

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0590c.6
(364 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
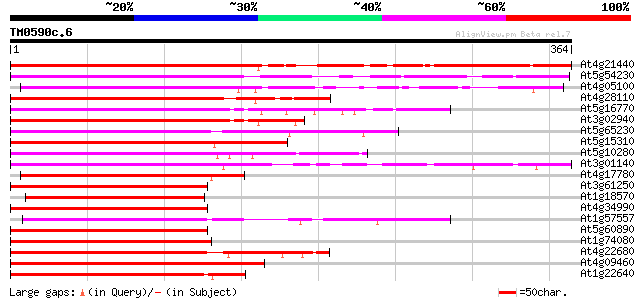
Score E
Sequences producing significant alignments: (bits) Value
At4g21440 myb-related protein M4 290 6e-79
At5g54230 Myb-related transcription factor-like protein 278 4e-75
At4g05100 MYB - like protein 273 1e-73
At4g28110 putative transcription factor (MYB41) 246 2e-65
At5g16770 putative transcription factor (MYB9) 233 1e-61
At3g02940 MYB family transcription factor like protein 231 4e-61
At5g65230 transcription factor-like protein 224 4e-59
At5g15310 myb-related protein - like 222 2e-58
At5g10280 putative transcription factor MYB92 221 6e-58
At3g01140 putative Myb-related transcription factor 219 1e-57
At4g17780 MYB transcription factor like protein 207 6e-54
At3g61250 putative transcription factor (MYB17) 201 5e-52
At1g18570 unknown protein 199 2e-51
At4g34990 MYB-like protein 197 7e-51
At1g57557 unknown protein 196 2e-50
At5g60890 Myb transcription factor homolog (ATR1) 194 5e-50
At1g74080 putative transcription factor 194 5e-50
At4g22680 myb-like protein 193 1e-49
At4g09460 DNA-binding protein 193 1e-49
At1g22640 transcription factor like protein (MYB3) 193 1e-49
>At4g21440 myb-related protein M4
Length = 350
Score = 290 bits (743), Expect = 6e-79
Identities = 181/376 (48%), Positives = 233/376 (61%), Gaps = 39/376 (10%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
MA+S EKNG+KKGPWT EEDQKLVDYIQKHG+G WRTLPKNAGL+RCGKSCRLRW NY
Sbjct: 1 MARSPCCEKNGLKKGPWTSEEDQKLVDYIQKHGYGNWRTLPKNAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG+FS EEEE IIQLHS LGNKWS IAA L GRTDNEIKN+WNTHI+KKLL+MG
Sbjct: 61 LRPDIKRGRFSFEEEETIIQLHSFLGNKWSAIAARLPGRTDNEIKNFWNTHIRKKLLRMG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMER---------VVNPSQLQLLN 171
+DPVTH+PRLD+L ++SIL SSLYN+S+ N M+ +VNP ++L
Sbjct: 121 IDPVTHSPRLDLLDISSILASSLYNSSSHHMNMSRLMMDTNRRHHQQHPLVNP---EILK 177
Query: 172 LVTTLLSCQNTNQDVWNTNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVNEEN 231
L T+L S QN NQ++ +TQ Q + ++ +Q NQ E
Sbjct: 178 LATSLFS-QNQNQNL-------------VVDHDSRTQEKQTVYSQTGVNQYQTNQYFEN- 222
Query: 232 HCTTSNPLHILEPPQLMKTNLEHQISPIASPFNHQNPMQNLLQYNGGFTSNRKRPEPSSA 291
T + L PP N Q + + FN QNL+ + TS + PS
Sbjct: 223 --TITQELQSSMPP---FPNEARQFNNMDHHFNGFGE-QNLV--STSTTSVQDCYNPSFN 274
Query: 292 TQSFSSLLCN-SEGMPNFNLSSLLSSTPSSS-SPSTLNSSSSTTFVKGTNEDERDTY-GN 348
S S+ + + S +FN ++ + +TPSSS SP+TLNSS + T EDE ++Y N
Sbjct: 275 DYSSSNFVLDPSYSDQSFNFANSVLNTPSSSPSPTTLNSSYINSSSCST-EDEIESYCSN 333
Query: 349 MFMYNISNGLNDSGLL 364
+ ++I + L+ +G +
Sbjct: 334 LMKFDIPDFLDVNGFI 349
>At5g54230 Myb-related transcription factor-like protein
Length = 319
Score = 278 bits (710), Expect = 4e-75
Identities = 165/364 (45%), Positives = 218/364 (59%), Gaps = 49/364 (13%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M KS+S E++ VKKGPWTPEED+KLV YIQ HG GKWRTLPKNAGLKRCGKSCRLRW NY
Sbjct: 1 MGKSSSSEESEVKKGPWTPEEDEKLVGYIQTHGPGKWRTLPKNAGLKRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG+FSL+EEE IIQLH +LGNKWS IA +L GRTDNEIKNYWNTHIKKKLL+MG
Sbjct: 61 LRPDIKRGEFSLQEEETIIQLHRLLGNKWSAIAIHLPGRTDNEIKNYWNTHIKKKLLRMG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTTLLSCQ 180
+DPVTH PR+++LQL+S L SSL+ + +Q N P L N+ +L+
Sbjct: 121 IDPVTHCPRINLLQLSSFLTSSLFKSMSQPMN----------TPFDLTTSNINPDILNHL 170
Query: 181 NTNQDVWNTNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVNEENHCTTSNPLH 240
+ + T +YQ NQ QL N ++ Q TT L
Sbjct: 171 TASLNNVQTESYQPNQ-----------------QLQNDLNTDQ----------TTFTGLL 203
Query: 241 ILEPPQLMKTNLEHQISPIASPFNHQNPMQNLLQYNGGFTSNRKRPEPSSATQSFSSLLC 300
PP + N E+ + S +P N + G ++S + + ++F +
Sbjct: 204 NSTPPVQWQNNGEY-LGDYHSYTGTGDPSNNKVPQAGNYSSAAFVSDHINDGENFKA--- 259
Query: 301 NSEGMPNFNLSSLLSSTPSSSSPSTLNSSSSTTFVKGTNEDERDTYG-NMFMYNISNGLN 359
+N SS + + SSSS + LN SSST +V G +ED+R+++G +M M++ + N
Sbjct: 260 ------GWNFSSSMLAGTSSSSSTPLN-SSSTFYVNGGSEDDRESFGSDMLMFHHHHDHN 312
Query: 360 DSGL 363
++ L
Sbjct: 313 NNAL 316
>At4g05100 MYB - like protein
Length = 324
Score = 273 bits (697), Expect = 1e-73
Identities = 169/366 (46%), Positives = 214/366 (58%), Gaps = 70/366 (19%)
Query: 8 EKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKR 67
+KNG+KKGPWTPEEDQKL+DYI HG+G WRTLPKNAGL+RCGKSCRLRW NYLRPDIKR
Sbjct: 9 KKNGLKKGPWTPEEDQKLIDYINIHGYGNWRTLPKNAGLQRCGKSCRLRWTNYLRPDIKR 68
Query: 68 GKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHT 127
G+FS EEEE IIQLHS++GNKWS IAA L GRTDNEIKNYWNTHI+K+LLKMG+DPVTHT
Sbjct: 69 GRFSFEEEETIIQLHSIMGNKWSAIAARLPGRTDNEIKNYWNTHIRKRLLKMGIDPVTHT 128
Query: 128 PRLDVLQLASILNSSLYNNS-------AQFNNPGLFGM-----ERVVNPSQLQLLNLVTT 175
PRLD+L ++SIL+SS+YN+S Q N M + +VNP ++L L T+
Sbjct: 129 PRLDLLDISSILSSSIYNSSHHHHHHHQQHMNMSRLMMSDGNHQPLVNP---EILKLATS 185
Query: 176 LLSCQNTNQDVWNTNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVNEENHCTT 235
L S QN + N Q ++ Q+QT + P NEE
Sbjct: 186 LFSNQNHPNNTHENNTVNQTEVN-----QYQTGYNMP--------------GNEE----- 221
Query: 236 SNPLHILEPPQLMKTNLEHQISPIASPFNHQNPMQNLLQYNGGFTSNRKRPEPSSATQSF 295
L P TN + + P+ + +QN L Y+ + + EP + F
Sbjct: 222 ---LQSWFPIMDQFTNFQ-DLMPM------KTTVQNSLSYDDDCSKSNFVLEPYYS--DF 269
Query: 296 SSLLCNSEGMPNFNLSSLLSSTPSSSSPSTLNSSSSTTFVKGT--NEDERDTYGNMFMYN 353
+S+L T SSSP+ LNSSSST T EDE+++Y + + N
Sbjct: 270 ASVL-----------------TTPSSSPTPLNSSSSTYINSSTCSTEDEKESYYSDNITN 312
Query: 354 ISNGLN 359
S +N
Sbjct: 313 YSFDVN 318
>At4g28110 putative transcription factor (MYB41)
Length = 282
Score = 246 bits (627), Expect = 2e-65
Identities = 127/212 (59%), Positives = 152/212 (70%), Gaps = 15/212 (7%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S +KNGVKKGPWT EEDQKL+DYI+ HG G WRTLPKNAGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCDKNGVKKGPWTAEEDQKLIDYIRFHGPGNWRTLPKNAGLHRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG+FS EEEE IIQLHSV+GNKWS IAA L GRTDNEIKN+WNTHI+K+L++ G
Sbjct: 61 LRPDIKRGRFSFEEEETIIQLHSVMGNKWSAIAARLPGRTDNEIKNHWNTHIRKRLVRSG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGM----ERVVNPSQLQLLNLVTTL 176
+DPVTH+PRLD+L L+S+L SA FN P + ++NP L+L +L L
Sbjct: 121 IDPVTHSPRLDLLDLSSLL-------SALFNQPNFSAVATHASSLLNPDVLRLASL---L 170
Query: 177 LSCQNTNQDVWNTNNYQQNQLGGSTQWQHQTQ 208
L QN N V+ +N Q Q ++ Q Q
Sbjct: 171 LPLQNPN-PVYPSNLDQNLQTPNTSSESSQPQ 201
>At5g16770 putative transcription factor (MYB9)
Length = 336
Score = 233 bits (594), Expect = 1e-61
Identities = 138/315 (43%), Positives = 182/315 (56%), Gaps = 38/315 (12%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S ++NG+KKGPWT EED KL+D+IQKHGHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCDENGLKKGPWTQEEDDKLIDHIQKHGHGSWRALPKQAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG F+ EEE+ II LHS+LGNKWS+IA NL GRTDNEIKNYWNTH++KKLL+MG
Sbjct: 61 LRPDIKRGNFTEEEEQTIINLHSLLGNKWSSIAGNLPGRTDNEIKNYWNTHLRKKLLQMG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVV-----NPSQLQLLNLVTT 175
+DPVTH PR D L + + L + +A FN+ L + + V ++ QLL+ +
Sbjct: 121 IDPVTHRPRTDHLNVLAALPQLI--AAANFNS--LLNLNQNVQLDATTLAKAQLLHTMIQ 176
Query: 176 LLS--------------CQNTNQDVWNTNNYQQNQ--LGGSTQWQHQTQCSQPMQ----L 215
+LS QN+N +++ +Y +NQ G S + H + M +
Sbjct: 177 VLSTNNNTTNPSFSSSTMQNSNTNLFGQASYLENQNLFGQSQNFSHILEDENLMVKTQII 236
Query: 216 DNSIHAF----QPNQVNEENHCTTSNPLHILEPPQLMKTNLEHQISPIASPFNHQNPMQN 271
DN + +F QP ++ N S PL + P+ K + I H + N
Sbjct: 237 DNPLDSFSSPIQPGFQDDHN----SLPLLVPASPEESK-ETQRMIKNKDIVDYHHHDASN 291
Query: 272 LLQYNGGFTSNRKRP 286
N FT + P
Sbjct: 292 PSSSNSTFTQDHHHP 306
>At3g02940 MYB family transcription factor like protein
Length = 321
Score = 231 bits (590), Expect = 4e-61
Identities = 114/198 (57%), Positives = 146/198 (73%), Gaps = 11/198 (5%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S +++G+KKGPWTPEEDQKL+++I+KHGHG WR LPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCDESGLKKGPWTPEEDQKLINHIRKHGHGSWRALPKQAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG F+ EEE+ II LHS+LGNKWS+IA +L GRTDNEIKNYWNTHI+KKL++MG
Sbjct: 61 LRPDIKRGNFTAEEEQTIINLHSLLGNKWSSIAGHLPGRTDNEIKNYWNTHIRKKLIQMG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMER-----VVNPSQLQLLNLVTT 175
+DPVTH PR D L + + L L +A FNN L + + + ++ QLL+ +
Sbjct: 121 IDPVTHRPRTDHLNVLAALPQLL--AAANFNN--LLNLNQNIQLDATSVAKAQLLHSMIQ 176
Query: 176 LLSCQNTNQ--DVWNTNN 191
+LS NT+ D+ +T N
Sbjct: 177 VLSNNNTSSSFDIHHTTN 194
>At5g65230 transcription factor-like protein
Length = 310
Score = 224 bits (572), Expect = 4e-59
Identities = 123/264 (46%), Positives = 158/264 (59%), Gaps = 18/264 (6%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S S ++ G+KKGPW PEED KL++YI KHGH W LPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPSSDETGLKKGPWLPEEDDKLINYIHKHGHSSWSALPKLAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRGKFS EEEE I+ LH+VLGNKWS IA++L GRTDNEIKN+WNTH+KKKL++MG
Sbjct: 61 LRPDIKRGKFSAEEEETILNLHAVLGNKWSMIASHLPGRTDNEIKNFWNTHLKKKLIQMG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTTLLSCQ 180
DP+TH PR D + SSL + N GL +++ LLNL T + Q
Sbjct: 121 FDPMTHQPRTD------DIFSSLSQLMSLSNLRGLVDLQQQFPMEDQALLNLQTEMAKLQ 174
Query: 181 ------NTNQDVWNTNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVN-----E 229
+ + NN N L + T L + + F N + +
Sbjct: 175 LFQYLLQPSPAPMSINNINPNILNLLIKENSVTSNIDLGFLSSHLQDFNNNNLPSLKTLD 234
Query: 230 ENHCT-TSNPLHILEPPQLMKTNL 252
+NH + ++P+ + EPP L +T L
Sbjct: 235 DNHFSQNTSPIWLHEPPSLNQTML 258
>At5g15310 myb-related protein - like
Length = 326
Score = 222 bits (566), Expect = 2e-58
Identities = 105/182 (57%), Positives = 137/182 (74%), Gaps = 2/182 (1%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S +K G+KKGPWTPEEDQKL+ YI++HGHG WR+LP+ AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCDKLGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLHRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRGKF+L+EE+ IIQLH++LGN+WS IA +L RTDNEIKNYWNTH+KK+L+KMG
Sbjct: 61 LRPDIKRGKFNLQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLVKMG 120
Query: 121 LDPVTHTPRLD--VLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTTLLS 178
+DPVTH P+ + + L N+++ +++AQ+ + L R+ S+L L T S
Sbjct: 121 IDPVTHKPKNETPLSSLGLSKNAAILSHTAQWESARLEAEARLARESKLLHLQHYQTKTS 180
Query: 179 CQ 180
Q
Sbjct: 181 SQ 182
>At5g10280 putative transcription factor MYB92
Length = 334
Score = 221 bits (562), Expect = 6e-58
Identities = 116/260 (44%), Positives = 156/260 (59%), Gaps = 30/260 (11%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S + +G+KKGPWTP+ED+KLV+Y+QKHGH WR LPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPISDDSGLKKGPWTPDEDEKLVNYVQKHGHSSWRALPKLAGLNRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG+FS +EE+ I+ LHSVLGNKWSTIA L GRTDNEIKN+WNTH+KKKL++MG
Sbjct: 61 LRPDIKRGRFSPDEEQTILNLHSVLGNKWSTIANQLPGRTDNEIKNFWNTHLKKKLIQMG 120
Query: 121 LDPVTHTPRLDVL----QLASILNS-----------------SLYNNSAQFNNPGLF--- 156
DP+TH PR D+ QL S+ ++ ++ + LF
Sbjct: 121 FDPMTHRPRTDIFSGLSQLMSLSSNLRGFVDLQQQFPIDQEHTILKLQTEMAKLQLFQYL 180
Query: 157 ----GMERVVNPSQLQLLNLVTTLLSCQNTNQDVWNTNNYQQNQLGGSTQWQHQTQCSQP 212
M VNP+ L+L+ ++ S + T+ + +NN LG Q H +
Sbjct: 181 LQPSSMSNNVNPNDFDTLSLLNSIASFKETSNNT-TSNNLDLGFLGSYLQDFHSLPSLKT 239
Query: 213 MQLDNSIHAFQPNQVNEENH 232
+ + + P + ++NH
Sbjct: 240 LNSNMEPSSVFPQNL-DDNH 258
>At3g01140 putative Myb-related transcription factor
Length = 345
Score = 219 bits (559), Expect = 1e-57
Identities = 142/382 (37%), Positives = 201/382 (52%), Gaps = 56/382 (14%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S +K G+KKGPWTPEEDQKL+ YI++HGHG WR+LP+ AGL+RCGKSCRLRW NY
Sbjct: 1 MGRSPCCDKAGLKKGPWTPEEDQKLLAYIEEHGHGSWRSLPEKAGLQRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRGKF+++EE+ IIQLH++LGN+WS IA +L RTDNEIKNYWNTH+KK+L+KMG
Sbjct: 61 LRPDIKRGKFTVQEEQTIIQLHALLGNRWSAIATHLPKRTDNEIKNYWNTHLKKRLIKMG 120
Query: 121 LDPVTHTPRLDVLQLAS--ILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTTLLS 178
+DPVTH + + L ++ N++ ++ AQ+ + L R+ S+L L
Sbjct: 121 IDPVTHKHKNETLSSSTGQSKNAATLSHMAQWESARLEAEARLARESKLLHL-------- 172
Query: 179 CQNTNQDVWNTNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVNEENHCTTSNP 238
Q N NN ++ + Q TQ S + +PNQ N +
Sbjct: 173 -----QHYQNNNNLNKS---AAPQQHCFTQ-------KTSTNWTKPNQGNGDQQ------ 211
Query: 239 LHILEPPQLMKTNLEHQISPIASPFNHQNPMQNLLQYNGGFTSNRKRPEPSSATQSFSSL 298
LE P T E+ + P+ P + N N +S SS+T S SL
Sbjct: 212 ---LESPTSTVTFSENLLMPLGIPTDSSRNRNN----NNNESSAMIELAVSSSTSSDVSL 264
Query: 299 L------------CNSEGMPNFNLSSLLSSTPSSSSPSTLNSSSSTTFVKGTNE----DE 342
+ C S G+ S L+ + P+ N +T V +E ++
Sbjct: 265 VKEHEHDWIRQINCGSGGIGEGFTSLLIGDSVGRGLPTGKN--EATAGVGNESEYNYYED 322
Query: 343 RDTYGNMFMYNISNGLNDSGLL 364
Y N + + + +DS +
Sbjct: 323 NKNYWNSILNLVDSSPSDSATM 344
>At4g17780 MYB transcription factor like protein
Length = 745
Score = 207 bits (528), Expect = 6e-54
Identities = 98/153 (64%), Positives = 118/153 (77%), Gaps = 8/153 (5%)
Query: 8 EKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKR 67
+ GVKKGPW PEED KL YI ++G+G WR+LPK AGL RCGKSCRLRW NYLRPDI+R
Sbjct: 9 QDKGVKKGPWLPEEDDKLTAYINENGYGNWRSLPKLAGLNRCGKSCRLRWMNYLRPDIRR 68
Query: 68 GKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHT 127
GKFS EE I++LH++LGNKWS IA +L GRTDNEIKNYWNTH++KKLL+MG+DPVTH
Sbjct: 69 GKFSDGEESTIVRLHALLGNKWSKIAGHLPGRTDNEIKNYWNTHMRKKLLQMGIDPVTHE 128
Query: 128 PR-------LDVLQ-LASILNSSLYNNSAQFNN 152
PR LDV Q LA+ +N+ + N+ NN
Sbjct: 129 PRTNDLSPILDVSQMLAAAINNGQFGNNNLLNN 161
>At3g61250 putative transcription factor (MYB17)
Length = 299
Score = 201 bits (511), Expect = 5e-52
Identities = 89/128 (69%), Positives = 107/128 (83%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ +K G+KKGPWTPEED+ LV +I+K+GHG WRTLPK AGL RCGKSCRLRW NY
Sbjct: 1 MGRTPCCDKIGLKKGPWTPEEDEVLVAHIKKNGHGSWRTLPKLAGLLRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG F+ +EE+ +IQLH++LGN+W+ IAA L GRTDNEIKN WNTH+KK+LL MG
Sbjct: 61 LRPDIKRGPFTADEEKLVIQLHAILGNRWAAIAAQLPGRTDNEIKNLWNTHLKKRLLSMG 120
Query: 121 LDPVTHTP 128
LDP TH P
Sbjct: 121 LDPRTHEP 128
>At1g18570 unknown protein
Length = 352
Score = 199 bits (507), Expect = 2e-51
Identities = 87/116 (75%), Positives = 101/116 (87%)
Query: 11 GVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRGKF 70
G+KKG WTPEEDQKL+ Y+ +HG G WRTLP+ AGLKRCGKSCRLRWANYLRPDIKRG+F
Sbjct: 12 GLKKGAWTPEEDQKLLSYLNRHGEGGWRTLPEKAGLKRCGKSCRLRWANYLRPDIKRGEF 71
Query: 71 SLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTH 126
+ +EE +II LH++ GNKWS IA L GRTDNEIKNYWNTHIKK+L+K G+DPVTH
Sbjct: 72 TEDEERSIISLHALHGNKWSAIARGLPGRTDNEIKNYWNTHIKKRLIKKGIDPVTH 127
>At4g34990 MYB-like protein
Length = 274
Score = 197 bits (501), Expect = 7e-51
Identities = 86/128 (67%), Positives = 104/128 (81%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK+ KG WT EED KL+ YI+ HG G WR+LP++AGL+RCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKDHTNKGAWTKEEDDKLISYIKAHGEGCWRSLPRSAGLQRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG F+LEE++ II+LHS+LGNKWS IA L GRTDNEIKNYWNTH+K+KLL+ G
Sbjct: 61 LRPDLKRGNFTLEEDDLIIKLHSLLGNKWSLIATRLPGRTDNEIKNYWNTHVKRKLLRKG 120
Query: 121 LDPVTHTP 128
+DP TH P
Sbjct: 121 IDPATHRP 128
>At1g57557 unknown protein
Length = 314
Score = 196 bits (497), Expect = 2e-50
Identities = 111/282 (39%), Positives = 157/282 (55%), Gaps = 42/282 (14%)
Query: 9 KNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANYLRPDIKRG 68
K ++KG W+PEED+KL++YI KHGHG W ++PK AGL+RCGKSCRLRW NYLRPD+KRG
Sbjct: 9 KQKLRKGLWSPEEDEKLLNYITKHGHGCWSSVPKLAGLERCGKSCRLRWINYLRPDLKRG 68
Query: 69 KFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMGLDPVTHTP 128
FS EE+ I++LH+VLGN+WS IAA L GRTDNEIKN WN+ IKKKL+K G+DP+TH P
Sbjct: 69 AFSSEEQNLIVELHAVLGNRWSQIAARLPGRTDNEIKNLWNSCIKKKLMKKGIDPITHKP 128
Query: 129 RLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTTLLSCQNTNQDVW- 187
+ ++ N S NNS F++ TNQD++
Sbjct: 129 ---LSEVGKETNRSDNNNSTSFSS----------------------------ETNQDLFV 157
Query: 188 -NTNNYQQNQLGGSTQWQHQTQCSQPMQLDNSIHAFQPNQVNEENHCTTSN--PLHILEP 244
T+++ + Q + S + L NS+ + P Q N ++ ++ +
Sbjct: 158 KKTSDFAEYS-------AFQKEESNSVSLRNSLSSMIPTQFNIDDGSVSNAGFDTQVCVK 210
Query: 245 PQLMKTNLEHQISPIASPFNHQNPMQNLLQYNGGFTSNRKRP 286
P ++ + S S +H N + + N G TS+ P
Sbjct: 211 PSIILLPPPNNTSSTVSGQDHVNVSEPNWESNSGTTSHLNNP 252
>At5g60890 Myb transcription factor homolog (ATR1)
Length = 295
Score = 194 bits (494), Expect = 5e-50
Identities = 84/128 (65%), Positives = 106/128 (82%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ ++ G+KKG WTPEEDQKL+ Y+ HG G WRTLP+ AGLKRCGKSCRLRWANY
Sbjct: 1 MVRTPCCKEEGIKKGAWTPEEDQKLIAYLHLHGEGGWRTLPEKAGLKRCGKSCRLRWANY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG+FS EE++ II+LH++ GNKW+ IA +L GRTDNEIKNYWNT++KK+L + G
Sbjct: 61 LRPDIKRGEFSPEEDDTIIKLHALKGNKWAAIATSLAGRTDNEIKNYWNTNLKKRLKQKG 120
Query: 121 LDPVTHTP 128
+D +TH P
Sbjct: 121 IDAITHKP 128
>At1g74080 putative transcription factor
Length = 333
Score = 194 bits (494), Expect = 5e-50
Identities = 87/131 (66%), Positives = 104/131 (78%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M ++ G+KKG WT EEDQKL+ Y+Q+HG G WRTLP AGLKRCGKSCRLRWANY
Sbjct: 1 MVRTPCCRAEGLKKGAWTQEEDQKLIAYVQRHGEGGWRTLPDKAGLKRCGKSCRLRWANY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPDIKRG+FS +EE++II LH++ GNKWS IA + RTDNEIKN+WNTHIKK L+K G
Sbjct: 61 LRPDIKRGEFSQDEEDSIINLHAIHGNKWSAIARKIPRRTDNEIKNHWNTHIKKCLVKKG 120
Query: 121 LDPVTHTPRLD 131
+DP+TH LD
Sbjct: 121 IDPLTHKSLLD 131
>At4g22680 myb-like protein
Length = 266
Score = 193 bits (491), Expect = 1e-49
Identities = 106/219 (48%), Positives = 140/219 (63%), Gaps = 23/219 (10%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M + +K GVKKGPWT EED+KL+++I +GH WR LPK AGL+RCGKSCRLRW NY
Sbjct: 1 MGRQPCCDKLGVKKGPWTVEEDKKLINFILTNGHCCWRALPKLAGLRRCGKSCRLRWTNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG S +EE+ +I LH+ LGNKWS IA+ L GRTDNEIKN+WNTHIKKKLLKMG
Sbjct: 61 LRPDLKRGLLSHDEEQLVIDLHANLGNKWSKIASRLPGRTDNEIKNHWNTHIKKKLLKMG 120
Query: 121 LDPVTHTPRLDVLQLASILN---SSLYNNSAQFNNPGLFGMERVVNPSQLQLLNLVTT-- 175
+DP+TH P LN S++ N+ +NP +E ++ +++ TT
Sbjct: 121 IDPMTHQP----------LNQEPSNIDNSKTIPSNPDDVSVEPKTTNTKYVEISVTTTEE 170
Query: 176 ----LLSCQNTNQDVWN---TNNYQQNQLGGSTQWQHQT 207
++ QN++ D N N Y ++L S W +T
Sbjct: 171 ESSSTVTDQNSSMDNENHLIDNIYDDDEL-FSYLWSDET 208
>At4g09460 DNA-binding protein
Length = 236
Score = 193 bits (491), Expect = 1e-49
Identities = 92/165 (55%), Positives = 117/165 (70%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK KG WT EEDQ+LVDYI+ HG G WR+LPK+AGL RCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKAHTNKGAWTKEEDQRLVDYIRNHGEGCWRSLPKSAGLLRCGKSCRLRWINY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG F+ +E++ II+LHS+LGNKWS IA L GRTDNEIKNYWNTHIK+KLL G
Sbjct: 61 LRPDLKRGNFTDDEDQIIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLLSHG 120
Query: 121 LDPVTHTPRLDVLQLASILNSSLYNNSAQFNNPGLFGMERVVNPS 165
+DP TH + ++S + + N++ +++ L + N S
Sbjct: 121 IDPQTHRQINESKTVSSQVVVPIQNDAVEYSFSNLAVKPKTENSS 165
>At1g22640 transcription factor like protein (MYB3)
Length = 257
Score = 193 bits (490), Expect = 1e-49
Identities = 95/156 (60%), Positives = 114/156 (72%), Gaps = 5/156 (3%)
Query: 1 MAKSNSGEKNGVKKGPWTPEEDQKLVDYIQKHGHGKWRTLPKNAGLKRCGKSCRLRWANY 60
M +S EK + KG WT EEDQ LVDYI+KHG G WR+LP+ AGL+RCGKSCRLRW NY
Sbjct: 1 MGRSPCCEKAHMNKGAWTKEEDQLLVDYIRKHGEGCWRSLPRAAGLQRCGKSCRLRWMNY 60
Query: 61 LRPDIKRGKFSLEEEEAIIQLHSVLGNKWSTIAANLQGRTDNEIKNYWNTHIKKKLLKMG 120
LRPD+KRG F+ EE+E II+LHS+LGNKWS IA L GRTDNEIKNYWNTHIK+KLL G
Sbjct: 61 LRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIKRKLLSRG 120
Query: 121 LDPVTHTPRL---DVLQLASILNSSLYNNSAQFNNP 153
+DP +H RL V+ +S+ N + F+ P
Sbjct: 121 IDPNSH--RLINESVVSPSSLQNDVVETIHLDFSGP 154
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.311 0.128 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,266,206
Number of Sequences: 26719
Number of extensions: 425328
Number of successful extensions: 1813
Number of sequences better than 10.0: 226
Number of HSP's better than 10.0 without gapping: 184
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 1400
Number of HSP's gapped (non-prelim): 322
length of query: 364
length of database: 11,318,596
effective HSP length: 101
effective length of query: 263
effective length of database: 8,619,977
effective search space: 2267053951
effective search space used: 2267053951
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0590c.6


