQTIDGFYGMESLEMSN 290
+ + +AN SN Q F NF + +T++ F G
Sbjct: 241 EGF--SMNSILSANTNSPLLNTSNDNQWFGNFQA----------ETVNLFSG-------- 280
Query: 291 VSSTNTPSAEST-SWGDMSSLVYS 313
+ST+T + +ST SW D+SSLVYS
Sbjct: 281 -ASTSTSADQSTISWEDISSLVYS 303
>gb|AAC83623.1| putative transcription factor [Arabidopsis thaliana]
gi|11273952|pir||T51673 myb-related transcription factor
MYB68 [imported] - Arabidopsis thaliana
Length = 376
Score = 261 bits (667), Expect = 2e-68
Identities = 164/366 (44%), Positives = 215/366 (57%), Gaps = 55/366 (15%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK YI++ GTGGNWIALPQKIGL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDAKLKDYIENSGTGGNWIALPQKIGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEEDNIICNLYV+IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL K
Sbjct: 61 YLRPNIKHGGFSEEEDNIICNLYVTIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLNK 120
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVASG-----VNTTHAAFYWPAEYSIP-----M 170
QRKE Q +AR M +++ Q ++G +N + WP +P +
Sbjct: 121 QRKEFQ-EARMKQEMVMMKRQQQGQGQGQSNGSTDLYLNNMFGSSPWPLLPQLPPPHHQI 179
Query: 171 PVANASIQDYDFHNQT-----------SLKSLLINKQGGVRFSYD--HQQPYSNAITATA 217
P+ +++ T +LK+++ ++ R + D HQ +N +
Sbjct: 180 PLGMMEPTSCNYYQTTPSCNLEQKPLITLKNMVKIEEEQERTNPDHHHQDSVTNPFDFSF 239
Query: 218 NS--QDPCDIYPASTMNMINPVAMVS---NANIGGTT-----CQQSNVFQ--GFENFTSD 265
+ DP + Y S A++S N+ + T+ QQ + Q G NF ++
Sbjct: 240 SQLLLDP-NYYLGSGGGGEGDFAIMSSSTNSPLPNTSSDQHPSQQQEILQWFGSSNFQTE 298
Query: 266 FSELVCVNPQTIDGFYGMESLEMSNV-----------------SSTNTPSAEST-SWGDM 307
+ +N + +E++E + V +T+T + +ST SW D+
Sbjct: 299 AINDMFINNNNNNNIVNLETIENTKVYGDASVAGAAVRAALAGGTTSTSADQSTISWEDI 358
Query: 308 SSLVYS 313
+SLV S
Sbjct: 359 TSLVNS 364
>emb|CAA16681.1| myb - related protein [Arabidopsis thaliana]
gi|10177337|dbj|BAB10686.1| transcription factor-like
protein [Arabidopsis thaliana]
gi|29029100|gb|AAO64929.1| At5g65790 [Arabidopsis
thaliana] gi|15239158|ref|NP_201380.1| myb family
transcription factor (MYB68) [Arabidopsis thaliana]
gi|41619504|gb|AAS10117.1| MYB transcription factor
[Arabidopsis thaliana] gi|7446168|pir||T05891
myb-related protein homolog F6H11.100 - Arabidopsis
thaliana
Length = 374
Score = 260 bits (664), Expect = 4e-68
Identities = 127/176 (72%), Positives = 141/176 (79%), Gaps = 6/176 (3%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEEDAKLK YI++ GTGGNWIALPQKIGL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDAKLKDYIENSGTGGNWIALPQKIGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEEDNIICNLYV+IGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL K
Sbjct: 61 YLRPNIKHGGFSEEEDNIICNLYVTIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLNK 120
Query: 121 QRKEQQAQARRVSTMKQEIKRETDQNLMVASG-----VNTTHAAFYWPAEYSIPMP 171
QRKE Q +AR M +++ Q ++G +N + WP +P P
Sbjct: 121 QRKEFQ-EARMKQEMVMMKRQQQGQGQGQSNGSTDLYLNNMFGSSPWPLLPQLPPP 175
>gb|AAT85046.1| P-type R2R3 Myb proteind [Oryza sativa (japonica cultivar-group)]
Length = 321
Score = 256 bits (654), Expect = 6e-67
Identities = 136/222 (61%), Positives = 157/222 (70%), Gaps = 18/222 (8%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA+VK+GPWSPEEDAKLK+YI+ +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKASVKKGPWSPEEDAKLKSYIEQNGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED II +LY+SIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL GK
Sbjct: 61 YLRPNIKHGGFSEEEDRIILSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLFGK 120
Query: 121 QRKEQQAQARRVS----------TMKQEIKRETDQNLMVASGVNTT---HAAFYWPAE-Y 166
Q ++ Q Q + ++ +KQE R ++ +A+G N T H A P
Sbjct: 121 QSRKDQRQQQHLARQAAAAASDLQIKQEASRGANEADGLAAGANYTWHHHHAMAVPVHPM 180
Query: 167 SIPMPVANASIQDYDFHNQTSLKSLLINKQGGVRFSYDHQQP 208
S PM V + D S++ LL K GG F+ P
Sbjct: 181 SAPMVVEGGRVGD---DVDESIRKLLF-KLGGNPFAASPAPP 218
>gb|AAL84760.1| typical P-type R2R3 Myb protein [Sorghum bicolor]
Length = 157
Score = 248 bits (633), Expect = 2e-64
Identities = 118/147 (80%), Positives = 126/147 (85%), Gaps = 9/147 (6%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA VK+GPWSPEEDAKLK+YI+ +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKATVKKGPWSPEEDAKLKSYIEQNGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED II +LY+SIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL GK
Sbjct: 61 YLRPNIKHGGFSEEEDRIILSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLFGK 120
Query: 121 QRKE---------QQAQARRVSTMKQE 138
Q ++ QQA A MKQE
Sbjct: 121 QSRKDQRQHQFMRQQAAAANDGMMKQE 147
>gb|AAQ54875.1| P-type R2R3 Myb protein [Sorghum bicolor]
Length = 157
Score = 248 bits (633), Expect = 2e-64
Identities = 118/147 (80%), Positives = 126/147 (85%), Gaps = 9/147 (6%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA VK+GPWSPEEDAKLK+YI+ +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKATVKKGPWSPEEDAKLKSYIEQNGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED II +LY+SIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKL GK
Sbjct: 61 YLRPNIKHGGFSEEEDRIILSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLFGK 120
Query: 121 QRKE---------QQAQARRVSTMKQE 138
Q ++ QQA A MKQE
Sbjct: 121 QSRKDQRQHQFMRQQAAAANDGMMKQE 147
>ref|XP_468166.1| R2R3-MYB transcription factor-like [Oryza sativa (japonica
cultivar-group)] gi|47497161|dbj|BAD19209.1| R2R3-MYB
transcription factor-like [Oryza sativa (japonica
cultivar-group)]
Length = 563
Score = 246 bits (627), Expect = 9e-64
Identities = 110/131 (83%), Positives = 125/131 (94%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA+VK+GPWSPEEDAKLKAYI+ +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 199 MGRAPCCDKASVKKGPWSPEEDAKLKAYIEENGTGGNWIALPQKIGLKRCGKSCRLRWLN 258
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHG F+EEE++IIC+LY+SIGSRWSIIAAQLPGRTDNDIKNYWNT+LKKKLLGK
Sbjct: 259 YLRPNIKHGDFTEEEEHIICSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTKLKKKLLGK 318
Query: 121 QRKEQQAQARR 131
+ ++A+A +
Sbjct: 319 RAPSRRARANQ 329
>dbj|BAB08797.1| Myb-related transcription factor-like protein [Arabidopsis
thaliana] gi|15242793|ref|NP_200570.1| myb family
transcription factor (MYB36) [Arabidopsis thaliana]
gi|41619474|gb|AAS10110.1| MYB transcription factor
[Arabidopsis thaliana]
Length = 333
Score = 245 bits (626), Expect = 1e-63
Identities = 126/213 (59%), Positives = 150/213 (70%), Gaps = 21/213 (9%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEED KLK YID +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDVKLKDYIDKYGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED II +LY+SIGSRWSIIAAQLPGRTDNDIKNYWNT+LKKKLLG+
Sbjct: 61 YLRPNIKHGGFSEEEDRIILSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTKLKKKLLGR 120
Query: 121 QRKEQQAQARRVST---MKQEIKRETDQNL-----------MVASGVNTTHAAFY----- 161
Q++ + + ST + ++ QNL M + + ++FY
Sbjct: 121 QKQMNRQDSITDSTENNLSNNNNNKSPQNLSNSALERLQLHMQLQNLQSPFSSFYNNPIL 180
Query: 162 WPAEYSIPMPVANASIQDYDFHNQTSLKSLLIN 194
WP + P+ + + Q+ +Q S L +N
Sbjct: 181 WPKLH--PLLQSTTTNQNPKLASQESFHPLGVN 211
>gb|AAC83600.1| putative transcription factor [Arabidopsis thaliana]
gi|11358686|pir||T51650 probable transcription factor
MYB36 [imported] - Arabidopsis thaliana
Length = 333
Score = 245 bits (626), Expect = 1e-63
Identities = 126/213 (59%), Positives = 150/213 (70%), Gaps = 21/213 (9%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVK+GPWSPEED KLK YID +GTGGNWIALPQKIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKKGPWSPEEDVKLKDYIDKYGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHGGFSEEED II +LY+SIGSRWSIIAAQLPGRTDNDIKNYWNT+LKKKLLG+
Sbjct: 61 YLRPNIKHGGFSEEEDRIILSLYISIGSRWSIIAAQLPGRTDNDIKNYWNTKLKKKLLGR 120
Query: 121 QRKEQQAQARRVST---MKQEIKRETDQNL-----------MVASGVNTTHAAFY----- 161
Q++ + + ST + ++ QNL M + + ++FY
Sbjct: 121 QKQMNRQDSITDSTENNLSNNNNNKSPQNLSNSALEKLQLHMQLQNLQSPFSSFYNNPIL 180
Query: 162 WPAEYSIPMPVANASIQDYDFHNQTSLKSLLIN 194
WP + P+ + + Q+ +Q S L +N
Sbjct: 181 WPKLH--PLLQSTTTNQNPKLASQESFHPLGVN 211
>ref|XP_480985.1| putative P-type R2R3 Myb protein [Oryza sativa (japonica
cultivar-group)] gi|40253739|dbj|BAD05679.1| putative
P-type R2R3 Myb protein [Oryza sativa (japonica
cultivar-group)]
Length = 316
Score = 235 bits (599), Expect = 2e-60
Identities = 103/122 (84%), Positives = 115/122 (93%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA VK+GPWSPEEDA LK YI+ HGTGGNWIALP KIGLKRCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKATVKKGPWSPEEDAMLKNYIEEHGTGGNWIALPHKIGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHG F+ EED+IIC+LY+SIGSRWSIIAAQLPGRTDND+KNYWNT+LKK+LLG+
Sbjct: 61 YLRPNIKHGDFTPEEDSIICSLYISIGSRWSIIAAQLPGRTDNDVKNYWNTKLKKRLLGR 120
Query: 121 QR 122
++
Sbjct: 121 RK 122
>dbj|BAD33914.1| R2R3-MYB transcription factor-like protein [Oryza sativa (japonica
cultivar-group)] gi|50726229|dbj|BAD33806.1| R2R3-MYB
transcription factor-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 319
Score = 232 bits (592), Expect = 1e-59
Identities = 106/155 (68%), Positives = 125/155 (80%), Gaps = 2/155 (1%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA VK+GPW+PEEDA LKAY+D HGTGGNWIALP KIGL RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKATVKKGPWAPEEDAALKAYVDAHGTGGNWIALPHKIGLNRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNI+HGGF+E+ED +IC+LY++IGSRW+ IAAQLPGRTDNDIKNYWN++LK++LLG
Sbjct: 61 YLRPNIRHGGFTEDEDRLICSLYIAIGSRWATIAAQLPGRTDNDIKNYWNSKLKRRLLGG 120
Query: 121 QRKEQQAQARRV--STMKQEIKRETDQNLMVASGV 153
R+ + A R V T +N M AS +
Sbjct: 121 GRRPRGAPPRLVLAGPGPAVTAAATSRNAMAASAI 155
>gb|AAL69334.1| blind [Lycopersicon esculentum]
Length = 315
Score = 229 bits (585), Expect = 6e-59
Identities = 102/119 (85%), Positives = 110/119 (91%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVKRGPWSPEEDAKLK +I GT GNWIALPQK GL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKRGPWSPEEDAKLKDFIHKFGTAGNWIALPQKAGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLG 119
YLRPNIKHG FS++ED +ICNLY +IGSRWSIIAAQLPGRTDNDIKNYWNT+LKKKL+G
Sbjct: 61 YLRPNIKHGDFSDDEDRVICNLYANIGSRWSIIAAQLPGRTDNDIKNYWNTKLKKKLMG 119
>ref|XP_475870.1| putative myb transctiption factor [Oryza sativa (japonica
cultivar-group)] gi|50878426|gb|AAT85200.1| putative
Myb-related transcription factor [Oryza sativa (japonica
cultivar-group)] gi|49328024|gb|AAT58725.1| putative myb
transctiption factor [Oryza sativa (japonica
cultivar-group)]
Length = 289
Score = 223 bits (568), Expect = 6e-57
Identities = 97/129 (75%), Positives = 114/129 (88%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA+VKRGPWSPEED +L++Y+ HG GGNWIALPQK GL RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKASVKRGPWSPEEDEQLRSYVQSHGIGGNWIALPQKAGLNRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP+IKHGG++E+ED+IIC+LY SIGSRWSIIA++LPGRTDND+KNYWNT+LKKK +G
Sbjct: 61 YLRPDIKHGGYTEQEDHIICSLYNSIGSRWSIIASKLPGRTDNDVKNYWNTKLKKKAMGA 120
Query: 121 QRKEQQAQA 129
+ A A
Sbjct: 121 VQPRAAASA 129
>gb|AAD31574.1| light-regulated myb protein, putative [Arabidopsis thaliana]
gi|15228049|ref|NP_181226.1| myb family transcription
factor (MYB38) [Arabidopsis thaliana]
gi|25294087|pir||H84785 probable MYB family
transcription factor [imported] - Arabidopsis thaliana
Length = 298
Score = 221 bits (564), Expect = 2e-56
Identities = 99/118 (83%), Positives = 108/118 (90%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVKRGPWSPEEDAKLK YI+ GTGGNWIALP K GL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKRGPWSPEEDAKLKDYIEKQGTGGNWIALPHKAGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRPNI+HG F+EEEDNII +L+ SIGSRWS+IAA L GRTDNDIKNYWNT+LKKKL+
Sbjct: 61 YLRPNIRHGDFTEEEDNIIYSLFASIGSRWSVIAAHLQGRTDNDIKNYWNTKLKKKLI 118
>gb|AAS10047.1| MYB transcription factor [Arabidopsis thaliana]
Length = 298
Score = 221 bits (564), Expect = 2e-56
Identities = 99/118 (83%), Positives = 108/118 (90%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKANVKRGPWSPEEDAKLK YI+ GTGGNWIALP K GL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKANVKRGPWSPEEDAKLKDYIEKQGTGGNWIALPHKAGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRPNI+HG F+EEEDNII +L+ SIGSRWS+IAA L GRTDNDIKNYWNT+LKKKL+
Sbjct: 61 YLRPNIRHGDFTEEEDNIIYSLFASIGSRWSVIAAHLQGRTDNDIKNYWNTKLKKKLI 118
>emb|CAB80443.1| myb transcription factor-like protein [Arabidopsis thaliana]
gi|4490723|emb|CAB38926.1| myb transcription factor-like
protein [Arabidopsis thaliana]
gi|15235710|ref|NP_195492.1| myb family transcription
factor (MYB87) [Arabidopsis thaliana]
gi|41619356|gb|AAS10084.1| MYB transcription factor
[Arabidopsis thaliana] gi|7438332|pir||T06025
transcription factor homolog T28I19.60 - Arabidopsis
thaliana
Length = 296
Score = 221 bits (563), Expect = 2e-56
Identities = 122/288 (42%), Positives = 168/288 (57%), Gaps = 21/288 (7%)
Query: 12 VKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLNYLRPNIKHGGF 71
VK+GPWS EEDA LK+YI+ HGTG NWI+LPQ+IG+KRCGKSCRLRWLNYLRPN+KHGGF
Sbjct: 3 VKKGPWSTEEDAVLKSYIEKHGTGNNWISLPQRIGIKRCGKSCRLRWLNYLRPNLKHGGF 62
Query: 72 SEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGKQRKEQQAQARR 131
++EED IIC+LY++IGSRWSIIA+QLPGRTDNDIKNYWNTRLKKKLL KQ K Q
Sbjct: 63 TDEEDYIICSLYITIGSRWSIIASQLPGRTDNDIKNYWNTRLKKKLLSKQGKAFHQQLNV 122
Query: 132 VSTMKQEIKRETDQNLMVASGVNTTHAAFYWPAEYSIPMPVANASIQDYDFHNQTSLKSL 191
+ + + NT + ++ V + S++ + Q+ +K+
Sbjct: 123 KFERGTTSSSSSQNQIQIFHDENT-------KSNQTLYNQVVDPSMRAFAMEEQSMIKNQ 175
Query: 192 LIN----KQGGVRFSYDHQ---QPYSNAITATANSQDPCDIYPASTMNMINPVAMVSNAN 244
++ + V F D+ Y + + + NS S++ N +S+
Sbjct: 176 ILEPFSWEPNKVLFDVDYDAAASSYHHHASPSLNSMS-----STSSIGTNNSSLQMSHYT 230
Query: 245 IGGTTCQQSNVF--QGFENFTSDFSELVCVNPQTIDGFYGMESLEMSN 290
+ Q ++F GFENF ++ + + N +GF G E L +N
Sbjct: 231 VNHNDHDQPDMFFMDGFENFQAELFDEIANNNTVENGFDGTEILINNN 278
>ref|NP_916974.1| putative Myb-related transcription factor-l [Oryza sativa (japonica
cultivar-group)] gi|19386683|dbj|BAB86065.1| myb-related
transcription factor LBM2-like [Oryza sativa (japonica
cultivar-group)] gi|20805028|dbj|BAB92703.1| myb-related
transcription factor LBM2-like [Oryza sativa (japonica
cultivar-group)]
Length = 318
Score = 219 bits (559), Expect = 7e-56
Identities = 95/125 (76%), Positives = 110/125 (88%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDKA+VKRGPWSPEED L++Y+ HGTGGNWIALPQK GL RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKASVKRGPWSPEEDELLRSYVRSHGTGGNWIALPQKAGLNRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRP+IKHGG++++ED IIC+LY SIGSRWSIIA++LPGRTDND+KNYWNT+LKKK +
Sbjct: 61 YLRPDIKHGGYTDQEDRIICSLYNSIGSRWSIIASKLPGRTDNDVKNYWNTKLKKKAMAM 120
Query: 121 QRKEQ 125
Q
Sbjct: 121 HHHHQ 125
>emb|CAC80101.1| R2R3-MYB transcription factor [Arabidopsis thaliana]
gi|9758402|dbj|BAB08873.1| myb-related transcription
factor-like [Arabidopsis thaliana]
gi|15237193|ref|NP_197691.1| myb family transcription
factor (MYB37) [Arabidopsis thaliana]
gi|51969156|dbj|BAD43270.1| myb like transcription
factor (MYB37) [Arabidopsis thaliana]
gi|41619420|gb|AAS10098.1| MYB transcription factor
[Arabidopsis thaliana]
Length = 329
Score = 218 bits (556), Expect = 1e-55
Identities = 100/156 (64%), Positives = 120/156 (76%), Gaps = 2/156 (1%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGRAPCCDK VKRGPWSPEED+KL+ YI+ +G GGNWI+ P K GL+RCGKSCRLRWLN
Sbjct: 1 MGRAPCCDKTKVKRGPWSPEEDSKLRDYIEKYGNGGNWISFPLKAGLRRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLLGK 120
YLRPNIKHG FSEEED II +L+ +IGSRWSIIAA LPGRTDNDIKNYWNT+L+KKLL
Sbjct: 61 YLRPNIKHGDFSEEEDRIIFSLFAAIGSRWSIIAAHLPGRTDNDIKNYWNTKLRKKLLSS 120
Query: 121 QRKEQQA--QARRVSTMKQEIKRETDQNLMVASGVN 154
+ + ++ + Q++KR T + +S N
Sbjct: 121 SSDSSSSAMASPYLNPISQDVKRPTSPTTIPSSSYN 156
>gb|AAL84761.1| typical P-type R2R3 Myb protein [Sorghum bicolor]
Length = 203
Score = 203 bits (517), Expect = 5e-51
Identities = 84/118 (71%), Positives = 105/118 (88%)
Query: 1 MGRAPCCDKANVKRGPWSPEEDAKLKAYIDHHGTGGNWIALPQKIGLKRCGKSCRLRWLN 60
MGR PCCD+A VKRGPWSPEED L++Y+ HG+GGNWIA+P+K GLKRCGKSCRLRWLN
Sbjct: 1 MGRTPCCDRAAVKRGPWSPEEDEALRSYVQRHGSGGNWIAMPKKAGLKRCGKSCRLRWLN 60
Query: 61 YLRPNIKHGGFSEEEDNIICNLYVSIGSRWSIIAAQLPGRTDNDIKNYWNTRLKKKLL 118
YLRP+I+HGGF+ EED +I +LY +GS+WS+IA+Q+ GRTDND+KN+WNT+LKK+LL
Sbjct: 61 YLRPDIRHGGFTAEEDAVILSLYTQLGSKWSLIASQMEGRTDNDVKNHWNTKLKKRLL 118
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.130 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 570,670,612
Number of Sequences: 2540612
Number of extensions: 23666852
Number of successful extensions: 62129
Number of sequences better than 10.0: 1386
Number of HSP's better than 10.0 without gapping: 1296
Number of HSP's successfully gapped in prelim test: 90
Number of HSP's that attempted gapping in prelim test: 59100
Number of HSP's gapped (non-prelim): 1854
length of query: 325
length of database: 863,360,394
effective HSP length: 128
effective length of query: 197
effective length of database: 538,162,058
effective search space: 106017925426
effective search space used: 106017925426
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0181.1


