

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
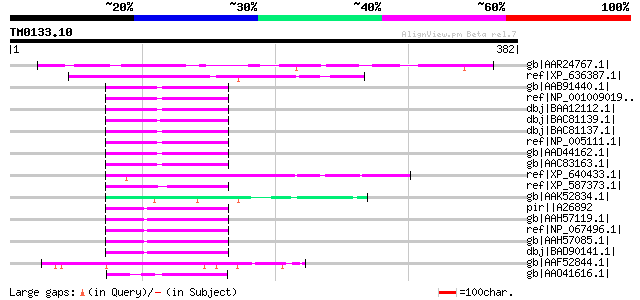
Query= TM0133.10
(382 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR24767.1| At5g21280 [Arabidopsis thaliana] gi|38454174|gb|A... 136 9e-31
ref|XP_636387.1| hypothetical protein DDB0188286 [Dictyostelium ... 61 6e-08
gb|AAB91440.1| CAGH45 [Homo sapiens] 59 3e-07
ref|NP_001009019.1| trinucleotide repeat containing 11 [Pan trog... 59 3e-07
dbj|BAA12112.1| Product has a CAG repeat region similar to mouse... 59 3e-07
dbj|BAC81139.1| TNRC11 [Pongo pygmaeus] 59 3e-07
dbj|BAC81137.1| TNRC11 [Homo sapiens] 59 3e-07
ref|NP_005111.1| mediator of RNA polymerase II transcription, su... 59 3e-07
gb|AAD44162.1| OPA-containing protein [Homo sapiens] 59 3e-07
gb|AAC83163.1| OPA-containing protein [Homo sapiens] 59 3e-07
ref|XP_640433.1| hypothetical protein DDB0205116 [Dictyostelium ... 57 1e-06
ref|XP_587373.1| PREDICTED: similar to OPA-containing protein, p... 55 3e-06
gb|AAK52834.1| cytoplasmic polyadenylation element-binding prote... 55 3e-06
pir||A26892 Mopa box protein - mouse (fragment) gi|387503|gb|AAA... 55 4e-06
gb|AAH57119.1| Tnrc11 protein [Mus musculus] 55 4e-06
ref|NP_067496.1| trinucleotide repeat containing 11 (THR-associa... 55 4e-06
gb|AAH57085.1| Tnrc11 protein [Mus musculus] 55 4e-06
dbj|BAD90141.1| mKIAA0192 protein [Mus musculus] 55 4e-06
gb|AAF52844.1| CG4722-PA [Drosophila melanogaster] gi|17136672|r... 55 4e-06
gb|AAO41616.1| CG12071-PB, isoform B [Drosophila melanogaster] g... 54 8e-06
>gb|AAR24767.1| At5g21280 [Arabidopsis thaliana] gi|38454174|gb|AAR20781.1|
At5g21280 [Arabidopsis thaliana]
gi|13374857|emb|CAC34491.1| putative protein
[Arabidopsis thaliana] gi|29294058|gb|AAO73895.1|
proline-rich protein family [Arabidopsis thaliana]
gi|22326944|ref|NP_680186.1| hydroxyproline-rich
glycoprotein family protein [Arabidopsis thaliana]
Length = 302
Score = 136 bits (343), Expect = 9e-31
Identities = 122/351 (34%), Positives = 156/351 (43%), Gaps = 99/351 (28%)
Query: 22 RNLEEEPPFSGAYMRSLVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQP 81
R+ ++EP S Y+RSLVKQ ++ ++ T G G Q+Q
Sbjct: 3 RSNKQEPRVSSTYIRSLVKQQ------LAYSTTMTTTTTTTTNDGSGGGKTQTQT----- 51
Query: 82 QQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQ 141
HKKQVRRRLHTSRPYQERLLNMAEARREIVTALK HRASM++A+ Q
Sbjct: 52 --HKKQVRRRLHTSRPYQERLLNMAEARREIVTALKQHRASMRQATRIPPPQ-------- 101
Query: 142 RAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGDFSSYPPAFVPNSYPVPVA 201
P PQ + FS PP
Sbjct: 102 -PPPPPQ-------------------------------PLNLFSPPPP------------ 117
Query: 202 SPIAPPPLMAENP--NFILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSNSFSSATSSSQ 259
P P P NP NF+LPNQPLGLNLNF DFN+ I S+T+ SS+S S+++SSS
Sbjct: 118 -PPPPDPFSWTNPSLNFLLPNQPLGLNLNFQDFNDF---IQTSSTTSSSSSSSTSSSSSS 173
Query: 260 EVPSVELSQGDGISSLVNSTESNAASQVNAGLHAAMDEEAIAEIRSLGEQYQMEWNDTMN 319
P+ + + S+ S + A +A Q N N
Sbjct: 174 IFPT---------NPHIYSSPSPPPTFTTATSDSA-------------PQLPSSSNGENN 211
Query: 320 LVKSACWFK-FLRNME-HGAPEAN----TEGDAYHNFDQPLEFPAWLNANE 364
+V SA W + L+ +E PE E D + F +EFP+WLN E
Sbjct: 212 VVTSAWWSELMLKTVEPEIKPETEEVIVVEDDVFPKFSDVMEFPSWLNQTE 262
>ref|XP_636387.1| hypothetical protein DDB0188286 [Dictyostelium discoideum]
gi|60464759|gb|EAL62883.1| hypothetical protein
DDB0188286 [Dictyostelium discoideum]
Length = 977
Score = 60.8 bits (146), Expect = 6e-08
Identities = 60/232 (25%), Positives = 102/232 (43%), Gaps = 21/232 (9%)
Query: 45 NDQGCVVNSDASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLN 104
N+ G +N++ +++ + + + + Q QQ QQ + Q+ L P + L +
Sbjct: 613 NNNGNNINNNNNNNNNSSNQIIQQQQQVQQQQMQQQQMMMQNQIIPTLPMPSPQSQVLQS 672
Query: 105 MAEARREIVTALKFHRASMKEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSR 164
+ ++++ ++ ++ +QQQQQQ QQQQQQ+ Q Q Q + + +
Sbjct: 673 QFQPQQQMQQQMQMQYQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ----QQQQQQQQQ 728
Query: 165 RNPRIYP--------SCNTNFSSYMGDFSSYPP-AFVPNSYPVPVASPIAPPPLMAENPN 215
+ +IY + + N SS GDF P PV V+ P P ++ +P+
Sbjct: 729 QQQQIYQQNLNSNSGNSSPNISSINGDFVRSPNYNNKKRPPPVNVSRPEFPVTPLSGSPS 788
Query: 216 FILPNQPLGLNLNFHDFNNLDATIHLSNTSLSSNSFSSATSSSQEVPSVELS 267
P Q NLN + NN N S +S+S+S S VPS LS
Sbjct: 789 H-SPAQSPHYNLNNGNNNN-------GNGSSNSSSYSGNQSPLGLVPSPSLS 832
Score = 33.9 bits (76), Expect = 8.5
Identities = 25/93 (26%), Positives = 35/93 (36%), Gaps = 25/93 (26%)
Query: 54 DASSHGQNLTKHGKIGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIV 113
D S H Q H QQ QQ Q H +Q +++ H + Q+
Sbjct: 60 DNSDHEQQQQHH-------HHQQQQQQQQHHHQQQQQQQHHQQQQQQ------------- 99
Query: 114 TALKFHRASMKEASEQQQQQQLQQQQQQRAPDS 146
H ++ QQQQQQ QQ + AP +
Sbjct: 100 -----HHQQQQQHHHQQQQQQQQQLDKSYAPSN 127
>gb|AAB91440.1| CAGH45 [Homo sapiens]
Length = 652
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 533 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 589
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 590 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 622
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 567 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 626
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 627 AALVRQLQQQLSNTQPQ----PSTNIFGRY 652
>ref|NP_001009019.1| trinucleotide repeat containing 11 [Pan troglodytes]
gi|33354109|dbj|BAC81138.1| TNRC11 [Pan troglodytes]
Length = 2027
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1908 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 1964
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1965 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1997
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1942 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2001
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2002 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2027
>dbj|BAA12112.1| Product has a CAG repeat region similar to mouse mopa box protein. It
also retains the DNA topoisomerase II motif and a
transmembrane domain at N-terminal region.~expressed
ubiquitously [Homo sapiens]
Length = 2124
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 2005 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 2061
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 2062 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2094
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 2039 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2098
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2099 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2124
>dbj|BAC81139.1| TNRC11 [Pongo pygmaeus]
Length = 2028
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1908 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ--QQQQQQQQQQQQQQQQQQQ 1965
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1966 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1998
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1943 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2002
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2003 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2028
>dbj|BAC81137.1| TNRC11 [Homo sapiens]
Length = 2027
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1908 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 1964
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1965 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1997
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1942 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2001
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2002 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2027
>ref|NP_005111.1| mediator of RNA polymerase II transcription, subunit 12 homolog [Homo
sapiens] gi|4530439|gb|AAD22033.1| thyroid hormone
receptor-associated protein complex component TRAP230
[Homo sapiens] gi|28558814|sp|Q93074|MED12_HUMAN Mediator
of RNA polymerase II transcription subunit 12 (Thyroid
hormone receptor-associated protein complex 230 kDa
component) (Trap230) (Activator-recruited cofactor 240
KDa component) (ARC240) (CAG repeat protein 45)
(OPA-containing protein) (Trinucleotide repeat-containing
gene 11 protein)
Length = 2212
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 2093 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 2149
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 2150 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2182
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 2127 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 2186
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 2187 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2212
>gb|AAD44162.1| OPA-containing protein [Homo sapiens]
Length = 2023
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1904 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 1960
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1961 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1993
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1938 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 1997
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 1998 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2023
>gb|AAC83163.1| OPA-containing protein [Homo sapiens]
Length = 2023
Score = 58.5 bits (140), Expect = 3e-07
Identities = 29/93 (31%), Positives = 52/93 (55%), Gaps = 3/93 (3%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q+++L + +++ + + ++ +QQQQ
Sbjct: 1904 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQILRQQQQQQQ---QQQQQQQQQQQQQQQQQQ 1960
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 1961 QHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1993
Score = 39.3 bits (90), Expect = 0.20
Identities = 27/90 (30%), Positives = 46/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ A + + + +F R +++ +QQQ
Sbjct: 1938 QQQQQQQQQQQQQQQQQQQQQQQQHQQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 1997
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 1998 AALVRQLQQQLSNTQPQ----PSTNIFGRY 2023
>ref|XP_640433.1| hypothetical protein DDB0205116 [Dictyostelium discoideum]
gi|60468439|gb|EAL66444.1| hypothetical protein
DDB0205116 [Dictyostelium discoideum]
Length = 611
Score = 56.6 bits (135), Expect = 1e-06
Identities = 56/239 (23%), Positives = 104/239 (43%), Gaps = 14/239 (5%)
Query: 73 QSQQSTQQPQQHKK---------QVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASM 123
QS QS QQPQQ Q +++ + Q++ + + +++ + +
Sbjct: 180 QSSQSPQQPQQQYNSYNPYQSYVQQQQQQQQQQQQQQQQQQIPQYQQQTSPPPPNYYEHI 239
Query: 124 KEASEQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNFSSYMGD 183
++ +QQQQQQ QQQQQQ+ Q Q Q + + + + S + S +
Sbjct: 240 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHQQQQRQQSPSLYNSYHSTQ 299
Query: 184 FSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFNNLDATIHLSN 243
PP+++ + SP P N N LP+QP N N +F N T +SN
Sbjct: 300 QPLPPPSYLQHMQQQQQQSPSPSPQYQQTNSN-SLPSQPPQPNYNLQNFIN---TPIISN 355
Query: 244 TSLSSNSFSSATSSSQEVPSVELSQGDGISSLVNSTESNAASQVNAGLHAAMDEEAIAE 302
S ++N+ + +S+ + + ++Q + + + S + S +N + A D ++E
Sbjct: 356 NSNNNNTNINGFTSNAGIGGI-INQMNKKTLENDKLLSESFSDLNILMEKAKDMVTLSE 413
>ref|XP_587373.1| PREDICTED: similar to OPA-containing protein, partial [Bos taurus]
Length = 825
Score = 55.5 bits (132), Expect = 3e-06
Identities = 29/93 (31%), Positives = 51/93 (54%), Gaps = 6/93 (6%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + R Q+++L + +++ + ++ +QQQQ
Sbjct: 709 QQQQQQQQQQQQQQQQQQQQYHIRQQQQQILRQQQQQQQQ------QQQQQQQQQQQQQQ 762
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
Q QQQQQQ AP PQ P F + G ++++
Sbjct: 763 QAHQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 795
Score = 39.3 bits (90), Expect = 0.20
Identities = 26/90 (28%), Positives = 47/90 (51%), Gaps = 5/90 (5%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
+ QQ QQ QQ ++Q +++ + +Q++ A + + + +F R +++ +QQQ
Sbjct: 740 RQQQQQQQQQQQQQQQQQQQQQQQAHQQQQQQAAPPQPQPQSQPQFQRQGLQQTQQQQQT 799
Query: 133 QQL-QQQQQQRAPDSPQLSQNPSFDQDGRY 161
L +Q QQQ + PQ PS + GRY
Sbjct: 800 AALVRQLQQQLSNTQPQ----PSTNIFGRY 825
Score = 35.0 bits (79), Expect = 3.8
Identities = 22/94 (23%), Positives = 44/94 (46%), Gaps = 16/94 (17%)
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
+Q QQ QQ ++Q +++ + Q++ ++ + +++I+ +QQQQ
Sbjct: 701 EQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQIL----------------RQQQQ 744
Query: 135 LQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPR 168
QQQQQQ+ Q Q + Q + + P+
Sbjct: 745 QQQQQQQQQQQQQQQQQQQAHQQQQQQAAPPQPQ 778
>gb|AAK52834.1| cytoplasmic polyadenylation element-binding protein isoform 1
[Aplysia californica]
Length = 687
Score = 55.5 bits (132), Expect = 3e-06
Identities = 54/216 (25%), Positives = 85/216 (39%), Gaps = 39/216 (18%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAE--ARREIVTALKFHRASMKEASEQQ 130
Q QQ TQQ QQ ++Q +++ H + Q++LL + A+++ + + K+ +QQ
Sbjct: 58 QQQQQTQQQQQQQQQQQQQQHLQQVQQQQLLKQQQQQAQQQQIQQQLLQQQQQKQQLQQQ 117
Query: 131 QQQQLQQQQQ--------------QRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP---SC 173
QQQ+ QQQQ Q+ P S + PS + P +C
Sbjct: 118 QQQEQLQQQQLQLQQQLQQQLQHIQKEPSSHTYTPGPSPELQSVLNYANVPLSKSAAFNC 177
Query: 174 NTNFSSYMGDFSSYPPAFVPNSYPVPVASPIAPPPLMAENPNFILPNQPLGLNLNFHDFN 233
N + S +G P PV SP+ P P + + N P N N
Sbjct: 178 NNSSSYSVG--------------PTPVQSPVTPSPAASA----VTVNSPSYGNFQLFGEN 219
Query: 234 NLDATIHLSNTSLSSNSFSSATSSSQEVPSVELSQG 269
D+T + S + S + S P V +S G
Sbjct: 220 AFDSTTPFQSDGTSQSHSRSLANDSD--PMVVMSPG 253
Score = 33.9 bits (76), Expect = 8.5
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 4/75 (5%)
Query: 78 TQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVT----ALKFHRASMKEASEQQQQQ 133
+Q PQ + + + QE + + E R I +L+ + S ++ QQQQQ
Sbjct: 2 SQSPQTVDQAISVKTDYEDNQQEHIPSNFEIFRRINALLDNSLEANNVSCSQSQSQQQQQ 61
Query: 134 QLQQQQQQRAPDSPQ 148
Q QQQQQQ+ Q
Sbjct: 62 QTQQQQQQQQQQQQQ 76
>pir||A26892 Mopa box protein - mouse (fragment) gi|387503|gb|AAA39860.1| mopa
protein
Length = 139
Score = 55.1 bits (131), Expect = 4e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 19 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 76
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 77 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 109
Score = 39.3 bits (90), Expect = 0.20
Identities = 26/110 (23%), Positives = 48/110 (43%), Gaps = 13/110 (11%)
Query: 68 IGKACQSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEAS 127
+G+ Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ +
Sbjct: 3 LGQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ-------------QQM 49
Query: 128 EQQQQQQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYPSCNTNF 177
+QQQQQ QQQQQQ+ Q Q + + P+ P F
Sbjct: 50 LRQQQQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQPQF 99
>gb|AAH57119.1| Tnrc11 protein [Mus musculus]
Length = 2158
Score = 55.1 bits (131), Expect = 4e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 2038 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 2095
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 2096 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2128
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 2027 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 2077
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 2078 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 2116
Score = 35.8 bits (81), Expect = 2.2
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 2024 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 2072
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 2073 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 2107
>ref|NP_067496.1| trinucleotide repeat containing 11 (THR-associated protein) [Mus
musculus] gi|3426322|gb|AAC83164.1| OPA-containing
protein 1 [Mus musculus]
Length = 2074
Score = 55.1 bits (131), Expect = 4e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 1954 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 2011
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 2012 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 2044
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 1943 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 1993
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 1994 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 2032
Score = 35.8 bits (81), Expect = 2.2
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 1940 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 1988
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 1989 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 2023
>gb|AAH57085.1| Tnrc11 protein [Mus musculus]
Length = 866
Score = 55.1 bits (131), Expect = 4e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 746 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 803
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 804 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 836
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 735 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 785
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 786 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 824
Score = 35.8 bits (81), Expect = 2.2
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 732 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 780
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 781 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 815
>dbj|BAD90141.1| mKIAA0192 protein [Mus musculus]
Length = 1146
Score = 55.1 bits (131), Expect = 4e-06
Identities = 29/93 (31%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ H + Q++ M +++ + + ++ +QQQQ
Sbjct: 1026 QQQQQQQQQQQQQQQQQQQYHIRQQQQQQ--QMLRQQQQQQQQQQQQQQQQQQQQQQQQQ 1083
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRR 165
QQ QQQQQ AP PQ P F + G ++++
Sbjct: 1084 QQPHQQQQQAAPPQPQPQSQPQFQRQGLQQTQQ 1116
Score = 45.8 bits (107), Expect = 0.002
Identities = 28/99 (28%), Positives = 46/99 (46%), Gaps = 9/99 (9%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ ++ + +++ M +QQQQ
Sbjct: 1015 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQQQQ---------QQMLRQQQQQQQ 1065
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNPRIYP 171
QQ QQQQQQ+ Q Q P Q + P+ P
Sbjct: 1066 QQQQQQQQQQQQQQQQQQQQPHQQQQQAAPPQPQPQSQP 1104
Score = 35.8 bits (81), Expect = 2.2
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 11/95 (11%)
Query: 73 QSQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQ 132
Q QQ QQ QQ ++Q +++ + Q++ R++ ++ +QQ
Sbjct: 1012 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQYHIRQQ-----------QQQQQMLRQQ 1060
Query: 133 QQLQQQQQQRAPDSPQLSQNPSFDQDGRYKSRRNP 167
QQ QQQQQQ+ Q Q Q + + + P
Sbjct: 1061 QQQQQQQQQQQQQQQQQQQQQQQQQPHQQQQQAAP 1095
>gb|AAF52844.1| CG4722-PA [Drosophila melanogaster] gi|17136672|ref|NP_476837.1|
CG4722-PA [Drosophila melanogaster]
gi|15292273|gb|AAK93405.1| LD45157p [Drosophila
melanogaster]
Length = 696
Score = 55.1 bits (131), Expect = 4e-06
Identities = 65/252 (25%), Positives = 105/252 (40%), Gaps = 59/252 (23%)
Query: 25 EEEPPFSGA---YMRS--LVKQLRTNDQGCVVNSDASSHGQNLTKHGKIGKA-----CQS 74
++EP + G Y RS L + Q S+ + + + + G + A ++
Sbjct: 379 QQEPLYGGTRSLYCRSPTLTRSNLNRSQSVYAKSNTAINRDIVPRPGPLVPAQSLYPMRT 438
Query: 75 QQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQQ 134
QQ QQ QQ ++QV +S + + N + R E + ++ ++ +QQQQQQ
Sbjct: 439 QQQQQQQQQQQQQVAPAPQSSHLQNQNVQNQMQQRSESIYGMRGSMRGQQQPIQQQQQQQ 498
Query: 135 LQQQQQQRAPD----------SPQLSQNPS----------------FDQDGRYK-SRRNP 167
QQQ QQ+ P+ PQ+ +P DG +K RR+P
Sbjct: 499 QQQQLQQQQPNMGVQQQQMQPPPQMMSDPQQQPQGFQPVYGTRTNPTPMDGNHKYDRRDP 558
Query: 168 -RIY------------PSCNTNFSSYMGDFSSYPPAFVPNSYPVPVASPI---APPPLMA 211
++Y S ++++ SY G + PPA P+ P P P+ APPP
Sbjct: 559 QQMYGVTGPRNRGQSAQSDDSSYGSYHGS-AVTPPARHPSVEPSPPPPPMLMYAPPP--- 614
Query: 212 ENPNFILPNQPL 223
PN P QP+
Sbjct: 615 -QPNAAHP-QPI 624
>gb|AAO41616.1| CG12071-PB, isoform B [Drosophila melanogaster]
gi|28571998|ref|NP_788769.1| CG12071-PB, isoform B
[Drosophila melanogaster]
Length = 578
Score = 53.9 bits (128), Expect = 8e-06
Identities = 30/91 (32%), Positives = 52/91 (56%), Gaps = 11/91 (12%)
Query: 74 SQQSTQQPQQHKKQVRRRLHTSRPYQERLLNMAEARREIVTALKFHRASMKEASEQQQQQ 133
S Q QQPQQH +Q + + Q++ ++A ++ A + H+ +++ +QQQQQ
Sbjct: 399 SHQQQQQPQQHPQQQQHQ-------QQQQQHVAHGKK----AARKHQQHLQQQQQQQQQQ 447
Query: 134 QLQQQQQQRAPDSPQLSQNPSFDQDGRYKSR 164
Q QQQQQQ+ +P + N S + +G +S+
Sbjct: 448 QQQQQQQQQQTSAPMYNNNSSSNHNGNNQSQ 478
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.312 0.126 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 638,624,027
Number of Sequences: 2540612
Number of extensions: 27176045
Number of successful extensions: 307673
Number of sequences better than 10.0: 3235
Number of HSP's better than 10.0 without gapping: 1661
Number of HSP's successfully gapped in prelim test: 1702
Number of HSP's that attempted gapping in prelim test: 196322
Number of HSP's gapped (non-prelim): 28174
length of query: 382
length of database: 863,360,394
effective HSP length: 130
effective length of query: 252
effective length of database: 533,080,834
effective search space: 134336370168
effective search space used: 134336370168
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0133.10


