

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
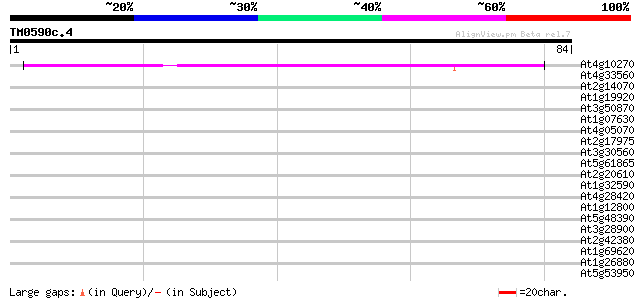
Query= TM0590c.4
(84 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10270 probable wound-induced protein 54 2e-08
At4g33560 unknown protein 37 0.002
At2g14070 unknown protein 32 0.056
At1g19920 ATP sulfurylase 28 1.1
At3g50870 transcription factor-like protein 27 1.4
At1g07630 unknown protein 27 1.4
At4g05070 unknown protein 27 1.8
At2g17975 putative protein 27 1.8
At3g30560 hypothetical protein 27 2.3
At5g61865 unknown protein 26 3.1
At2g20610 putative tyrosine aminotransferase 26 3.1
At1g32590 hypothetical protein, 5' partial 26 3.1
At4g28420 tyrosine transaminase-like protein 26 4.0
At1g12800 unknown protein 26 4.0
At5g48390 putative protein 25 5.2
At3g28900 60S ribosomal protein L34, putative 25 5.2
At2g42380 bZip transcription factor AtbZip34 25 5.2
At1g69620 putative 60S ribosomal protein L34 25 5.2
At1g26880 60s ribosomal protein L34 25 5.2
At5g53950 CUC2 (dbj|BAA19529.1) 25 6.8
>At4g10270 probable wound-induced protein
Length = 90
Score = 53.5 bits (127), Expect = 2e-08
Identities = 33/86 (38%), Positives = 49/86 (56%), Gaps = 10/86 (11%)
Query: 3 AATRSLIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKR 62
A T ++ +GAVEALKD+LG+ R NY LRS+ Q + N G S++++ + V
Sbjct: 7 AWTVAVSIGAVEALKDQLGLC--RWNYILRSVNQHLRNNVRSVSQGKRFSSSSVSAAVTS 64
Query: 63 SNQ--------EFMRKVMDLNCWGPS 80
S + E +R VM L+CWGP+
Sbjct: 65 SGESEKAKKAEESLRTVMYLSCWGPN 90
>At4g33560 unknown protein
Length = 95
Score = 37.0 bits (84), Expect = 0.002
Identities = 29/84 (34%), Positives = 43/84 (50%), Gaps = 12/84 (14%)
Query: 10 VGAVEALKDKLGVYNRRRNYALRSLQQRVVKN-------NNITPNGNSSSAAAIPSKVKR 62
+ AVE LKD+ GV R NY R L + + + +P +SSSA +I SK
Sbjct: 15 IAAVEVLKDQ-GV--ARWNYLFRLLHKEAMARVRTITVPSRPSPPTSSSSATSIRSKPLS 71
Query: 63 SN--QEFMRKVMDLNCWGPSTTKF 84
+ + K M L+C+GP+T +F
Sbjct: 72 TTPFETSFEKAMGLSCFGPTTVRF 95
>At2g14070 unknown protein
Length = 196
Score = 32.0 bits (71), Expect = 0.056
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 11/70 (15%)
Query: 26 RRNYALRSLQQ------RVVKNNNITPNGNSSSAAAIPSKVKRSN-----QEFMRKVMDL 74
R NY LR + R + + P+ SSS+++ VK ++ + M +VM L
Sbjct: 127 RWNYPLRFFNKDVRARLRAIAVTSRPPSSASSSSSSSADLVKENHPMPKSEASMERVMCL 186
Query: 75 NCWGPSTTKF 84
+C+GP+T +F
Sbjct: 187 SCFGPTTVRF 196
>At1g19920 ATP sulfurylase
Length = 476
Score = 27.7 bits (60), Expect = 1.1
Identities = 15/49 (30%), Positives = 26/49 (52%), Gaps = 3/49 (6%)
Query: 26 RRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSK---VKRSNQEFMRKV 71
+RN ++S+ + VK++ I P+G +P VK++ E M KV
Sbjct: 47 KRNLTMQSVSKMTVKSSLIDPDGGELVELIVPETEIGVKKAESETMPKV 95
>At3g50870 transcription factor-like protein
Length = 295
Score = 27.3 bits (59), Expect = 1.4
Identities = 19/67 (28%), Positives = 28/67 (41%), Gaps = 3/67 (4%)
Query: 2 SAATRSLIVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVK 61
+AAT + +VGA D+ G +N N + NNN TP + S +P
Sbjct: 191 TAATGNTVVGAAPVQTDQYGHHNSGYNNYHAATNN---NNNNGTPWAHHHSTQRVPCNYP 247
Query: 62 RSNQEFM 68
+ FM
Sbjct: 248 ANEIRFM 254
>At1g07630 unknown protein
Length = 662
Score = 27.3 bits (59), Expect = 1.4
Identities = 18/58 (31%), Positives = 29/58 (49%), Gaps = 8/58 (13%)
Query: 9 IVGAVEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNG-NSSSAAAIPSKVKRSNQ 65
IV ++++KD N+ +RS + R + N N+T N N SS ++ V NQ
Sbjct: 205 IVAPIKSVKDS-------DNWGIRSEKSRNLHNENLTVNSLNFSSEVSLDDDVSLENQ 255
>At4g05070 unknown protein
Length = 87
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/29 (44%), Positives = 17/29 (57%)
Query: 50 SSSAAAIPSKVKRSNQEFMRKVMDLNCWG 78
SS A S+ R +E +R VM L+CWG
Sbjct: 57 SSRFGAAASERLRQAEESLRTVMFLSCWG 85
>At2g17975 putative protein
Length = 268
Score = 26.9 bits (58), Expect = 1.8
Identities = 13/28 (46%), Positives = 17/28 (60%), Gaps = 5/28 (17%)
Query: 25 RRRNYALRSL-----QQRVVKNNNITPN 47
R RNYA RS Q R++ +NN +PN
Sbjct: 15 RNRNYAFRSFCNRCKQPRLIMDNNTSPN 42
>At3g30560 hypothetical protein
Length = 1473
Score = 26.6 bits (57), Expect = 2.3
Identities = 13/61 (21%), Positives = 25/61 (40%)
Query: 13 VEALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRKVM 72
++ALK K G Y + + + V + GN+ + K K+ + K++
Sbjct: 203 IDALKPKPGDYAKFQQLYVMDTDNEVDNRIEVMSKGNNGKTEGVKQKFKKETVSALLKML 262
Query: 73 D 73
D
Sbjct: 263 D 263
>At5g61865 unknown protein
Length = 417
Score = 26.2 bits (56), Expect = 3.1
Identities = 14/40 (35%), Positives = 20/40 (50%)
Query: 17 KDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAI 56
K K VYNRRR + L + + + + P G S SA +
Sbjct: 375 KQKTLVYNRRRTRSQALLSSQTARPDALIPLGLSPSARTV 414
>At2g20610 putative tyrosine aminotransferase
Length = 462
Score = 26.2 bits (56), Expect = 3.1
Identities = 16/56 (28%), Positives = 33/56 (58%), Gaps = 6/56 (10%)
Query: 15 ALKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRK 70
AL D GV+ + L+S++Q N ++TP+ + AA+P+ ++++++ F K
Sbjct: 289 ALNDPEGVFETTK--VLQSIKQ----NLDVTPDPATIIQAALPAILEKADKNFFAK 338
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 26.2 bits (56), Expect = 3.1
Identities = 14/64 (21%), Positives = 32/64 (49%), Gaps = 5/64 (7%)
Query: 13 VEALKDKLGVYNRRRNYALRSLQQRVVK-----NNNITPNGNSSSAAAIPSKVKRSNQEF 67
VE ++D+ G++ +R YA +++ ++ N I P + A A+ ++ N++
Sbjct: 1019 VEVIQDERGIFINQRKYAAEIIKKYGMEGCNSVKNPIVPGQKLTKAGAVSRYMESPNEQH 1078
Query: 68 MRKV 71
+ V
Sbjct: 1079 LLAV 1082
>At4g28420 tyrosine transaminase-like protein
Length = 389
Score = 25.8 bits (55), Expect = 4.0
Identities = 11/35 (31%), Positives = 21/35 (59%)
Query: 36 QRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRK 70
Q + +N +ITP+ + AA+P + ++N+E K
Sbjct: 298 QSIQQNLDITPDATTIVQAALPEILGKANKELFAK 332
>At1g12800 unknown protein
Length = 767
Score = 25.8 bits (55), Expect = 4.0
Identities = 19/55 (34%), Positives = 27/55 (48%), Gaps = 2/55 (3%)
Query: 16 LKDKLGVYNRRRNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRK 70
L+D L VY+R + L S + +K N + N NS I S R N+E + K
Sbjct: 519 LEDLLMVYDREKQKFLSSFVGQKIKVNVVMANRNSRK--LIFSMRPRENEEEVEK 571
>At5g48390 putative protein
Length = 1337
Score = 25.4 bits (54), Expect = 5.2
Identities = 12/32 (37%), Positives = 15/32 (46%)
Query: 32 RSLQQRVVKNNNITPNGNSSSAAAIPSKVKRS 63
R Q RV NNN N + IP ++ RS
Sbjct: 430 RGAQPRVTDNNNNINNNKTGYGRVIPERLTRS 461
>At3g28900 60S ribosomal protein L34, putative
Length = 120
Score = 25.4 bits (54), Expect = 5.2
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 4/27 (14%)
Query: 22 VYNRRRNYALRSLQQRVVKNNNITPNG 48
VY R +YA +S Q R+VK TP G
Sbjct: 6 VYRSRHSYATKSNQHRIVK----TPGG 28
>At2g42380 bZip transcription factor AtbZip34
Length = 300
Score = 25.4 bits (54), Expect = 5.2
Identities = 10/27 (37%), Positives = 16/27 (59%)
Query: 40 KNNNITPNGNSSSAAAIPSKVKRSNQE 66
KNNN+ P G+SS+ + + N+E
Sbjct: 102 KNNNVGPTGSSSNTSTPSNSFNDDNKE 128
>At1g69620 putative 60S ribosomal protein L34
Length = 119
Score = 25.4 bits (54), Expect = 5.2
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 4/27 (14%)
Query: 22 VYNRRRNYALRSLQQRVVKNNNITPNG 48
VY R +YA +S Q R+VK TP G
Sbjct: 6 VYRSRHSYATKSNQHRIVK----TPGG 28
>At1g26880 60s ribosomal protein L34
Length = 120
Score = 25.4 bits (54), Expect = 5.2
Identities = 13/27 (48%), Positives = 16/27 (59%), Gaps = 4/27 (14%)
Query: 22 VYNRRRNYALRSLQQRVVKNNNITPNG 48
VY R +YA +S Q R+VK TP G
Sbjct: 6 VYRSRHSYATKSNQHRIVK----TPGG 28
>At5g53950 CUC2 (dbj|BAA19529.1)
Length = 375
Score = 25.0 bits (53), Expect = 6.8
Identities = 17/57 (29%), Positives = 28/57 (48%)
Query: 27 RNYALRSLQQRVVKNNNITPNGNSSSAAAIPSKVKRSNQEFMRKVMDLNCWGPSTTK 83
RN + +S + +N N P SSSA+ + S V + + V +N W P+T +
Sbjct: 300 RNVSTQSNFRSFQENFNQFPYFGSSSASTMTSAVNLPSFQGGGGVSGMNYWLPATAE 356
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.126 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,594,265
Number of Sequences: 26719
Number of extensions: 51710
Number of successful extensions: 219
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 201
Number of HSP's gapped (non-prelim): 28
length of query: 84
length of database: 11,318,596
effective HSP length: 60
effective length of query: 24
effective length of database: 9,715,456
effective search space: 233170944
effective search space used: 233170944
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0590c.4


