

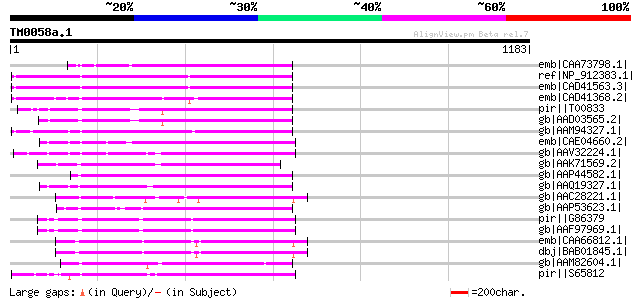
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0058a.1
(1183 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulg... 348 5e-94
ref|NP_912383.1| putative reverse transcriptase [Oryza sativa (j... 254 2e-65
emb|CAD41563.3| OSJNBa0006A01.18 [Oryza sativa (japonica cultiva... 246 3e-63
emb|CAD41368.2| OSJNBa0088A01.7 [Oryza sativa (japonica cultivar... 243 4e-62
pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabi... 242 6e-62
gb|AAD03565.2| putative non-LTR retroelement reverse transcripta... 241 8e-62
gb|AAM94327.1| putative reverse transcriptase [Sorghum bicolor] 239 3e-61
emb|CAE04660.2| OSJNBa0061G20.16 [Oryza sativa (japonica cultiva... 239 4e-61
gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cult... 238 9e-61
gb|AAK71569.2| putative reverse transcriptase [Oryza sativa (jap... 236 3e-60
gb|AAP44582.1| putative reverse transcriptase [Oryza sativa (jap... 234 2e-59
gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultiva... 232 5e-59
gb|AAC28221.1| similar to reverse transcriptases (PFam: rvt.hmm,... 231 1e-58
gb|AAP53623.1| putative non-LTR retroelement reverse transcripta... 229 3e-58
pir||G86379 protein F5A9.24 [imported] - Arabidopsis thaliana gi... 229 4e-58
gb|AAF97969.1| F21J9.30 [Arabidopsis thaliana] 229 4e-58
emb|CAA66812.1| non-ltr retrotransposon reverse transcriptase-li... 228 9e-58
dbj|BAB01845.1| non-LTR retroelement reverse transcriptase-like ... 228 1e-57
gb|AAM82604.1| putative AP endonuclease/reverse transcriptase [B... 223 3e-56
pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW1... 222 5e-56
>emb|CAA73798.1| reverse transcriptase [Beta vulgaris subsp. vulgaris]
gi|7484649|pir||T14619 reverse transcriptase - beet
retrotransposon (fragment)
Length = 507
Score = 348 bits (894), Expect = 5e-94
Identities = 183/514 (35%), Positives = 290/514 (55%), Gaps = 12/514 (2%)
Query: 133 DVGEWCLA-GDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFTWR 191
D G CL GD+N ER S ++SS + R+F FI NM L++I FTW
Sbjct: 1 DRGSPCLIMGDYNETLIPAER--GSHLISS--QGARDFQNFINNMGLIEISPIKGFFTWF 56
Query: 192 RANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGPKPFRVLNC 251
R Q++S++DR V W +P+ +L R SDHCPLL +WGPKPFR LNC
Sbjct: 57 RG--QSKSKLDRLFVQPEWLTSFPSLKLNILKRSPSDHCPLLAFSSHQNWGPKPFRFLNC 114
Query: 252 WLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQK 311
WL P+ + + W + +KL+ L+ +L+ N++ FG + + +K
Sbjct: 115 WLTHPQCMKIISEIWTKNHNSS-----VPDKLRTLKEALKQWNQKEFGIIDDNIKHCEEK 169
Query: 312 INLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHS 371
I+ LD L EL E D + W L+ ES+ + +W+ EGD N+ +FH+
Sbjct: 170 IHFLDNIANSRLLCTNELQERDDAQLNLWTWLKRSESYWALNSRARWIKEGDSNTRYFHT 229
Query: 372 TINWKRRADSIVGLMVDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTITDR 431
+R + + + +G +P +K +FQ F RP +G+ F+ +++
Sbjct: 230 LATIRRHKNHLQYIKTEGKNISKPGEIKEEAVAYFQHIFAEEFSTRPVFNGLNFNKLSES 289
Query: 432 DNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA 491
+ + L+ PF EI +AV C K+PGPDGFNF+FI+ W+V+ D+ K V +FW G+
Sbjct: 290 ECSSLIAPFTHEEIDEAVDSCDAQKAPGPDGFNFKFIKSSWEVIKKDIYKIVEDFWASGS 349
Query: 492 *PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFT 551
P G N++FI LIPK + P+ N+FRPIS++GC+YK++ K+ A RL+ V+ +I Q +
Sbjct: 350 LPHGCNSAFIALIPKTQHPRGFNEFRPISMVGCLYKIIAKILARRLQRVMDHLIGPYQSS 409
Query: 552 FVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKW 611
F+ R +LD V++ +E++ + K ++ K+D+ KA++S+ W +LE+++ +MNF W
Sbjct: 410 FIKGRQILDGVLIASELIDSCRRMKNEAVVLKLDFHKAFDSISWNYLEWVLCQMNFPLVW 469
Query: 612 IGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
WI C+ SA+ +VL+NGSPS F +++GLRQG
Sbjct: 470 RSWIRSCVMSASAAVLINGSPSSVFKLQRGLRQG 503
>ref|NP_912383.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|29893671|gb|AAP06925.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 870
Score = 254 bits (648), Expect = 2e-65
Identities = 189/643 (29%), Positives = 296/643 (45%), Gaps = 10/643 (1%)
Query: 5 NVRGLGVGGNAKKRVVRELVCQQHVELLCIQETKCQVVDRRMCGLL*GDSEFEWKATPAV 64
NVRGL A+ V EL+ L C QETK Q +D + L G ++ PA
Sbjct: 11 NVRGLN--SPARCNAVHELMLDTKCTLACFQETKLQSIDDGLARFLGGYKLDKFAFKPAR 68
Query: 65 NRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDA 124
GG+L +W+S F ++E G + + ++ +Y P K +
Sbjct: 69 GTRGGILLLWSSGDFDVQEVRIGRFSISAQITDIRSGVTFLLTGVYGPTRHHLKDSFLRH 128
Query: 125 LCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLT 184
+ R + G W + GDFN + A ++ S+ L+ MR F I +L +IPL
Sbjct: 129 IRRLRPSPGEG-WLILGDFNMIYRARDKNNSNLNLA----RMRRFRAVIDRCELHEIPLQ 183
Query: 185 GRRFTWRRANDQAQS-RIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGP 243
R+FTW +A ++DR + W +P+ L SDHCPL++ P
Sbjct: 184 NRKFTWSNERRKATLVKLDRCFCNEEWDLAFPHHVLHALPTGPSDHCPLVLSNPQGPHRP 243
Query: 244 KPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKT 303
+ F+ N W P V+ W + L +KL+ LR K + + K
Sbjct: 244 RTFKFENFWTRVPGFKDKVKDIWSQPSSHTEPMHRLNQKLQRTAVGLRDWTKGICSEAKL 303
Query: 304 RREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGD 363
+ + I LDV +E L P E L + R+ + + Q + + + EGD
Sbjct: 304 QFHMALNVIQRLDVAQEHRELSPPETRLRAALKRKVLRLASIERARKKQASRVTHIREGD 363
Query: 364 LNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGV 423
N+ FFH +N +RR ++I L D W ++ IHD F F RP
Sbjct: 364 ANTKFFHLKVNARRRRNTIQRLKKDSGWAVTHGEKESTIHDHFAS-FMGRPGPRPNELNW 422
Query: 424 QFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAV 483
+ IT D L PF E E+ +A+ E K+PGPDGF F + W+++ D++
Sbjct: 423 ETLDITLIDLATLGEPFTEEEVHRAIKEMPADKAPGPDGFTGAFFKVCWEIIKDDILLVF 482
Query: 484 REFW-MRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIG 542
+ +R A N++ IVLIPK + ++D+RPISLI I K+ K+ A+RLR +
Sbjct: 483 SSIYNLRCAHLNLLNSANIVLIPKKDGAESVSDYRPISLIHSIAKIFAKMLALRLRPHMH 542
Query: 543 KVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMM 602
++I Q F+ R++ D+ + + + +R + FK+D KA++SV W +L ++
Sbjct: 543 ELISVNQSAFIKGRSIHDNYLFVRNMTRRYHRLRRAMLLFKLDITKAFDSVRWDYLLALL 602
Query: 603 GRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
R F +WI W+ L+S+T VL+NGSP +GLRQG
Sbjct: 603 QRKGFPTRWIDWLGALLSSSTSQVLLNGSPGQRIKHGRGLRQG 645
>emb|CAD41563.3| OSJNBa0006A01.18 [Oryza sativa (japonica cultivar-group)]
Length = 1189
Score = 246 bits (628), Expect = 3e-63
Identities = 187/643 (29%), Positives = 294/643 (45%), Gaps = 10/643 (1%)
Query: 5 NVRGLGVGGNAKKRVVRELVCQQHVELLCIQETKCQVVDRRMCGLL*GDSEFEWKATPAV 64
NVRGL A+ VV EL+ L C QETK Q +D + L G ++ PA
Sbjct: 11 NVRGLN--SPARCNVVHELMLDTKCTLACFQETKLQSIDDGLARFLGGYKLDKFAFKPAR 68
Query: 65 NRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDA 124
GG+L +W+S F ++E G + + ++ +Y P K +
Sbjct: 69 GTRGGILLLWSSGDFDVQEVRIGRFSISAQITDIRSGVTFLLTGVYGPTRHHLKDSFLRH 128
Query: 125 LCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLT 184
+ R + G W + GDFN + A ++ S+ L+ MR F I +L +IPL
Sbjct: 129 IRRLRPSPGEG-WLILGDFNMIYRARDKNNSNLNLA----RMRRFRAVIDRCELHEIPLQ 183
Query: 185 GRRFTWRRANDQAQS-RIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGP 243
R+FTW +A ++DR + W +P+ L SDH PL++ P
Sbjct: 184 NRKFTWSNERRKATLVKLDRCFCNEEWDLAFPHHVLHALPTGPSDHYPLVLSNPQGPHRP 243
Query: 244 KPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKT 303
+ F+ N W P V+ W + L +KL+ LR K + + K
Sbjct: 244 RTFKFENFWTRVPGFKDKVKDIWSQPSSHTEPMHRLNQKLQHTAVGLRDWTKGICSEAKL 303
Query: 304 RREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGD 363
+ + I LDV +E L P E L + + + + Q + + + EGD
Sbjct: 304 QFHMALNVIQRLDVAQEHRELTPLETRLRAALKRKVLGLASIERARKKQASRVTHIREGD 363
Query: 364 LNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGV 423
N+ FFH +N +RR ++I L D W + IHD F F RP
Sbjct: 364 ANTKFFHLKVNARRRRNTIQRLKKDSGWAVTHGDKELTIHDHFAS-FMGRPGPRPNELNW 422
Query: 424 QFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAV 483
+ IT D L PF E E+ +A+ E K+PGPDGF F + W+++ D++
Sbjct: 423 ETLDITPIDLATLGEPFTEEEVHKAIKEMPADKAPGPDGFTGAFFKVCWEIIKDDILLVF 482
Query: 484 REFW-MRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIG 542
+ +R A N++ IVLIPK + ++D+RPISLI I K+ K+ A+RLR +
Sbjct: 483 SSIYNLRCAHLNLLNSANIVLIPKKDGAESVSDYRPISLIHSIAKIFAKMLALRLRPHMH 542
Query: 543 KVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMM 602
++I Q F+ R++ D+ + + + ++R + FK+D KA++SV W +L ++
Sbjct: 543 ELISVNQSAFIKGRSIHDNYLFVRNTIRRYHRQRRAMLLFKLDITKAFDSVRWDYLLALL 602
Query: 603 GRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
R F +WI W+ L+S+ VL+NGSP +GLRQG
Sbjct: 603 QRKGFPTRWIDWLGALLSSSISQVLLNGSPGQRIKHGRGLRQG 645
>emb|CAD41368.2| OSJNBa0088A01.7 [Oryza sativa (japonica cultivar-group)]
gi|50928243|ref|XP_473649.1| OSJNBa0088A01.7 [Oryza
sativa (japonica cultivar-group)]
Length = 1324
Score = 243 bits (619), Expect = 4e-62
Identities = 186/654 (28%), Positives = 297/654 (44%), Gaps = 32/654 (4%)
Query: 5 NVRGLGVGGNAKKRVVRELVCQQHVELLCIQETKCQVVDRRMCGLL*GDSEFEWKATPAV 64
NVRGL AK V+ EL+ + C QETK + +D + L G + PA
Sbjct: 463 NVRGLN--SPAKCTVLHELMVDTACSIACFQETKLRAIDDVLARYLGGYKLDSYAIKPAR 520
Query: 65 NRGGGLLCIWNSK-LFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWD 123
GG+L +WNS + + E + + A I +Y P K
Sbjct: 521 GTRGGILLLWNSNSINITEVRLRRYSMTAKVSLLHTADS-FTISAVYGPTRHREKEIFLR 579
Query: 124 ALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPL 183
+ + + W L GDFN + A ++ S+ L R MR F I + +L +I L
Sbjct: 580 QIRRLK-PANNEPWLLFGDFNMIYRACDKSNSNINL----RRMRRFRAAINHCELKEIHL 634
Query: 184 TGRRFTW-RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWG 242
R+FTW R+DRF + +W + + L+ SDHCPLL+
Sbjct: 635 QNRKFTWTNERRHPTMVRLDRFFCNENWDLAFGHHILHTLSSGTSDHCPLLLSNPQGPQR 694
Query: 243 PKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLK 302
P+ F+ N W P V+ +W+ + L +KL +L+ + + ++K
Sbjct: 695 PRTFKFENFWTKLPGFRTEVQLAWERTSTHHEPLHRLADKLSHTALALKKWARGICSEVK 754
Query: 303 TRREKVVQKINLLDVKEEEVGLQPEEL---VEMRDLLIEFWRVLRYQESFLCQKACMKWL 359
+ + I LDV +E L E + ++ L+ + + R ++ Q + + +
Sbjct: 755 LKFHLALDVIQRLDVAQENRDLTQAEFRLRLGLKRRLLGYAAIERARKK---QASRITNI 811
Query: 360 VEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAIHDFFQK-------KFKS 412
EGD N+ +FH +N ++R + I L W + I D F++ +
Sbjct: 812 KEGDANTKYFHRKVNARKRRNHIFRLRRQNGWAVTHAEKEGVIADHFRQVMGRPEPRTSD 871
Query: 413 VAWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFW 472
+ W P LD + D T L PF E EI++A+ E G K+PGPDGF +F + W
Sbjct: 872 LNW--PSLD------LAPTDLTRLEEPFSEAEIRKAINEMPGDKAPGPDGFTGKFFKSCW 923
Query: 473 DVLSGDVVKAVREFW-MRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPK 531
D++ DV+ A MR N++ +VLIPK + +NDFRPISLI K+ K
Sbjct: 924 DIIQVDVIAAFNALHNMRSVHLNLLNSANVVLIPKKDGAEGINDFRPISLIHGFAKLSSK 983
Query: 532 LFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYN 591
+ AIRLR ++ +I Q F+ R++ D+ + + ++ +RP + FK+D KA++
Sbjct: 984 VLAIRLRPLMQSLISPNQSAFIRGRSIHDNFMYVRNMVRRYHKTRRPILLFKLDISKAFD 1043
Query: 592 SVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
SV W +L ++ KW WI G L+++T + +NG P KGLRQG
Sbjct: 1044 SVRWDYLLALLQNRGLPQKWRDWISGLLSTSTSKITLNGIPGETIRHWKGLRQG 1097
>pir||T00833 RNA-directed DNA polymerase homolog T13L16.7 - Arabidopsis thaliana
(fragment)
Length = 1365
Score = 242 bits (617), Expect = 6e-62
Identities = 190/654 (29%), Positives = 296/654 (45%), Gaps = 61/654 (9%)
Query: 18 RVVRELVCQQHVELLCIQETKCQ--VVDRRMCGLL*GDSEFEWKATPAVNRGGGLLCIWN 75
R ++E+ ++L + ETK V + C L +F P R GGL W
Sbjct: 17 RYLKEMKKDHFPDILFLMETKNSQDFVYKVFCWL---GYDFIHTVEPE-GRSGGLAIFWK 72
Query: 76 SKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRA-MWDALCAWRGTCDV 134
S L + D LM +++ + I+ + R +W+ L + G
Sbjct: 73 SHLEIEFLYADK----NLMDLQVSSRNKVWFISCVYGLPVTHMRPKLWEHLNSI-GLKRA 127
Query: 135 GEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVF---IANMDLVDIPLTGRRFTWR 191
WCL GDFN +R +E+ G RR F F + N + ++ TG FTW
Sbjct: 128 EAWCLIGDFNDIRSNDEKLGGP------RRSPSSFQCFEHMLLNCSMHELGSTGNSFTWG 181
Query: 192 -RANDQ-AQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVA-DWGPKPFRV 248
NDQ Q ++DR + +W ++PN L + SDH P+L++ + FR
Sbjct: 182 GNRNDQWVQCKLDRCFGNPAWFSIFPNAHQWFLEKFGSDHRPVLVKFTNDNELFRGQFRY 241
Query: 249 LNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKV 308
DDP + +SW QG + F ++ RR
Sbjct: 242 DKRLDDDPYCIEVIHRSWNSAMSQGT-------------------HSSFFSLIECRRAIS 282
Query: 309 VQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRY-----------QESFLCQKACMK 357
V K + + + ++L + + I W + Y +E F QK+ K
Sbjct: 283 VWKHSSDTNAQSRIKRLRKDLDAEKSIQIPCWPRIEYIKDQLSLAYGDEELFWRQKSRQK 342
Query: 358 WLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAI-HDFFQKKFKSVAWL 416
WL GD N+GFFH+T++ +R + + L+ + E K I FF+ F S L
Sbjct: 343 WLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSFFENLFTSTYIL 402
Query: 417 RPK--LDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDV 474
L+G+Q + +T N L+ E+E+ AV+ +PGPDGF F Q+ WD+
Sbjct: 403 THNNHLEGLQ-AKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGFTALFFQQHWDL 461
Query: 475 LSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFA 534
+ ++ + F+ G P+ N + I LIPK+ PQ ++D RPISL +YK++ K+
Sbjct: 462 VKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSVLYKIISKILT 521
Query: 535 IRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKAYN 591
RL+ + ++ Q FV R + D+++V +E++H + R S + FK D KAY+
Sbjct: 522 QRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYD 581
Query: 592 SVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
VEW FLE MM + F KWI WIM C+ S + SVL+NG P G +G+RQG
Sbjct: 582 RVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQG 635
>gb|AAD03565.2| putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana] gi|25411819|pir||H84557 hypothetical protein
At2g17910 [imported] - Arabidopsis thaliana
Length = 1344
Score = 241 bits (616), Expect = 8e-62
Identities = 179/604 (29%), Positives = 277/604 (45%), Gaps = 55/604 (9%)
Query: 66 RGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRA-MWDA 124
R GGL W S L + D LM +++ + I+ + R +W+
Sbjct: 42 RSGGLAIFWKSHLEIEFLYADK----NLMDLQVSSRNKVWFISCVYGLPVTHMRPKLWEH 97
Query: 125 LCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVF---IANMDLVDI 181
L + G WCL GDFN +R +E+ G RR F F + N + ++
Sbjct: 98 LNSI-GLKRAEAWCLIGDFNDIRSNDEKLGGP------RRSPSSFQCFEHMLLNCSMHEL 150
Query: 182 PLTGRRFTWR-RANDQ-AQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVA 239
TG FTW NDQ Q ++DR + +W ++PN L + SDH P+L++
Sbjct: 151 GSTGNSFTWGGNRNDQWVQCKLDRCFGNPAWFSIFPNAHQWFLEKFGSDHRPVLVKFTND 210
Query: 240 -DWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVF 298
+ FR DDP + +SW QG + F
Sbjct: 211 NELFRGQFRYDKRLDDDPYCIEVIHRSWNSAMSQGT-------------------HSSFF 251
Query: 299 GDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRY-----------QE 347
++ RR V K + + + ++L + + I W + Y +E
Sbjct: 252 SLIECRRAISVWKHSSDTNAQSRIKRLRKDLDAEKSIQIPCWPRIEYIKDQLSLAYGDEE 311
Query: 348 SFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWEDEPVRVKAAI-HDFF 406
F QK+ KWL GD N+GFFH+T++ +R + + L+ + E K I FF
Sbjct: 312 LFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDENDQEFTRNSDKGKIASSFF 371
Query: 407 QKKFKSVAWLRPK--LDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFN 464
+ F S L L+G+Q + +T N L+ E+E+ AV+ +PGPDGF
Sbjct: 372 ENLFTSTYILTHNNHLEGLQ-AKVTSEMNHNLIQEVTELEVYNAVFSINKESAPGPDGFT 430
Query: 465 FRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGC 524
F Q+ WD++ ++ + F+ G P+ N + I LIPK+ PQ ++D RPISL
Sbjct: 431 ALFFQQHWDLVKHQILTEIFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSV 490
Query: 525 IYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IF 581
+YK++ K+ RL+ + ++ Q FV R + D+++V +E++H + R S +
Sbjct: 491 LYKIISKILTQRLKKHLPAIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMA 550
Query: 582 FKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKG 641
FK D KAY+ VEW FLE MM + F KWI WIM C+ S + SVL+NG P G +G
Sbjct: 551 FKTDMSKAYDRVEWPFLETMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRG 610
Query: 642 LRQG 645
+RQG
Sbjct: 611 IRQG 614
>gb|AAM94327.1| putative reverse transcriptase [Sorghum bicolor]
Length = 1998
Score = 239 bits (611), Expect = 3e-61
Identities = 189/650 (29%), Positives = 292/650 (44%), Gaps = 25/650 (3%)
Query: 5 NVRGLGVGGNAKKRVVRELVCQQHVELLCIQETKCQVVD----RRMCGLL*GDSEFE-WK 59
NVRGL K +R + V+++C+QETK D R +C +F+ ++
Sbjct: 814 NVRGLN--SKEKWNPIRNKIVDLAVDIICLQETKKDSFDIAFLRNIC-----PPDFDRFE 866
Query: 60 ATPAVNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKR 119
P V GG+L W SKLF E + + ++ N+Y+PC +GKR
Sbjct: 867 YLPLVGASGGILTAWKSKLFSGELIFSNEYAISIQMSSMHNNDSWLLTNVYAPCTDEGKR 926
Query: 120 AMWDALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLV 179
+ + +V +W + GDFN +R E+R G S EM FN I+++ L
Sbjct: 927 QFVNWFKNIQTPHEV-DWLVVGDFNLMRKHEDRNKEGGDSS----EMFLFNSAISSLGLN 981
Query: 180 DIPLTGRRFTWRRA-NDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVV 238
+I L GR++TW ++ +ID S SW + + S + SDH P L++
Sbjct: 982 EIILQGRKYTWSNMQSNPLLQKIDWTFTSNSWNIKFLDTSVKGYDMTPSDHAPCLVKVST 1041
Query: 239 ADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVF 298
K FR NCWL P + W A + K K LR +L+ +
Sbjct: 1042 KIPKTKVFRFENCWLRLPGFQQIISDVWNVPTHHLDQAKSITAKFKKLRKALKE-KQASM 1100
Query: 299 GDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKW 358
LKT KV I L++ EE L E L + +L Q+ + Q+ +KW
Sbjct: 1101 AHLKTVISKVKLVIQFLELLEEYRDLSMLEWNFRAILKKKLTELLEQQKIYWRQRGAIKW 1160
Query: 359 LVEGDLNSGFFHSTINWKRRADSIVGLMV-DGMWEDEPVRVKAAIHDFFQKKFKSVAWLR 417
+ GD + FFH+ K R + + L DG + I + F+++ + R
Sbjct: 1161 IKLGDATTKFFHANATIKMRGNLVNQLQKQDGSILTAHKDKEQHIWEEFKERLGTKEETR 1220
Query: 418 PKLDGVQFSTITDRDNT--ELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVL 475
G+ TI N L PF E EI+ V KSPGPDGF+ F++ W +
Sbjct: 1221 ---FGINPDTILQASNDLETLEEPFTETEIESVVKLLPNDKSPGPDGFSNEFMKASWQTV 1277
Query: 476 SGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAI 535
D+ F R N SFI LIPK P +ND+RPISL+ K++ KL A
Sbjct: 1278 KVDIFNLCHAFHNHNVCLRSINTSFITLIPKSHNPVSVNDYRPISLLNTSMKLITKLLAN 1337
Query: 536 RLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEW 595
RL++ I ++ + Q+ F+ R + D + E +H ++ + K+D+EKA++ +
Sbjct: 1338 RLQMKITDLVHKNQYGFIRQRTIQDCLAWSFEYLHLCHHSRKAIVIIKLDFEKAFDKMNH 1397
Query: 596 GFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
+ +M F KW+ W+ S T +VL+NG+P H +G+RQG
Sbjct: 1398 KAMLTIMKAKGFGQKWLNWMKSIFTSGTSNVLLNGTPGKTIHCLRGVRQG 1447
>emb|CAE04660.2| OSJNBa0061G20.16 [Oryza sativa (japonica cultivar-group)]
Length = 1285
Score = 239 bits (610), Expect = 4e-61
Identities = 173/601 (28%), Positives = 279/601 (45%), Gaps = 36/601 (5%)
Query: 68 GGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCA 127
GGL W+ + V + ++ ++ R + + + I +Y ++ + MW L
Sbjct: 613 GGLALYWDESVSVDVKDIN-KRYIDAYVRLSSDEPQWHITFVYGEPRVENRHRMWSLLRT 671
Query: 128 WRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRR 187
R + + W + GDFN + E + + +M+ F + + DL D+ G
Sbjct: 672 IRQSSAL-PWMVIGDFNETLWQFEHFSKNPRCET---QMQNFRDALYDCDLQDLGFKGVP 727
Query: 188 FTWRRAND---QAQSRIDRFLVSTSWCDVWPNC--SHLVLNRDVSDHCPLLMRQVVADWG 242
T+ D + R+DR + W D++P SHLV SDH P+L+ +V D
Sbjct: 728 HTYDNRRDGWRNVKVRLDRAVADDKWRDLFPEAQVSHLV--SPCSDHSPILLEFIVKDTT 785
Query: 243 PKPFRVLN---CWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFG 299
+ L+ W +P +E++W + +K G + I V
Sbjct: 786 RPRQKCLHYEIVWEREPESVQVIEEAW------------INAGVKTDLGDINIALGRVMS 833
Query: 300 DL----KTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKAC 355
L KT+ + V +++ K E++ +R +L +E Q++
Sbjct: 834 ALRSWGKTKVKNVGRELEKARKKLEDLIASNAARSSIRQATDHMNEMLYREEMLWLQRSR 893
Query: 356 MKWLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDEPVRVKAAIHDFFQKKFKSVA 414
+ WL EGD N+ FFHS W+ + + I L + G ++ ++FQ+ +K+
Sbjct: 894 VNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIHSMTSVLETMATEYFQEVYKADP 953
Query: 415 WLRPKLDGVQFST-ITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWD 473
L P+ F +TD N +L F E EI QA+++ G KSPGPDGF RF QR W
Sbjct: 954 SLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPGPDGFPARFYQRNWG 1013
Query: 474 VLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLF 533
L D++ AVR F+ G P G N + IVLIPK P L D+RPISL +YKVV K
Sbjct: 1014 TLKSDIILAVRNFFQSGLMPEGVNDTAIVLIPKKDQPIDLKDYRPISLCNVVYKVVSKCL 1073
Query: 534 AIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKR---PSIFFKVDYEKAY 590
RLR ++ ++ + Q F+ R + D+ ++ E H + K+ + +K+D KAY
Sbjct: 1074 VNRLRPILDDLVSKEQSAFIQGRMITDNALLAFECFHSIQKNKKANSAACAYKLDLSKAY 1133
Query: 591 NSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLRR 650
+ V+W FLE + ++ F +W+ WIM C+ + SV NG+ F +GLRQG L
Sbjct: 1134 DRVDWRFLELALNKLGFAHRWVSWIMSCVTTVRYSVKFNGTLLRSFAPTRGLRQGDPLSP 1193
Query: 651 F 651
F
Sbjct: 1194 F 1194
>gb|AAV32224.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|45267888|gb|AAS55787.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 1936
Score = 238 bits (607), Expect = 9e-61
Identities = 186/658 (28%), Positives = 304/658 (45%), Gaps = 36/658 (5%)
Query: 10 GVGGNAKKRVVRELVCQQHVELLCIQETKCQVVDRRMCGLL*GDSEFEWKATPAVNRGGG 69
G+G A + +R L+ + +L+ + ET+ V +M L + + + + GG
Sbjct: 645 GLGNTATVQDLRALIQKAGSQLVFLCETRQSV--EKMSRLRRKLAFRGFVGVSSEGKSGG 702
Query: 70 LLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCAWR 129
L W+ + V + ++ ++ R + + I +Y ++ + MW L R
Sbjct: 703 LALYWDESVSVDVKDIN-KRYIDAYVRLSPDEPQWHITFVYGEPRVENRHRMWSLLRTIR 761
Query: 130 GTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFT 189
+ + W + GDFN + E + + +M+ F + + DL D+ G T
Sbjct: 762 QSSAL-PWMVIGDFNETLWQFEHFSKNPRCET---QMQNFRDALYDCDLQDLGFKGVPHT 817
Query: 190 WRRAND---QAQSRIDRFLVSTSWCDVWPNC--SHLVLNRDVSDHCPLLMRQVVADWGPK 244
+ D + R+DR + W D++P SHLV SDH P+L+ +V D
Sbjct: 818 YDNRRDGWRNVKVRLDRAVADDKWRDLFPEAQVSHLV--SPCSDHSPILLEFIVKDTTRP 875
Query: 245 PFRVLN---CWLDDPRLFPFVEQSW--KGMKIQ-GWGAYVLKEKLKGLRGSLRI*NKEVF 298
+ L+ W +P +E++W G+K G L + LR + K V
Sbjct: 876 RQKCLHYEIVWEREPESVQVIEEAWINAGVKTDLGDINIALGRVMSALRSWSKTKVKNVG 935
Query: 299 GDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKW 358
+L+ R+K+ I + + + + EM L +E Q++ + W
Sbjct: 936 KELEKARKKLEDLI-ASNAARSSIRQATDHMNEM----------LYREEMLWLQRSRVNW 984
Query: 359 LVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDEPVRVKAAIHDFFQKKFKSVAWLR 417
L EGD N+ FFHS W+ + + I L + G ++ ++FQ +K+ L
Sbjct: 985 LKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIHSTTSVLETMATEYFQGVYKADPSLN 1044
Query: 418 PKLDGVQFST-ITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLS 476
P+ F +TD N +L F E EI QA+++ G KSP PDGF RF QR W L
Sbjct: 1045 PESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPRPDGFPARFYQRNWGTLK 1104
Query: 477 GDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIR 536
D++ AVR F+ G P+G N + IVLIPK P L D+RPISL +YKVV K R
Sbjct: 1105 SDIILAVRNFFQSGLMPKGVNDTAIVLIPKKDQPIDLKDYRPISLCNVVYKVVSKCLVNR 1164
Query: 537 LRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKR---PSIFFKVDYEKAYNSV 593
LR ++ ++ + Q F+ R + D+ ++ E H + K+ + +K+D KAY+ V
Sbjct: 1165 LRPILDDLVSKEQSAFIQGRMITDNALLAFECFHSIQKNKKANSAACAYKLDLSKAYDRV 1224
Query: 594 EWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLRRF 651
+W FLE + ++ F +W+ WIM C+ + SV NG+ F +GLRQG L F
Sbjct: 1225 DWRFLELALNKLGFAHRWVSWIMLCVTTVRYSVKFNGTLLRSFAPTRGLRQGEPLSPF 1282
>gb|AAK71569.2| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|50918647|ref|XP_469720.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1833
Score = 236 bits (602), Expect = 3e-60
Identities = 173/574 (30%), Positives = 274/574 (47%), Gaps = 44/574 (7%)
Query: 64 VNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWD 123
V +GGGL W+ + V+ +S + L+ R + Y + MW
Sbjct: 42 VGKGGGLALFWDESMNVVLKSYNSRHIDVLITELDCCTWRATFV--YGEPRAQDRHLMWS 99
Query: 124 ALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRR----EMREFNVFIANMDLV 179
L R W + GDFN + E + SHR+ +MR+F ++ DL
Sbjct: 100 LLRRIRSNSG-DPWLMIGDFNEAMWQTEHK-------SHRKRSESQMRDFREVLSECDLH 151
Query: 180 DIPLTGRRFTW---RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQ 236
DI G +T+ +R + R+DR + S +W +P L SDH PLL+ +
Sbjct: 152 DIGFQGAPWTFCNMQREGRNVKVRLDRGVASPAWSSRFPQAVITHLTTPSSDHAPLLLER 211
Query: 237 VVADWGPKPFRVLN---CWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI* 293
+P +++ W + L ++++W M + +K+K L
Sbjct: 212 EETTLA-RPMKIMRYEEVWERESSLPEVIQEAWT-MGADASTLGDINDKMKVTMTKLVSW 269
Query: 294 NKEVFGDLKTR----REKV--VQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQE 347
+K+ G+++ + REK+ ++ I LLD + EV +EL EM L +E
Sbjct: 270 SKDKIGNVRKKIKDLREKLGELRNIGLLDT-DNEVHSVKKELEEM----------LHREE 318
Query: 348 SFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMV-DGMWEDEPVRVKAAIHDFF 406
+ Q++ + WL EGDLN+ +FH +W+ + + I L DG +K FF
Sbjct: 319 IWWKQRSRITWLKEGDLNTRYFHLKASWRAKKNKIKKLKKNDGSTTMNKKEMKEINRSFF 378
Query: 407 QKKFKSVAWLRP-KLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNF 465
Q+ + L P L + I+++ N +L+ PF EI A+++ G K+PGPDGF
Sbjct: 379 QQLYTKDDNLNPVNLLNMFHEKISEQMNADLIKPFTNEEISDALFQIGPLKAPGPDGFPA 438
Query: 466 RFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCI 525
RF+QR W +L G+V+ AVR F+ G N + IV+IPK + + + DFRPISL +
Sbjct: 439 RFLQRNWGLLKGEVIAAVRNFFEDEVMQEGVNDTVIVMIPKKNLAEDMKDFRPISLCNVV 498
Query: 526 YKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFF--- 582
YKVV K R+R ++ ++I E Q F+ R + D+ ++ E H K KR + F
Sbjct: 499 YKVVAKCLVNRMRPMLQEIISEAQSAFIPGRLITDNALIAFECFHSIKNCKRENQNFCAL 558
Query: 583 KVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIM 616
K+D KAY+ V+W FL +M R+ FC KW WIM
Sbjct: 559 KLDLSKAYDRVDWEFLNGVMERLGFCDKWRRWIM 592
>gb|AAP44582.1| putative reverse transcriptase [Oryza sativa (japonica
cultivar-group)] gi|34896522|ref|NP_909605.1| putative
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1556
Score = 234 bits (596), Expect = 2e-59
Identities = 157/512 (30%), Positives = 247/512 (47%), Gaps = 11/512 (2%)
Query: 139 LAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRRFTW-RRANDQA 197
+ G FN +R +E+ + + R FN I ++L ++ +GR FTW +Q
Sbjct: 918 VGGVFNILRNPQEKNNDN----FNDRWPFLFNAVIDTLNLRELESSGRSFTWANHLQNQT 973
Query: 198 QSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGPKP-FRVLNCWLDDP 256
++DR LVST + +P + +L R++SDH PLL A +P F+ WL
Sbjct: 974 FEKLDRILVSTEFETKFPLSTVSILTREISDHTPLLYNSGGASAAYQPQFKFELGWLRRD 1033
Query: 257 RLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLD 316
V++ W+G+ I+G + K++ LR LR K + G K +++++ K++ L
Sbjct: 1034 GFSDIVKEVWQGVSIEGTPLERWQRKIRRLRQFLRGWAKNITGAYKKEKKELLDKLDTLV 1093
Query: 317 VKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHSTINWK 376
K E L EL L +LR +E Q+A +K L+EGD N+ ++H N +
Sbjct: 1094 KKAEHTILNEFELNVKHVLNDRLAELLREEEIKWYQRAKVKHLLEGDANTKYYHLLANGR 1153
Query: 377 RRADSIVGLMVDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTITD---RDN 433
R I L ++K I +F+ F LD Q I +N
Sbjct: 1154 HRKTRIFQLQDGNEVITGDAQLKEHITKYFKTLFGPSQTSSIFLDESQVDDIRQVSAEEN 1213
Query: 434 TELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*P 493
L F E EIK A+++ +PGPDGF F Q FW ++ D++ +F +G+ P
Sbjct: 1214 ESLTADFTEEEIKCAIFQMKHNTAPGPDGFPPEFYQAFWTIIKDDLLALFSDF-QQGSLP 1272
Query: 494 RGS-NASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTF 552
S N I+L+PK K + + +RPI L+ +K+ K+ RL V KVI Q F
Sbjct: 1273 LNSLNFGTIILLPKQKDVRTIQQYRPICLLNVSFKIFTKVATNRLTSVAQKVIKPTQTAF 1332
Query: 553 VGNRNMLDSVVVLNEVMHEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWI 612
+ RN+++ ++L+E +HE KK+ + FK+D+EKAY+ V W FL+ + F KW
Sbjct: 1333 LPGRNIMEGAIILHETLHELHTKKQDGVIFKIDFEKAYDKVRWSFLQQTLRMKGFSHKWC 1392
Query: 613 GWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQ 644
W+ VS+ VN F +KGLRQ
Sbjct: 1393 NWVEKFTQGGNVSIKVNDQVGNYFQSKKGLRQ 1424
>gb|AAQ19327.1| bZIP-like protein [Oryza sativa (japonica cultivar-group)]
Length = 2367
Score = 232 bits (592), Expect = 5e-59
Identities = 164/592 (27%), Positives = 274/592 (45%), Gaps = 30/592 (5%)
Query: 68 GGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWDALCA 127
GGL W+ + V + ++ ++ + + + + +Y ++ + MW L
Sbjct: 795 GGLALYWDESVSVDVKDIN-KRYIDAYVQLSPEEPQWHVTFVYGEPRVENRHRMWSLLRT 853
Query: 128 WRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDLVDIPLTGRR 187
+ + W + GDFN + E + +M++F + + +L D+ G
Sbjct: 854 IHQSSSL-PWAVIGDFNETMWQFEHFSRT---PRGEPQMQDFRDVLQDCELHDLGFKGVP 909
Query: 188 FTWRRAND---QAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVVADWGPK 244
T+ + + R+DR + W D++ + L SDHCP+L+ VV D
Sbjct: 910 HTYDNKREGWRNVKVRLDRVVADDKWRDIYSTAQVVHLVSPCSDHCPILLNLVVKDPHQL 969
Query: 245 PFRVLN---CWLDDPRLFPFVEQSW--KGMKIQ-GWGAYVLKEKLKGLRGSLRI*NKEVF 298
+ L+ W +P +E++W G K G L + + LR R K V
Sbjct: 970 RQKCLHYEIVWEREPEATQVIEEAWVVAGEKADLGDINKALAKVMTALRSWSRAKVKNVG 1029
Query: 299 GDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACMKW 358
+L+ R+K+ + I + +R+ +L +E Q++ + W
Sbjct: 1030 RELEKARKKLAELIE-----------SNADRTVIRNATDHMNELLYREEMLWLQRSRVNW 1078
Query: 359 LVEGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSVAWLR 417
L + D N+ FFHS W+ + + I L + +++++ ++FQ + + L
Sbjct: 1079 LKDEDRNTKFFHSRAVWRAKKNKISKLRDANETVHSSTMKLESMATEYFQDVYTADPNLN 1138
Query: 418 PK-LDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLS 476
P+ + + +TD N +L F E EI QA+++ G KSPGPDGF RF QR W +
Sbjct: 1139 PETVTRLIQEKVTDIMNEKLCEDFTEDEISQAIFQIGPLKSPGPDGFPARFYQRNWGTIK 1198
Query: 477 GDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIR 536
D++ AVR F+ G P G N + IVLIPK + P L DFRPISL +YKVV K R
Sbjct: 1199 ADIIGAVRRFFQTGLMPEGVNDTAIVLIPKKEQPVDLRDFRPISLCNVVYKVVSKCLVNR 1258
Query: 537 LRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKR---PSIFFKVDYEKAYNSV 593
LR ++ ++ Q FV R + D+ ++ E H + K+ + +K+D KAY+ V
Sbjct: 1259 LRPILDDLVSVEQSAFVQGRMITDNALLAFECFHAMQKNKKANHAACAYKLDLSKAYDRV 1318
Query: 594 EWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQG 645
+W FLE M ++ F +W+ WIM C+ S V NG+ F +GLRQG
Sbjct: 1319 DWRFLEMAMNKLGFARRWVNWIMKCVTSVRYMVKFNGTLLQSFAPTRGLRQG 1370
>gb|AAC28221.1| similar to reverse transcriptases (PFam: rvt.hmm, score: 60.13)
[Arabidopsis thaliana] gi|7488321|pir||T01871
RNA-directed DNA polymerase homolog T24M8.8 -
Arabidopsis thaliana
Length = 1164
Score = 231 bits (588), Expect = 1e-58
Identities = 174/608 (28%), Positives = 280/608 (45%), Gaps = 51/608 (8%)
Query: 105 VIINIYSPCDIDGKRAMWDALCAWRGT-CDVGE-WCLAGDFNAVRYAEERRGSSGVLSSH 162
V+ +Y+ D ++ +W+ + + C + + W + GDFN + + E S G +
Sbjct: 2 VLSFVYASTDEVTRQILWNEIVDFSNDPCVIDKPWTVLGDFNQILHPSEHSTSDGF--NV 59
Query: 163 RREMREFNVFIANMDLVDIPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHL 220
R R F I L D+ G FTW +R+ ++DR LV+ W +P+ L
Sbjct: 60 DRPTRIFRETILLASLTDLSFRGNTFTWWNKRSRAPVAKKLDRILVNDKWTTTFPSSLGL 119
Query: 221 VLNRDVSDH--CPLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYV 278
D SDH C L + + KPFR N L D + W + G Y
Sbjct: 120 FGEPDFSDHSSCELSLMSA-SPRSKKPFRFNNFLLKDENFLSLICLKWFSTSVTGSAMYR 178
Query: 279 LKEKLKGLRGSLRI*NKEVFGDLKTRREK------VVQKINLLDVKEEEVGLQPEELVEM 332
+ KLK L+ +R +++ + D++ R ++ + Q + L ++ E +
Sbjct: 179 VSVKLKALKKVIRDFSRDNYSDIEKRTKEAHDALLLAQSVLLASPCPSNAAIEAETQRK- 237
Query: 333 RDLLIEFWRVLRYQE-SFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSI------VGL 385
WR+L E SF Q++ + WL EGD+NS +FH + ++ + I VG
Sbjct: 238 -------WRILAEAEASFFYQRSRVNWLREGDMNSSYFHKMASARQSLNHIHFLSDPVGD 290
Query: 386 MVDGMWEDEPVRVKAAIHDFFQKKFKSVAWLRPKLDGVQFSTI-----TDRDNTELVGPF 440
++G E V ++FQ S L P + S + + L PF
Sbjct: 291 RIEGQQNLENHCV-----EYFQSNLGSEQGL-PLFEQADISNLLSYRCSPAQQVSLDTPF 344
Query: 441 DEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASF 500
+IK A + K+ GPDGF+ F W ++ G+V +A+ EF+ G + NA+
Sbjct: 345 SSEQIKNAFFSLPRNKASGPDGFSPEFFCACWPIIGGEVTEAIHEFFTSGKLLKQWNATN 404
Query: 501 IVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLD 560
+VLIPK+ ++DFRPIS + +YKV+ KL RL+ + I Q F+ R L+
Sbjct: 405 LVLIPKITNASSMSDFRPISCLNTVYKVISKLLTDRLKDFLPAAISHSQSAFMPGRLFLE 464
Query: 561 SVVVLNEVMHEAKVKK-RPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCL 619
+V++ E++H K PS KVD KA++SV W F+ + +N K+ WI+ CL
Sbjct: 465 NVLLATELVHGYNKKNIAPSSMLKVDLRKAFDSVRWDFIVSALRALNVPEKFTCWILECL 524
Query: 620 NSATVSVLVNGSPSGEFHMEKGLRQG--------ILLRRFFS*LL-LKA*MGYSRMQLEL 670
++A+ SV++NG +G F KGLRQG +L FS LL + GY +
Sbjct: 525 STASFSVILNGHSAGHFWSSKGLRQGDPMSPYLFVLAMEVFSGLLQSRYTSGYIAYHPKT 584
Query: 671 GNLKVSRL 678
L++S L
Sbjct: 585 SQLEISHL 592
>gb|AAP53623.1| putative non-LTR retroelement reverse transcriptase [Oryza sativa
(japonica cultivar-group)] gi|37534068|ref|NP_921336.1|
putative non-LTR retroelement reverse transcriptase
[Oryza sativa (japonica cultivar-group)]
gi|19881727|gb|AAM01128.1| putative non-LTR retroelement
reverse transcriptase [Oryza sativa (japonica
cultivar-group)]
Length = 1228
Score = 229 bits (585), Expect = 3e-58
Identities = 172/556 (30%), Positives = 266/556 (46%), Gaps = 33/556 (5%)
Query: 106 IINIYSPCDIDGKRAMWDALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRRE 165
I+ IY P D + K+A D L W + GDFN R+A E+ ++ ++ R
Sbjct: 623 IMVIYGPVDNNLKQAFLDEL-NWHILYRSHHVVIGGDFNLYRFASEKSNTN----TNSRN 677
Query: 166 MREFNVFIANMDLVDIPLTGRRFTWRRANDQ-AQSRIDRFLVSTSWCDVWPNCSHLVLNR 224
M FN FIAN+DL +I +G +FTW D Q +DR LV+T W D++P + R
Sbjct: 678 MDMFNSFIANLDLREIYRSGPKFTWTNKQDSLVQEVLDRILVTTDWDDMYPTALLSSILR 737
Query: 225 DVSDHCPLLM---RQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGW----GAY 277
SDH PLL+ ++ K F+ + WL E+S+K + IQ +Y
Sbjct: 738 VGSDHIPLLLDTCEGIIVK--SKYFKFESAWL--------AEESFKELVIQRLLPRDNSY 787
Query: 278 VLK---EKLKGLRGSLRI*NKEVFGDLKTRREKVVQ-KINLLDVKEEEVGLQPEELVEMR 333
+L+ +K LR L K K R ++ +Q KI LD K L +E +
Sbjct: 788 ILEFWNKKQSSLRRFLVGWGKNNRSK-KLREKRTLQCKIEELDKKALTEELTSDEWNQRY 846
Query: 334 DLLIEFWRVLRYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVDGMWE- 392
+ + +E + ++ +W++EGD N+ FFH N ++R I LM DG E
Sbjct: 847 SWENDLEHIYEMEELYWHKRCGEQWILEGDRNTEFFHRVANGRKRKCHITSLM-DGETEL 905
Query: 393 DEPVRVKAAIHDFFQKKFKSVAWLRPKLD-GVQFS--TITDRDNTELVGPFDEVEIKQAV 449
++ I ++++ F+ V L GV + ++D L PF E+ + +
Sbjct: 906 TSKSEIREHIVEYYKTLFRDVPTANIHLSQGVWTNHLNLSDASKENLTRPFVMAELDKVI 965
Query: 450 WECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKI 509
E +PGPDGF+ F + FW + D+ + + + N I LIPK
Sbjct: 966 NEAKLNTAPGPDGFSIPFYRAFWPQVKNDLFEMLLMLHNEDLDLKRLNFGVISLIPKNNN 1025
Query: 510 PQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVM 569
P + FRPI ++ +K + K+ RL V +V+ + Q F+ R +L+ V+++EV+
Sbjct: 1026 PTDIKQFRPICVLNDCFKFLSKVVTNRLTEVADEVVSQTQTAFIPGRFILEGCVIIHEVL 1085
Query: 570 HEAKVKKRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVN 629
HE K K I K+D+EKAY+ V W FL +M R NF KWI WI C+ V + +N
Sbjct: 1086 HELKSKNSEGIILKIDFEKAYDKVNWEFLIEVMERKNFPKKWISWIKSCVMGGRVCININ 1145
Query: 630 GSPSGEFHMEKGLRQG 645
G + F +GLRQG
Sbjct: 1146 GERTDFFRTFRGLRQG 1161
>pir||G86379 protein F5A9.24 [imported] - Arabidopsis thaliana
gi|9945082|gb|AAG03119.1| F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 229 bits (584), Expect = 4e-58
Identities = 182/603 (30%), Positives = 283/603 (46%), Gaps = 37/603 (6%)
Query: 64 VNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWD 123
V + GGL +W S + V + VD + ++GA CV +Y D + W+
Sbjct: 28 VGKCGGLALLWKSSVQVDLKFVD-KNLMDAQVQFGAVNF-CVSC-VYGDPDRSKRSQAWE 84
Query: 124 ALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRR---EMREFNVFIANMDLVD 180
+ G +WC+ GDFN + + E+ G RR + + FN I DLV+
Sbjct: 85 RISRI-GVGRRDKWCMFGDFNDILHNGEKNGGP------RRSDLDCKAFNEMIKGCDLVE 137
Query: 181 IPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVV 238
+P G FTW RR + Q R+DR + W +P + L+ SDH P+L++ +
Sbjct: 138 MPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLIKLMS 197
Query: 239 A-DWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEV 297
+ D FR +L + + ++W K G V ++L+ R SL K+
Sbjct: 198 SQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGK-HGTNISVA-DRLRACRKSLSSWKKQ- 254
Query: 298 FGDLKTRREKVVQKINLLDVK-EEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACM 356
+ KIN L+ E+E L + L + + R +E++ QK+
Sbjct: 255 ------NNLNSLDKINQLEAALEKEQSLVWPIFQRVSVLKKDLAKAYREEEAYWKQKSRQ 308
Query: 357 KWLVEGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSVAW 415
KWL G+ NS +FH+ + R+ I L V+G + +F FKS
Sbjct: 309 KWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLFKSS-- 366
Query: 416 LRPKLDGVQFSTITDRD----NTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRF 471
P FS + R N LVG EIK+AV+ +PGPDG + F Q +
Sbjct: 367 -NPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHY 425
Query: 472 WDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPK 531
W + V V++F+ G P N + + LIPK + P + D RPISL +YK++ K
Sbjct: 426 WSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISK 485
Query: 532 LFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFF---KVDYEK 588
+ A RL+ + +++ + Q FV R + D+++V +E++H KV R S F K D K
Sbjct: 486 IMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSK 545
Query: 589 AYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILL 648
AY+ VEW +L ++ + F KW+ WIM C++S T SVL+N P G +++GLRQG L
Sbjct: 546 AYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPL 605
Query: 649 RRF 651
F
Sbjct: 606 SPF 608
>gb|AAF97969.1| F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 229 bits (584), Expect = 4e-58
Identities = 182/603 (30%), Positives = 283/603 (46%), Gaps = 37/603 (6%)
Query: 64 VNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWD 123
V + GGL +W S + V + VD + ++GA CV +Y D + W+
Sbjct: 25 VGKCGGLALLWKSSVQVDLKFVD-KNLMDAQVQFGAVNF-CVSC-VYGDPDRSKRSQAWE 81
Query: 124 ALCAWRGTCDVGEWCLAGDFNAVRYAEERRGSSGVLSSHRR---EMREFNVFIANMDLVD 180
+ G +WC+ GDFN + + E+ G RR + + FN I DLV+
Sbjct: 82 RISRI-GVGRRDKWCMFGDFNDILHNGEKNGGP------RRSDLDCKAFNEMIKGCDLVE 134
Query: 181 IPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQVV 238
+P G FTW RR + Q R+DR + W +P + L+ SDH P+L++ +
Sbjct: 135 MPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQTFLDFRGSDHRPVLIKLMS 194
Query: 239 A-DWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NKEV 297
+ D FR +L + + ++W K G V ++L+ R SL K+
Sbjct: 195 SQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGK-HGTNISVA-DRLRACRKSLSSWKKQ- 251
Query: 298 FGDLKTRREKVVQKINLLDVK-EEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKACM 356
+ KIN L+ E+E L + L + + R +E++ QK+
Sbjct: 252 ------NNLNSLDKINQLEAALEKEQSLVWPIFQRVSVLKKDLAKAYREEEAYWKQKSRQ 305
Query: 357 KWLVEGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSVAW 415
KWL G+ NS +FH+ + R+ I L V+G + +F FKS
Sbjct: 306 KWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNGNMQTSEAAKGEVAAAYFGNLFKSS-- 363
Query: 416 LRPKLDGVQFSTITDRD----NTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRF 471
P FS + R N LVG EIK+AV+ +PGPDG + F Q +
Sbjct: 364 -NPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEAVFSIKPASAPGPDGMSALFFQHY 422
Query: 472 WDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPK 531
W + V V++F+ G P N + + LIPK + P + D RPISL +YK++ K
Sbjct: 423 WSTVGNQVTSEVKKFFADGIMPAEWNYTHLCLIPKTQHPTEMVDLRPISLCSVLYKIISK 482
Query: 532 LFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPSIFF---KVDYEK 588
+ A RL+ + +++ + Q FV R + D+++V +E++H KV R S F K D K
Sbjct: 483 IMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSK 542
Query: 589 AYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILL 648
AY+ VEW +L ++ + F KW+ WIM C++S T SVL+N P G +++GLRQG L
Sbjct: 543 AYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPL 602
Query: 649 RRF 651
F
Sbjct: 603 SPF 605
>emb|CAA66812.1| non-ltr retrotransposon reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 893
Score = 228 bits (581), Expect = 9e-58
Identities = 172/609 (28%), Positives = 297/609 (48%), Gaps = 52/609 (8%)
Query: 105 VIINIYSPCDIDGKRAMWDALCAWR-GTCDVGE-WCLAGDFNAVRYAEERRGSSGVLSSH 162
V+ +Y+ + ++ +W+ L VG W + GDFN + E S + ++
Sbjct: 105 VVSIVYASNEEGTRKELWNELVQLALSPVVVGRSWIVLGDFNQILNPE-----SAINANI 159
Query: 163 RREMREFNVFIANMDLVDIPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHL 220
R++R F + + DL D+ G +TW + ++ +IDR LV+ W ++P+
Sbjct: 160 GRKIRAFRSCLLDSDLYDLVYKGSSYTWWNKCSSRPLAKKIDRILVNDHWNTLFPSAYAN 219
Query: 221 VLNRDVSDH--CPLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYV 278
D SDH C +++ V +PFR N +L +P + ++W + G Y
Sbjct: 220 FGEPDFSDHSSCEVVLDPAVLK-AKRPFRFFNYFLHNPDFLQLIRENWYSCNVSGSAMYR 278
Query: 279 LKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLL-- 336
+ +KLK L+ + ++E + D+ EK V + + + + + + L +V L
Sbjct: 279 VSKKLKHLKLPICCFSRENYSDI----EKRVSEAHAIVLHRQRITLTNPSVVHATLELEA 334
Query: 337 IEFWRVL-RYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDE 394
W++L + +ESF CQK+ + WL EGD N+ +FH + ++ ++I L+ D G +
Sbjct: 335 TRKWQILAKAEESFFCQKSSISWLYEGDNNTAYFHKMADMRKSINTINFLIDDFGERIET 394
Query: 395 PVRVKAAIHDFFQKKFKSVAWLRPKLDGVQ---------------FSTITDRDNTELVGP 439
+K I + F+S+ L GV+ F D+ N +L
Sbjct: 395 QQGIKEGIKEHSCNFFESL------LCGVEGENSLAQSDMNLLLSFRCSVDQIN-DLERS 447
Query: 440 FDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNAS 499
F +++I++A + K+ GPDG++ F + W V+ +V +AV+EF+ G + NA+
Sbjct: 448 FSDLDIQEAFFSLPRNKASGPDGYSSEFFKGVWFVVGPEVTEAVQEFFRSGQLLKQWNAT 507
Query: 500 FIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNML 559
+VLIPK+ + DFRPIS + +YKV+ KL RL+ ++ +VI Q F+ R +
Sbjct: 508 TLVLIPKITNSSKMTDFRPISCLNTLYKVIAKLLTSRLKKLLNEVISPSQSAFLPGRLLS 567
Query: 560 DSVVVLNEVMHEAKVKKRPSI-FFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGC 618
++V++ E++H K S KVD KA++SV W F+ + K++ WI C
Sbjct: 568 ENVLLATEIVHGYNTKNISSRGMLKVDLRKAFDSVRWDFIISAFRALAVPEKFVCWINQC 627
Query: 619 LNSATVSVLVNGSPSGEFHMEKGLRQG--------ILLRRFFS*LL-LKA*MGYSRMQLE 669
+++ SV+VNGS SG F KGLRQG +L FS LL + GY + +
Sbjct: 628 ISTPYFSVMVNGSSSGFFKSNKGLRQGDPLSPYLFVLAMEVFSSLLKARFDAGYIQYHPK 687
Query: 670 LGNLKVSRL 678
+L +S L
Sbjct: 688 TADLSISHL 696
>dbj|BAB01845.1| non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 893
Score = 228 bits (580), Expect = 1e-57
Identities = 172/609 (28%), Positives = 296/609 (48%), Gaps = 52/609 (8%)
Query: 105 VIINIYSPCDIDGKRAMWDALCAWR-GTCDVGE-WCLAGDFNAVRYAEERRGSSGVLSSH 162
V+ +Y+ + ++ +W+ L VG W + GDFN + E S + ++
Sbjct: 105 VVSIVYASNEEGTRKELWNELVQLALSPVVVGRSWIVLGDFNQILNPE-----SAINANI 159
Query: 163 RREMREFNVFIANMDLVDIPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHL 220
R++R F + + DL D+ G +TW + ++ +IDR LV+ W ++P+
Sbjct: 160 GRKIRAFRSCLLDSDLYDLVYKGSSYTWWNKCSSRPLAKKIDRILVNDHWNTLFPSAYAN 219
Query: 221 VLNRDVSDH--CPLLMRQVVADWGPKPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYV 278
D SDH C +++ V +PFR N +L +P + ++W + G Y
Sbjct: 220 FGEPDFSDHSSCEVVLDPAVLK-AKRPFRFFNYFLHNPDFLQLIRENWYSCNVSGSAMYR 278
Query: 279 LKEKLKGLRGSLRI*NKEVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLL-- 336
+ +KLK L+ + ++E + D+ EK V + + + + + + L +V L
Sbjct: 279 VSKKLKHLKLPICCFSRENYSDI----EKRVSEAHAIVLHRQRITLTNPSVVHATLELEA 334
Query: 337 IEFWRVL-RYQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDE 394
W++L + +ESF CQK+ + WL EGD N+ +FH + ++ ++I L+ D G +
Sbjct: 335 TRKWQILAKAEESFFCQKSSISWLYEGDNNTAYFHKMADMRKSINTINFLIDDFGERIET 394
Query: 395 PVRVKAAIHDFFQKKFKSVAWLRPKLDGVQ---------------FSTITDRDNTELVGP 439
+K I + F+S+ L GV+ F D+ N +L
Sbjct: 395 QQGIKEGIKEHSCNFFESL------LCGVEGENSLAQSDMNLLLSFRCSVDQIN-DLERS 447
Query: 440 FDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNAS 499
F +++I++A + K+ GPDG++ F + W V+ +V +AV+EF+ G + NA+
Sbjct: 448 FSDLDIQEAFFSLPRNKASGPDGYSSEFFKGVWFVVGPEVTEAVQEFFRSGQLLKQWNAT 507
Query: 500 FIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNML 559
+VLIPK+ + DFRPIS + +YKV+ KL RL+ ++ +VI Q F+ R +
Sbjct: 508 TLVLIPKITNSSKMTDFRPISCLNTLYKVIAKLLTSRLKKLLNEVISPSQSAFLPGRLLS 567
Query: 560 DSVVVLNEVMHEAKVKKRPSI-FFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGC 618
++V++ E++H K S KVD KA++SV W F+ + K++ WI C
Sbjct: 568 ENVLLATEIVHGYNTKNISSRGMLKVDLRKAFDSVRWDFIISAFRALAVPEKFVCWINQC 627
Query: 619 LNSATVSVLVNGSPSGEFHMEKGLRQG--------ILLRRFFS*LL-LKA*MGYSRMQLE 669
+++ SV+VNGS SG F KGLRQG +L FS LL + GY +
Sbjct: 628 ISTPYFSVMVNGSSSGFFKSNKGLRQGDPLSPYLFVLAMEVFSSLLKARFDAGYIHYHPK 687
Query: 670 LGNLKVSRL 678
+L +S L
Sbjct: 688 TADLSISHL 696
>gb|AAM82604.1| putative AP endonuclease/reverse transcriptase [Brassica napus]
Length = 1214
Score = 223 bits (568), Expect = 3e-56
Identities = 161/550 (29%), Positives = 261/550 (47%), Gaps = 34/550 (6%)
Query: 117 GKRAMWDAL--CAWRGTCDVGEWCLAGDFN-AVRYAEERRGSSGVLSSHRREMREFNVFI 173
G+R +W L A T W + GDFN ++ + G S + R M EF +
Sbjct: 116 GRRRLWSELELLAANQTTSDKPWIILGDFNQSLDPVDASTGGSRIT----RGMEEFRECL 171
Query: 174 ANMDLVDIPLTGRRFTW--RRANDQAQSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCP 231
++ D+P G +TW + N+ +IDR LV+ SW P + SDHCP
Sbjct: 172 LTSNISDLPFRGNHYTWWNNQENNPIAKKIDRILVNDSWLIASPLSYGSFCAMEFSDHCP 231
Query: 232 LLMRQVVADWGP-KPFRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSL 290
+ G KPF++ N + P + +W + QG + L +K K L+G++
Sbjct: 232 SCVNISNQSGGRNKPFKLSNFLMHHPEFIEKIRVTWDRLAYQGSAMFTLSKKSKFLKGTI 291
Query: 291 RI*NKEVFGDLKTRREKVVQKI-----NLLDVKEEEV-GLQPEELVEMRDLLIEFWRVLR 344
R N+E + L+ R + Q + NLL + GL+ E +L +
Sbjct: 292 RTFNREHYSGLEKRVVQAAQNLKTCQNNLLAAPSSYLAGLEKEAHRSWAELALA------ 345
Query: 345 YQESFLCQKACMKWLVEGDLNSGFFHSTINWKRRADSIVGLMVD-GMWEDEPVRVKAAIH 403
+E FLCQK+ + WL GD N+ FFH + +R + I L+ G + ++
Sbjct: 346 -EERFLCQKSRVLWLKCGDSNTTFFHRMMTARRAINEIHYLLDQTGRRIENTDELQTHCV 404
Query: 404 DFFQKKFKSVAWLRPKLDGVQFSTIT----DRDNTELV-GPFDEVEIKQAVWECGGTKSP 458
DFF++ F S + L Q +++T D + +L+ E +IK + KSP
Sbjct: 405 DFFKELFGSSSHLISAEGISQINSLTRFKCDENTRQLLEAEVSEADIKSEFFALPSNKSP 464
Query: 459 GPDGFNFRFIQRFWDVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRP 518
GPDG+ F ++ W ++ ++ AV+EF+ G N++ + ++PK + +FRP
Sbjct: 465 GPDGYTSEFFKKTWSIVGPSLIAAVQEFFRSGRLLGQWNSTAVTMVPKKPNADRITEFRP 524
Query: 519 ISLIGCIYKVVPKLFAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMH---EAKVK 575
IS IYKV+ KL A RL ++ I Q FV R + ++V++ E++ +A +
Sbjct: 525 ISCCNAIYKVISKLLARRLENILPLWISPSQSAFVKGRLLTENVLLATELVQGFGQANIS 584
Query: 576 KRPSIFFKVDYEKAYNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGE 635
R + KVD KA++SV WGF+ + N +++ WI C+ S + S+ V+GS G
Sbjct: 585 SRGVL--KVDLRKAFDSVGWGFIIETLKAANAPPRFVNWIKQCITSTSFSINVSGSLCGY 642
Query: 636 FHMEKGLRQG 645
F KGLRQG
Sbjct: 643 FKGSKGLRQG 652
>pir||S65812 RNA-directed DNA polymerase (EC 2.7.7.49) (clone DW15) -
Arabidopsis thaliana retrotransposon Ta11-1
gi|976278|gb|AAA75254.1| reverse transcriptase
Length = 1333
Score = 222 bits (566), Expect = 5e-56
Identities = 178/662 (26%), Positives = 302/662 (44%), Gaps = 39/662 (5%)
Query: 5 NVRGLGVGGNAKKRVVRELVCQQHVELLCIQETK-CQVVDRRMCGLL*GDSEFEWKATPA 63
N +GLG + + E+ E+L + ETK C V + L + F
Sbjct: 7 NCQGLGWSQDLTIPRLMEMRLSHFPEVLFLMETKNCSNVVVDLQEWLGYERVF---TVNP 63
Query: 64 VNRGGGLLCIWNSKLFVLEESVDGPGFLGLMGRWGAAQQRCVIINIYSPCDIDGKRAMWD 123
+ GGL W + ++ + D + ++G+ + Y C + G A D
Sbjct: 64 IGLSGGLALFWKKGVDIVIKYAD-KNLIDFQIQFGSHE-------FYVSC-VYGNPAFSD 114
Query: 124 ALCAWRGTCDVG-----EWCLAGDFNAVRYAEERRGSSGVLSSHRREMREFNVFIANMDL 178
W +G WC+ GDFN + + E+RG S F + + D+
Sbjct: 115 KHLVWEKITRIGINRKEPWCMLGDFNPILHNGEKRGGPRRGDS---SFLPFTDMLDSCDM 171
Query: 179 VDIPLTGRRFTWRRANDQA--QSRIDRFLVSTSWCDVWPNCSHLVLNRDVSDHCPLLMRQ 236
+++P G FTW ++ QSR+DR + +W +P + L++ SDH P+L+R
Sbjct: 172 LELPSIGNPFTWGGKTNEMWIQSRLDRCFGNKNWFRFFPISNQEFLDKRGSDHRPVLVRL 231
Query: 237 VVADWGPKP-FRVLNCWLDDPRLFPFVEQSWKGMKIQGWGAYVLKEKLKGLRGSLRI*NK 295
+ FR + P + + Q+W G + + VL +KLK R +L K
Sbjct: 232 TKTKEEYRGNFRFDKRLFNQPNVKETIVQAWNGSQ-RNENLLVL-DKLKHCRSALSRWKK 289
Query: 296 EVFGDLKTRREKVVQKINLLDVKEEEVGLQPEELVEMRDLLIEFWRVLRYQESFLCQKAC 355
E + TR + + L E+ G +LV L + + +E F QK+
Sbjct: 290 ENNINSSTRITQARAALEL----EQSSGFPRADLVF--SLKNDLCKANHDEEVFWSQKSR 343
Query: 356 MKWLVEGDLNSGFFHSTINWKRRADSIVGLM-VDGMWEDEPVRVKAAIHDFFQKKFKSV- 413
KW+ GD N+ FFH+++ R I L V+G++ + + A +F FKS
Sbjct: 344 AKWMHSGDKNTSFFHASVKDNRGKQHIDQLCDVNGLFHKDEMNKGAIAEAYFSDLFKSTD 403
Query: 414 -AWLRPKLDGVQFSTITDRDNTELVGPFDEVEIKQAVWECGGTKSPGPDGFNFRFIQRFW 472
+ + Q +T+ N L+ + EI++AV+ + +PG DGF F Q++W
Sbjct: 404 PSSFVDLFEDYQ-PRVTESMNNTLIAAVSKNEIREAVFAIRSSSAPGVDGFTGFFFQKYW 462
Query: 473 DVLSGDVVKAVREFWMRGA*PRGSNASFIVLIPKVKIPQLLNDFRPISLIGCIYKVVPKL 532
++ V K ++ F++ G P+ N + + L+PK K P + D RPISL +YK++ K+
Sbjct: 463 SIICLQVTKEIQNFFLLGYFPKSWNFTHLCLLPKKKKPDKMTDLRPISLCSVLYKIISKI 522
Query: 533 FAIRLRLVIGKVIDERQFTFVGNRNMLDSVVVLNEVMHEAKVKKRPS---IFFKVDYEKA 589
RL+ + ++ Q FV R + D++++ +EV+H + K S I K + KA
Sbjct: 523 MVRRLQPFLPDLVSPNQSAFVAERLIFDNILIAHEVVHGLRTHKSVSKGFIAIKSNMSKA 582
Query: 590 YNSVEWGFLEYMMGRMNFCAKWIGWIMGCLNSATVSVLVNGSPSGEFHMEKGLRQGILLR 649
++ VEW ++ ++ + F KW+GWIM ++S + SVL+N G +GLRQG L
Sbjct: 583 FDRVEWNYVRALLDALGFHQKWVGWIMFMISSVSYSVLINDKAFGNIVPSRGLRQGDPLS 642
Query: 650 RF 651
F
Sbjct: 643 PF 644
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.339 0.151 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,992,950,347
Number of Sequences: 2540612
Number of extensions: 85079128
Number of successful extensions: 227605
Number of sequences better than 10.0: 891
Number of HSP's better than 10.0 without gapping: 621
Number of HSP's successfully gapped in prelim test: 270
Number of HSP's that attempted gapping in prelim test: 225491
Number of HSP's gapped (non-prelim): 1239
length of query: 1183
length of database: 863,360,394
effective HSP length: 139
effective length of query: 1044
effective length of database: 510,215,326
effective search space: 532664800344
effective search space used: 532664800344
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 81 (35.8 bits)
Lotus: description of TM0058a.1


