

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0143c.1
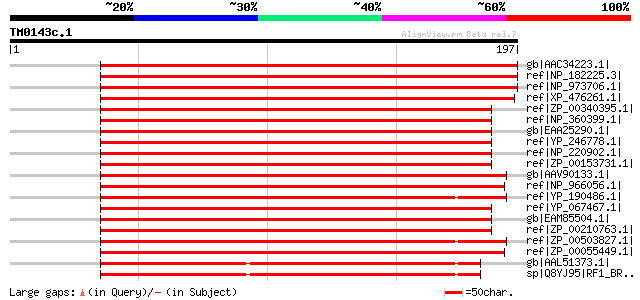
(197 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAC34223.1| putative peptide chain release factor [Arabidopsi... 262 5e-69
ref|NP_182225.3| peptide chain release factor, putative [Arabido... 262 5e-69
ref|NP_973706.1| peptide chain release factor, putative [Arabido... 262 5e-69
ref|XP_476261.1| putative peptide chain release factor [Oryza sa... 254 7e-67
ref|ZP_00340395.1| COG0216: Protein chain release factor A [Rick... 190 2e-47
ref|NP_360399.1| peptide chain release factor RF-1 [Rickettsia c... 189 4e-47
gb|EAA25290.1| peptide chain release factor RF-1 [Rickettsia sib... 189 4e-47
ref|YP_246778.1| Peptide chain release factor RF-1 [Rickettsia f... 189 4e-47
ref|NP_220902.1| PEPTIDE CHAIN RELEASE FACTOR 1 (prfA) [Ricketts... 188 8e-47
ref|ZP_00153731.1| COG0216: Protein chain release factor A [Rick... 187 1e-46
gb|AAV90133.1| protein chain release factor A [Zymomonas mobilis... 187 1e-46
ref|NP_966056.1| peptide chain release factor 1 [Wolbachia endos... 186 3e-46
ref|YP_190486.1| Bacterial Peptide Chain Release Factor 1 (RF-1)... 184 9e-46
ref|YP_067467.1| peptide chain release factor 1 (RF-1) [Ricketts... 183 2e-45
gb|EAM85504.1| Peptide chain release factor 1 [Ehrlichia chaffee... 183 3e-45
ref|ZP_00210763.1| COG0216: Protein chain release factor A [Ehrl... 183 3e-45
ref|ZP_00503827.1| Peptide chain release factor 1 [Clostridium t... 182 4e-45
ref|ZP_00055449.1| COG0216: Protein chain release factor A [Magn... 182 4e-45
gb|AAL51373.1| Bacterial Peptide Chain Release Factor 1 (RF-1) [... 182 4e-45
sp|Q8YJ95|RF1_BRUME Peptide chain release factor 1 (RF-1) 182 4e-45
>gb|AAC34223.1| putative peptide chain release factor [Arabidopsis thaliana]
gi|7443589|pir||T02185 probale translation releasing
factor RF-1 F14M4.15 - Arabidopsis thaliana
Length = 395
Score = 262 bits (669), Expect = 5e-69
Identities = 125/162 (77%), Positives = 153/162 (94%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ SV + + +VDVQL+NEDL++DTYRSGGSGGQHANTTNSAVRI H+PTG++V+IQ
Sbjct: 233 TSAISVAILPQADEVDVQLRNEDLRIDTYRSGGSGGQHANTTNSAVRIIHLPTGMMVSIQ 292
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQHMN+AKALK+LCA+LYE+ERLR+ S+RSKLRS+QIGSGDRS RIRTYNFPQGRVT
Sbjct: 293 DERSQHMNRAKALKVLCARLYEIERLRIQSSRSKLRSDQIGSGDRSGRIRTYNFPQGRVT 352
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTSS 197
DHRVGIT+H+I+D+MQGENLD+FIDALLLR+EMDA+ASF+S+
Sbjct: 353 DHRVGITHHAIEDMMQGENLDMFIDALLLRQEMDAIASFSST 394
>ref|NP_182225.3| peptide chain release factor, putative [Arabidopsis thaliana]
Length = 413
Score = 262 bits (669), Expect = 5e-69
Identities = 125/162 (77%), Positives = 153/162 (94%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ SV + + +VDVQL+NEDL++DTYRSGGSGGQHANTTNSAVRI H+PTG++V+IQ
Sbjct: 251 TSAISVAILPQADEVDVQLRNEDLRIDTYRSGGSGGQHANTTNSAVRIIHLPTGMMVSIQ 310
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQHMN+AKALK+LCA+LYE+ERLR+ S+RSKLRS+QIGSGDRS RIRTYNFPQGRVT
Sbjct: 311 DERSQHMNRAKALKVLCARLYEIERLRIQSSRSKLRSDQIGSGDRSGRIRTYNFPQGRVT 370
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTSS 197
DHRVGIT+H+I+D+MQGENLD+FIDALLLR+EMDA+ASF+S+
Sbjct: 371 DHRVGITHHAIEDMMQGENLDMFIDALLLRQEMDAIASFSST 412
>ref|NP_973706.1| peptide chain release factor, putative [Arabidopsis thaliana]
Length = 348
Score = 262 bits (669), Expect = 5e-69
Identities = 125/162 (77%), Positives = 153/162 (94%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ SV + + +VDVQL+NEDL++DTYRSGGSGGQHANTTNSAVRI H+PTG++V+IQ
Sbjct: 186 TSAISVAILPQADEVDVQLRNEDLRIDTYRSGGSGGQHANTTNSAVRIIHLPTGMMVSIQ 245
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQHMN+AKALK+LCA+LYE+ERLR+ S+RSKLRS+QIGSGDRS RIRTYNFPQGRVT
Sbjct: 246 DERSQHMNRAKALKVLCARLYEIERLRIQSSRSKLRSDQIGSGDRSGRIRTYNFPQGRVT 305
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTSS 197
DHRVGIT+H+I+D+MQGENLD+FIDALLLR+EMDA+ASF+S+
Sbjct: 306 DHRVGITHHAIEDMMQGENLDMFIDALLLRQEMDAIASFSST 347
>ref|XP_476261.1| putative peptide chain release factor [Oryza sativa (japonica
cultivar-group)] gi|48926665|gb|AAT47454.1| putative
peptide chain release factor [Oryza sativa (japonica
cultivar-group)]
Length = 350
Score = 254 bits (650), Expect = 7e-67
Identities = 125/161 (77%), Positives = 146/161 (90%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ SV + + +VDVQL+NEDL++DTYRSGGSGGQ NTT+SAVRITH+PTG +VAIQ
Sbjct: 190 TSAVSVAILPQADEVDVQLRNEDLRIDTYRSGGSGGQSVNTTDSAVRITHVPTGTVVAIQ 249
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DERSQH NKAKALK+L A+LYE+ER RLH +RSKLRSEQIGSGDRSERIRTYNFPQGRVT
Sbjct: 250 DERSQHQNKAKALKVLRARLYEIERHRLHMDRSKLRSEQIGSGDRSERIRTYNFPQGRVT 309
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVASFTS 196
DHRVGIT+HSI+DVM+GE+LD+FIDALLLR EMDA+ASF S
Sbjct: 310 DHRVGITHHSIEDVMEGESLDIFIDALLLRYEMDAIASFAS 350
>ref|ZP_00340395.1| COG0216: Protein chain release factor A [Rickettsia akari str.
Hartford]
Length = 355
Score = 190 bits (482), Expect = 2e-47
Identities = 90/152 (59%), Positives = 121/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER R+ R QIGSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRNKEQERADSRRGQIGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD FI+AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKNGQLDEFIEALIAEDE 348
>ref|NP_360399.1| peptide chain release factor RF-1 [Rickettsia conorii str. Malish
7] gi|15619858|gb|AAL03300.1| peptide chain release
factor RF-1 [Rickettsia conorii str. Malish 7]
gi|25299679|pir||B97795 peptide chain release factor
RF-1 [imported] - Rickettsia conorii (strain Malish 7)
gi|20139335|sp|Q92HK9|RF1_RICCN Peptide chain release
factor 1 (RF-1)
Length = 355
Score = 189 bits (480), Expect = 4e-47
Identities = 88/152 (57%), Positives = 122/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER + R+ R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRKKEQERADSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD F++AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKNGQLDEFVEALIADDE 348
>gb|EAA25290.1| peptide chain release factor RF-1 [Rickettsia sibirica 246]
gi|34580401|ref|ZP_00141881.1| peptide chain release
factor RF-1 [Rickettsia sibirica 246]
Length = 355
Score = 189 bits (480), Expect = 4e-47
Identities = 88/152 (57%), Positives = 122/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER + R+ R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRKKEQERADSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD F++AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKNGQLDEFVEALIADDE 348
>ref|YP_246778.1| Peptide chain release factor RF-1 [Rickettsia felis URRWXCal2]
gi|67004687|gb|AAY61613.1| Peptide chain release factor
RF-1 [Rickettsia felis URRWXCal2]
Length = 355
Score = 189 bits (480), Expect = 4e-47
Identities = 88/152 (57%), Positives = 122/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER + R+ R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRKKEQERADSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD F++AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKNGQLDEFVEALIADDE 348
>ref|NP_220902.1| PEPTIDE CHAIN RELEASE FACTOR 1 (prfA) [Rickettsia prowazekii str.
Madrid E] gi|3861078|emb|CAA14978.1| PEPTIDE CHAIN
RELEASE FACTOR 1 (prfA) [Rickettsia prowazekii]
gi|7443569|pir||H71656 translation releasing factor RF-1
prfA RP529 [similarity] - Rickettsia prowazekii
gi|6225942|sp|Q9ZD21|RF1_RICPR Peptide chain release
factor 1 (RF-1)
Length = 355
Score = 188 bits (477), Expect = 8e-47
Identities = 89/152 (58%), Positives = 121/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +D++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEGIDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A+LYE +R + RS R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARLYEEKRRQKEQERSDSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD FI+AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKHGQLDEFIEALIANDE 348
>ref|ZP_00153731.1| COG0216: Protein chain release factor A [Rickettsia rickettsii]
Length = 355
Score = 187 bits (476), Expect = 1e-46
Identities = 88/152 (57%), Positives = 122/152 (79%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD++++++DL++DTYR+ G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEEVDIKIEDKDLRIDTYRASGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A++YE ER + R+ R Q+GSGDRSERIRTYNFPQGRV+
Sbjct: 257 DEKSQHKNKAKALKILRARIYEEERRKKEQARADSRRGQVGSGDRSERIRTYNFPQGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD F++AL+ +E
Sbjct: 317 DHRIHLTLYKIDEVVKNGQLDEFVEALIADDE 348
>gb|AAV90133.1| protein chain release factor A [Zymomonas mobilis subsp. mobilis
ZM4] gi|56552405|ref|YP_163244.1| protein chain release
factor A [Zymomonas mobilis subsp. mobilis ZM4]
Length = 358
Score = 187 bits (475), Expect = 1e-46
Identities = 89/158 (56%), Positives = 118/158 (74%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VDV + DL++D +RS G GGQ NTT+SAVRITHIP+GI+V+ Q
Sbjct: 198 TSAATVAVLPEAEEVDVDIDERDLRIDIFRSSGPGGQSVNTTDSAVRITHIPSGIVVSQQ 257
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKA+K+L A+LYE ER RLHS R+ R +GSGDRSERIRTYNFPQGRVT
Sbjct: 258 DEKSQHKNKAKAMKVLRARLYERERERLHSERAGQRKSMVGSGDRSERIRTYNFPQGRVT 317
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T H + +++ G LD I AL+ +E + +AS
Sbjct: 318 DHRINLTLHRLPEILAGPGLDEVISALIAEDEAERLAS 355
>ref|NP_966056.1| peptide chain release factor 1 [Wolbachia endosymbiont of
Drosophila melanogaster] gi|42409878|gb|AAS13990.1|
peptide chain release factor 1 [Wolbachia endosymbiont
of Drosophila melanogaster]
gi|58698355|ref|ZP_00373269.1| peptide chain release
factor 1 [Wolbachia endosymbiont of Drosophila
ananassae] gi|58535109|gb|EAL59194.1| peptide chain
release factor 1 [Wolbachia endosymbiont of Drosophila
ananassae]
Length = 359
Score = 186 bits (472), Expect = 3e-46
Identities = 86/157 (54%), Positives = 120/157 (75%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E+ +VD +++ +DL++D YRS G GGQ NTT+SAVR+TH+PTGI+V Q
Sbjct: 200 TSAATVAILPEVEEVDFKIEEKDLRIDVYRSSGPGGQSVNTTDSAVRVTHLPTGIVVIQQ 259
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALK+L A+LYE+ER + RS +R QIGSGDRSERIRTYNFPQ R+T
Sbjct: 260 DEKSQHKNKAKALKVLRARLYEIERQKKEMERSTMRKSQIGSGDRSERIRTYNFPQSRIT 319
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVA 192
DHR+ +T H ++ +++ LD FI+AL+ R E + +A
Sbjct: 320 DHRINLTSHRLEQIIKEGELDEFIEALISRNEAERLA 356
>ref|YP_190486.1| Bacterial Peptide Chain Release Factor 1 (RF-1) [Gluconobacter
oxydans 621H] gi|58000936|gb|AAW59830.1| Bacterial
Peptide Chain Release Factor 1 (RF-1) [Gluconobacter
oxydans 621H]
Length = 353
Score = 184 bits (468), Expect = 9e-46
Identities = 88/158 (55%), Positives = 122/158 (76%), Gaps = 1/158 (0%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VDV + ++DL++D YR+ G+GGQH N T SAVR+TH+P+GI+VA+Q
Sbjct: 194 TSTVTVAVLPEAEEVDVTVNDDDLRIDVYRASGAGGQHVNKTESAVRVTHMPSGIVVAMQ 253
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
+E+SQH NKAKA+KIL A+LYE ER +LH+ R+ R Q+G+GDRSERIRTYNFPQGRVT
Sbjct: 254 EEKSQHKNKAKAMKILRARLYERERAQLHATRAADRKSQVGTGDRSERIRTYNFPQGRVT 313
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+ +T + ID +M GE D IDAL E+ + +A+
Sbjct: 314 DHRINLTLYKIDRIMGGE-FDEIIDALTREEQTELLAA 350
>ref|YP_067467.1| peptide chain release factor 1 (RF-1) [Rickettsia typhi str.
Wilmington] gi|51460022|gb|AAU03985.1| peptide chain
release factor 1 (RF-1) [Rickettsia typhi str.
Wilmington] gi|61214605|sp|Q68WK7|RF1_RICTY Peptide
chain release factor 1 (RF-1)
Length = 355
Score = 183 bits (465), Expect = 2e-45
Identities = 89/152 (58%), Positives = 119/152 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E VD++++++DL++DTYRS G+GGQH NTT+SAVRITHIPTGI VA+Q
Sbjct: 197 TSAATVAVLPEAEGVDIKIEDKDLRIDTYRSSGAGGQHVNTTDSAVRITHIPTGITVALQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NKAKALKIL A+LYE +R + RS R Q+GSGDRSERIRTYNF GRV+
Sbjct: 257 DEKSQHKNKAKALKILRARLYEEKRRQKEQERSDSRRWQVGSGDRSERIRTYNFLHGRVS 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ID+V++ LD FI+AL+ +E
Sbjct: 317 DHRINLTLYKIDEVVKHGQLDEFIEALIANDE 348
>gb|EAM85504.1| Peptide chain release factor 1 [Ehrlichia chaffeensis str. Sapulpa]
Length = 311
Score = 183 bits (464), Expect = 3e-45
Identities = 85/152 (55%), Positives = 119/152 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + EI +VD+++ +DL++D YRS G GGQ NTT+SAVRITHIP+GI+V Q
Sbjct: 151 TSAATVAVLPEIEEVDLKIDEKDLRIDVYRSSGPGGQSVNTTDSAVRITHIPSGIVVIQQ 210
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NK+KALK+L A+LY +E+ + + S++R QIGSGDRSERIRTYNFPQ R+T
Sbjct: 211 DEKSQHKNKSKALKVLRARLYNLEKQKRDAEISQMRKSQIGSGDRSERIRTYNFPQSRIT 270
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + +DD+M+ NLD FI+AL+ +E
Sbjct: 271 DHRINLTLYRLDDIMKEGNLDEFIEALIAEDE 302
>ref|ZP_00210763.1| COG0216: Protein chain release factor A [Ehrlichia canis str. Jake]
Length = 359
Score = 183 bits (464), Expect = 3e-45
Identities = 86/152 (56%), Positives = 118/152 (77%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + EI +VD+++ +DL++D YRS G GGQ NTT+SAVRITHIP+GI+V Q
Sbjct: 199 TSAATVAVLPEIEEVDLKIDEKDLRIDVYRSSGPGGQSVNTTDSAVRITHIPSGIVVIQQ 258
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
DE+SQH NK KALK+L A+LY +E+ + S S++R QIGSGDRSERIRTYNFPQ R+T
Sbjct: 259 DEKSQHKNKNKALKVLRARLYNLEKQKRDSEISQMRKSQIGSGDRSERIRTYNFPQSRIT 318
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREE 187
DHR+ +T + ++D+M+ NLD FIDAL+ +E
Sbjct: 319 DHRINLTLYRLEDIMKEGNLDEFIDALIAEDE 350
>ref|ZP_00503827.1| Peptide chain release factor 1 [Clostridium thermocellum ATCC
27405] gi|67851543|gb|EAM47107.1| Peptide chain release
factor 1 [Clostridium thermocellum ATCC 27405]
Length = 360
Score = 182 bits (463), Expect = 4e-45
Identities = 83/158 (52%), Positives = 122/158 (76%), Gaps = 1/158 (0%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E ++DV++ DL++DTYR+ G+GGQH N T+SA+RITH+PTG++V Q
Sbjct: 197 TSTVTVAVLPEAEEIDVEINPNDLRIDTYRASGAGGQHINKTDSAIRITHLPTGLVVCCQ 256
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
D+RSQH NK KA+K+L +KLYEM R + H+ ++ R Q+G+GDRSERIRTYNFPQGRVT
Sbjct: 257 DQRSQHKNKEKAMKVLRSKLYEMAREQQHNEIAQERKSQVGTGDRSERIRTYNFPQGRVT 316
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVAS 193
DHR+G+T + IDD++ G+ +D IDAL+ ++ +A+
Sbjct: 317 DHRIGLTLYKIDDILDGD-IDEIIDALITTDQASKLAN 353
>ref|ZP_00055449.1| COG0216: Protein chain release factor A [Magnetospirillum
magnetotacticum MS-1]
Length = 354
Score = 182 bits (463), Expect = 4e-45
Identities = 89/157 (56%), Positives = 116/157 (73%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +VD+QL + DL+ D YRS GSGGQ NTT+SAVR+THIPTG+ VA Q
Sbjct: 195 TSAATVAIMPEAEEVDIQLNDSDLRFDVYRSQGSGGQSVNTTDSAVRVTHIPTGLAVACQ 254
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
E+SQH NKA ALK+L A+LYE ER + R+ R Q+GSGDRSERIRTYNFPQGRVT
Sbjct: 255 QEKSQHKNKATALKLLRARLYERERSAKDAERAAARKSQVGSGDRSERIRTYNFPQGRVT 314
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALLLREEMDAVA 192
DHR+ +T + ID VM G+ LD I+AL+ ++ + +A
Sbjct: 315 DHRINLTLYKIDAVMSGDALDELIEALVAADQAERLA 351
>gb|AAL51373.1| Bacterial Peptide Chain Release Factor 1 (RF-1) [Brucella
melitensis 16M] gi|17986475|ref|NP_539109.1| Bacterial
Peptide Chain Release Factor 1 (RF-1) [Brucella
melitensis 16M] gi|25299697|pir||AB3276 bacterial
peptide chain release factor 1 (RF-1) [imported] -
Brucella melitensis (strain 16M)
Length = 377
Score = 182 bits (463), Expect = 4e-45
Identities = 86/148 (58%), Positives = 120/148 (80%), Gaps = 2/148 (1%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +D++++NED+++DT R+ G+GGQH NTT+SAVRITHIPTGI+V +Q
Sbjct: 217 TSAATVAVLPEAEDIDIEIRNEDIRIDTMRASGAGGQHVNTTDSAVRITHIPTGIMV-VQ 275
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
E+SQH N+A+A++IL A+LY+MER + S RS+ R Q+GSGDRSERIRTYNFPQGRVT
Sbjct: 276 AEKSQHQNRARAMQILRARLYDMERQKAESERSQARRSQVGSGDRSERIRTYNFPQGRVT 335
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALL 183
DHR+ +T + +D VM+GE LD +DAL+
Sbjct: 336 DHRINLTLYKLDRVMEGE-LDELVDALI 362
>sp|Q8YJ95|RF1_BRUME Peptide chain release factor 1 (RF-1)
Length = 359
Score = 182 bits (463), Expect = 4e-45
Identities = 86/148 (58%), Positives = 120/148 (80%), Gaps = 2/148 (1%)
Query: 36 SSSGSVGLTNEIMKVDVQLKNEDLKVDTYRSGGSGGQHANTTNSAVRITHIPTGIIVAIQ 95
+S+ +V + E +D++++NED+++DT R+ G+GGQH NTT+SAVRITHIPTGI+V +Q
Sbjct: 199 TSAATVAVLPEAEDIDIEIRNEDIRIDTMRASGAGGQHVNTTDSAVRITHIPTGIMV-VQ 257
Query: 96 DERSQHMNKAKALKILCAKLYEMERLRLHSNRSKLRSEQIGSGDRSERIRTYNFPQGRVT 155
E+SQH N+A+A++IL A+LY+MER + S RS+ R Q+GSGDRSERIRTYNFPQGRVT
Sbjct: 258 AEKSQHQNRARAMQILRARLYDMERQKAESERSQARRSQVGSGDRSERIRTYNFPQGRVT 317
Query: 156 DHRVGITYHSIDDVMQGENLDVFIDALL 183
DHR+ +T + +D VM+GE LD +DAL+
Sbjct: 318 DHRINLTLYKLDRVMEGE-LDELVDALI 344
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.128 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 282,125,676
Number of Sequences: 2540612
Number of extensions: 10591077
Number of successful extensions: 33533
Number of sequences better than 10.0: 797
Number of HSP's better than 10.0 without gapping: 764
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 29031
Number of HSP's gapped (non-prelim): 2028
length of query: 197
length of database: 863,360,394
effective HSP length: 121
effective length of query: 76
effective length of database: 555,946,342
effective search space: 42251921992
effective search space used: 42251921992
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0143c.1


