

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0371a.1
(409 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
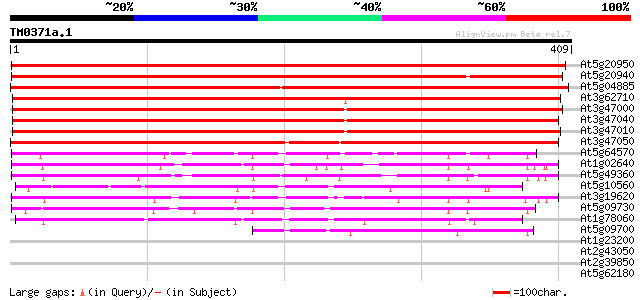
Score E
Sequences producing significant alignments: (bits) Value
At5g20950 beta-D-glucan exohydrolase-like protein 601 e-172
At5g20940 beta-glucosidase - like protein 568 e-162
At5g04885 unknown protein 564 e-161
At3g62710 beta-D-glucan exohydrolase-like protein 462 e-130
At3g47000 beta-D-glucan exohydrolase - like protein 447 e-126
At3g47040 beta-D-glucan exohydrolase - like protein 442 e-124
At3g47010 beta-D-glucan exohydrolase - like protein 427 e-120
At3g47050 beta-D-glucan exohydrolase - like protein 422 e-118
At5g64570 beta-xylosidase 100 1e-21
At1g02640 beta-xylosidase, putative 100 1e-21
At5g49360 xylosidase 97 2e-20
At5g10560 beta-xylosidase - like protein 93 3e-19
At3g19620 beta-xylosidase, putative 92 7e-19
At5g09730 beta-xylosidase - like protein 89 3e-18
At1g78060 putative protein 74 1e-13
At5g09700 beta-glucosidase - like protein 52 8e-07
At1g23200 putative pectinesterase 30 2.4
At2g43050 putative pectinesterase 30 3.1
At2g39850 putative serine protease 29 4.1
At5g62180 putative protein 29 5.4
>At5g20950 beta-D-glucan exohydrolase-like protein
Length = 624
Score = 601 bits (1549), Expect = e-172
Identities = 279/404 (69%), Positives = 338/404 (83%)
Query: 2 KVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGI 61
KV ACAKH+VGDGGT+ GID NNTVID GL +HMPGY+++++KGVATIMVSYS+WNG+
Sbjct: 220 KVAACAKHFVGDGGTVRGIDENNTVIDSKGLFGIHMPGYYNAVNKGVATIMVSYSAWNGL 279
Query: 62 KMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
+MH N +++TGFLKN L F+GFVISD++GIDRIT+PPH N++YSV AG+SAGIDM M+P
Sbjct: 280 RMHANKELVTGFLKNKLKFRGFVISDWQGIDRITTPPHLNYSYSVYAGISAGIDMIMVPY 339
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRK 181
+TEFI++++ ++ KLIP+SRIDDA+KRIL VKF MG+FE P AD S LG ++HR+
Sbjct: 340 NYTEFIDEISSQIQKKLIPISRIDDALKRILRVKFTMGLFEEPLADLSFANQLGSKEHRE 399
Query: 182 LAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGN 241
LAREAVRKSLVLLKNG + KP+LPL KK KILVAG HADNLGYQCGGWTI WQG++GN
Sbjct: 400 LAREAVRKSLVLLKNGKTGAKPLLPLPKKSGKILVAGAHADNLGYQCGGWTITWQGLNGN 459
Query: 242 NHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNL 301
+H GTTIL A+KNTV P T V+Y +NPD FV+S F YAIVVVGE PYAEM GD+ NL
Sbjct: 460 DHTVGTTILAAVKNTVAPTTQVVYSQNPDANFVKSGKFDYAIVVVGEPPYAEMFGDTTNL 519
Query: 302 TIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYG 361
TI PGP I NVC ++ CVV+++SGRP+VI+PY++ IDALVA WLPG+EGQGVAD L+G
Sbjct: 520 TISDPGPSIIGNVCGSVKCVVVVVSGRPVVIQPYVSTIDALVAAWLPGTEGQGVADALFG 579
Query: 362 DYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTK 405
DYGFTGKL RTWFKSV QLPMNVGD HYDPL+PFGFGL+T+P K
Sbjct: 580 DYGFTGKLARTWFKSVKQLPMNVGDRHYDPLYPFGFGLTTKPYK 623
>At5g20940 beta-glucosidase - like protein
Length = 626
Score = 568 bits (1464), Expect = e-162
Identities = 273/402 (67%), Positives = 330/402 (81%), Gaps = 2/402 (0%)
Query: 2 KVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGI 61
KV ACAKH+VGDGGT+ G++ NNTVI+ +GL+ +HMP Y +++KGVAT+MVSYSS NG+
Sbjct: 226 KVAACAKHFVGDGGTLRGMNANNTVINSNGLLGIHMPAYHDAVNKGVATVMVSYSSINGL 285
Query: 62 KMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPK 121
KMH N +ITGFLKN L F+G VISD+ G+D+I +P AN+++SV A +AG+DMFM
Sbjct: 286 KMHANKKLITGFLKNKLKFRGIVISDYLGVDQINTPLGANYSHSVYAATTAGLDMFMGSS 345
Query: 122 FFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRK 181
T+ I++LT VK K IPMSRIDDAVKRIL VKF MG+FENP AD+SL LG ++HR+
Sbjct: 346 NLTKLIDELTSQVKRKFIPMSRIDDAVKRILRVKFTMGLFENPIADHSLAKKLGSKEHRE 405
Query: 182 LAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGN 241
LAREAVRKSLVLLKNG +A+KP+LPL KK KILVAGTHADNLGYQCGGWTI WQG++GN
Sbjct: 406 LAREAVRKSLVLLKNGENADKPLLPLPKKANKILVAGTHADNLGYQCGGWTITWQGLNGN 465
Query: 242 NHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNL 301
N GTTIL A+K TVDP+T VIY +NPDT FV++ F YAIV VGE PYAE GDS NL
Sbjct: 466 NLTIGTTILAAVKKTVDPKTQVIYNQNPDTNFVKAGDFDYAIVAVGEKPYAEGFGDSTNL 525
Query: 302 TIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLYG 361
TI PGP TI NVC ++ CVV+++SGRP+V++ ++ IDALVA WLPG+EGQGVADVL+G
Sbjct: 526 TISEPGPSTIGNVCASVKCVVVVVSGRPVVMQ--ISNIDALVAAWLPGTEGQGVADVLFG 583
Query: 362 DYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQP 403
DYGFTGKL RTWFK+VDQLPMNVGDPHYDPL+PFGFGL T+P
Sbjct: 584 DYGFTGKLARTWFKTVDQLPMNVGDPHYDPLYPFGFGLITKP 625
>At5g04885 unknown protein
Length = 665
Score = 564 bits (1453), Expect = e-161
Identities = 263/407 (64%), Positives = 325/407 (79%), Gaps = 1/407 (0%)
Query: 1 EKVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNG 60
+KV ACAKHYVGDGGT G++ NNTV D GL+ VHMP Y ++ KGV+T+MVSYSSWNG
Sbjct: 224 DKVAACAKHYVGDGGTTRGVNENNTVTDLHGLLSVHMPAYADAVYKGVSTVMVSYSSWNG 283
Query: 61 IKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIP 120
KMH N ++ITG+LK TL FKGFVISD++G+D+I++PPH ++T SV A + AGIDM M+P
Sbjct: 284 EKMHANTELITGYLKGTLKFKGFVISDWQGVDKISTPPHTHYTASVRAAIQAGIDMVMVP 343
Query: 121 KFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHR 180
FTEF+NDLT LVKN IP++RIDDAV+RIL VKF MG+FENP ADYS LG Q HR
Sbjct: 344 FNFTEFVNDLTTLVKNNSIPVTRIDDAVRRILLVKFTMGLFENPLADYSFSSELGSQAHR 403
Query: 181 KLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSG 240
LAREAVRKSLVLLKNG + P+LPL +K KILVAGTHADNLGYQCGGWTI WQG SG
Sbjct: 404 DLAREAVRKSLVLLKNG-NKTNPMLPLPRKTSKILVAGTHADNLGYQCGGWTITWQGFSG 462
Query: 241 NNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMN 300
N + GTT+L+A+K+ VD T V+++ENPD EF++SN F+YAI+ VGE PYAE GDS
Sbjct: 463 NKNTRGTTLLSAVKSAVDQSTEVVFRENPDAEFIKSNNFAYAIIAVGEPPYAETAGDSDK 522
Query: 301 LTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAGWLPGSEGQGVADVLY 360
LT+ PGP I++ C+A+ CVV++ISGRPLV+EPY+A IDALVA WLPG+EGQG+ D L+
Sbjct: 523 LTMLDPGPAIISSTCQAVKCVVVVISGRPLVMEPYVASIDALVAAWLPGTEGQGITDALF 582
Query: 361 GDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQPTKAI 407
GD+GF+GKLP TWF++ +QLPM+ GD HYDPLF +G GL T+ +I
Sbjct: 583 GDHGFSGKLPVTWFRNTEQLPMSYGDTHYDPLFAYGSGLETESVASI 629
>At3g62710 beta-D-glucan exohydrolase-like protein
Length = 650
Score = 462 bits (1188), Expect = e-130
Identities = 231/416 (55%), Positives = 299/416 (71%), Gaps = 17/416 (4%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGIK 62
V CAKH+VGDGGTINGI+ NNTV D L +HMP + ++ KG+A+IM SYSS NG+K
Sbjct: 235 VAGCAKHFVGDGGTINGINENNTVADNATLFGIHMPPFEIAVKKGIASIMASYSSLNGVK 294
Query: 63 MHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPKF 122
MH N MIT +LKNTL F+GFVISD+ GID+IT +N+TYS+EA ++AGIDM M+P
Sbjct: 295 MHANRAMITDYLKNTLKFQGFVISDWLGIDKITPIEKSNYTYSIEASINAGIDMVMVPWA 354
Query: 123 FTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSL-VGYLGIQKHRK 181
+ E++ LT LV IPMSRIDDAV+RIL VKF +G+FEN AD L G + HR+
Sbjct: 355 YPEYLEKLTNLVNGGYIPMSRIDDAVRRILRVKFSIGLFENSLADEKLPTTEFGSEAHRE 414
Query: 182 LAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGN 241
+ REAVRKS+VLLKNG + I+PL KKV KI+VAG HA+++G+QCGG+++ WQG +G
Sbjct: 415 VGREAVRKSMVLLKNGKTDADKIVPLPKKVKKIVVAGRHANDMGWQCGGFSLTWQGFNGT 474
Query: 242 NH--------------LEGTTILTAIKNTVDPETTVIYKENPDTEFVESNG-FSYAIVVV 286
++GTTIL AI+ VDP T V+Y E P+ + + + +Y IVVV
Sbjct: 475 GEDMPTNTKHGLPTGKIKGTTILEAIQKAVDPTTEVVYVEEPNQDTAKLHADAAYTIVVV 534
Query: 287 GEHPYAEMEGDSMNLTIPSPGPETITNVC-EAITCVVIIISGRPLVIEPYLALIDALVAG 345
GE PYAE GDS L I PGP+T+++ C + C+VI+++GRPLVIEPY+ ++DAL
Sbjct: 535 GETPYAETFGDSPTLGITKPGPDTLSHTCGSGMKCLVILVTGRPLVIEPYIDMLDALAVA 594
Query: 346 WLPGSEGQGVADVLYGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLST 401
WLPG+EGQGVADVL+GD+ FTG LPRTW K V QLPMNVGD +YDPL+PFG+G+ T
Sbjct: 595 WLPGTEGQGVADVLFGDHPFTGTLPRTWMKHVTQLPMNVGDKNYDPLYPFGYGIKT 650
>At3g47000 beta-D-glucan exohydrolase - like protein
Length = 608
Score = 447 bits (1149), Expect = e-126
Identities = 219/403 (54%), Positives = 295/403 (72%), Gaps = 3/403 (0%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGIK 62
V+AC KH+VGDGGT GI+ NT+ + L ++H+P Y +++GV+T+M SYSSWNG +
Sbjct: 206 VVACVKHFVGDGGTDKGINEGNTIASYEELEKIHIPPYLKCLAQGVSTVMASYSSWNGTR 265
Query: 63 MHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPKF 122
+H + ++T LK L FKGF++SD+EG+DR++ P +N+ Y ++ V+AGIDM M+P
Sbjct: 266 LHADRFLLTEILKEKLGFKGFLVSDWEGLDRLSEPQGSNYRYCIKTAVNAGIDMVMVPFK 325
Query: 123 FTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRKL 182
+ +FI D+T LV++ IPM+RI+DAV+RIL VKF+ G+F +P D SL+ +G ++HR+L
Sbjct: 326 YEQFIQDMTDLVESGEIPMARINDAVERILRVKFVAGLFGHPLTDRSLLPTVGCKEHREL 385
Query: 183 AREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGNN 242
A+EAVRKSLVLLK+G +A+KP LPL + +ILV GTHAD+LGYQCGGWT W G+SG
Sbjct: 386 AQEAVRKSLVLLKSGKNADKPFLPLDRNAKRILVTGTHADDLGYQCGGWTKTWFGLSGRI 445
Query: 243 HLEGTTILTAIKNTVDPETTVIYKENPDTE-FVESNGFSYAIVVVGEHPYAEMEGDSMNL 301
+ GTT+L AIK V ET VIY++ P E S GFSYAIV VGE PYAE GD+ L
Sbjct: 446 TI-GTTLLDAIKEAVGDETEVIYEKTPSKETLASSEGFSYAIVAVGEPPYAETMGDNSEL 504
Query: 302 TIPSPGPETITNVCEAITCVVIIISGRPLVIEP-YLALIDALVAGWLPGSEGQGVADVLY 360
IP G + +T V E I +VI+ISGRP+V+EP L +ALVA WLPG+EGQGVADV++
Sbjct: 505 RIPFNGTDIVTAVAEIIPTLVILISGRPVVLEPTVLEKTEALVAAWLPGTEGQGVADVVF 564
Query: 361 GDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLSTQP 403
GDY F GKLP +WFK V+ LP++ YDPLFPFGFGL+++P
Sbjct: 565 GDYDFKGKLPVSWFKHVEHLPLDAHANSYDPLFPFGFGLNSKP 607
>At3g47040 beta-D-glucan exohydrolase - like protein
Length = 636
Score = 442 bits (1138), Expect = e-124
Identities = 219/400 (54%), Positives = 294/400 (72%), Gaps = 3/400 (0%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGIK 62
V+ACAKH+VGDGGT GI+ NT++ + L ++H+ Y + +++GV+T+M SYSSWNG K
Sbjct: 231 VVACAKHFVGDGGTDKGINEGNTIVSYEELEKIHLAPYLNCLAQGVSTVMASYSSWNGSK 290
Query: 63 MHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPKF 122
+H+++ ++T LK L FKGFVISD+E ++R++ P +N+ V+ V+AG+DM M+P
Sbjct: 291 LHSDYFLLTELLKQKLGFKGFVISDWEALERLSEPFGSNYRNCVKISVNAGVDMVMVPFK 350
Query: 123 FTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRKL 182
+ +FI DLT LV++ + MSRIDDAV+RIL VKF+ G+FE+P D SL+G +G ++HR+L
Sbjct: 351 YEQFIKDLTDLVESGEVTMSRIDDAVERILRVKFVAGLFEHPLTDRSLLGTVGCKEHREL 410
Query: 183 AREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGNN 242
ARE+VRKSLVLLKNG ++EKP LPL + V +ILV GTHAD+LGYQCGGWT W G+SG
Sbjct: 411 ARESVRKSLVLLKNGTNSEKPFLPLDRNVKRILVTGTHADDLGYQCGGWTKAWFGLSGRI 470
Query: 243 HLEGTTILTAIKNTVDPETTVIYKENPDTEFVES-NGFSYAIVVVGEHPYAEMEGDSMNL 301
+ GTT+L AIK V +T VIY++ P E + S FSYAIV VGE PYAE GD+ L
Sbjct: 471 TI-GTTLLDAIKEAVGDKTEVIYEKTPSEETLASLQRFSYAIVAVGETPYAETLGDNSEL 529
Query: 302 TIPSPGPETITNVCEAITCVVIIISGRPLVIEP-YLALIDALVAGWLPGSEGQGVADVLY 360
TIP G + +T + E I +V++ SGRPLV+EP L +ALVA WLPG+EGQG+ DV++
Sbjct: 530 TIPLNGNDIVTALAEKIPTLVVLFSGRPLVLEPLVLEKAEALVAAWLPGTEGQGMTDVIF 589
Query: 361 GDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLS 400
GDY F GKLP +WFK VDQLP+ YDPLFP GFGL+
Sbjct: 590 GDYDFEGKLPVSWFKRVDQLPLTADANSYDPLFPLGFGLN 629
>At3g47010 beta-D-glucan exohydrolase - like protein
Length = 609
Score = 427 bits (1099), Expect = e-120
Identities = 212/401 (52%), Positives = 293/401 (72%), Gaps = 3/401 (0%)
Query: 3 VIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNGIK 62
VIACAKH+VGDGGT G+ NT+ + L ++H+ Y + I++GV+T+M S+SSWNG +
Sbjct: 207 VIACAKHFVGDGGTEKGLSEGNTITSYEDLEKIHVAPYLNCIAQGVSTVMASFSSWNGSR 266
Query: 63 MHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIPKF 122
+H+++ ++T LK L FKGF++SD++G++ I+ P +N+ V+ G++AGIDM M+P
Sbjct: 267 LHSDYFLLTEVLKQKLGFKGFLVSDWDGLETISEPEGSNYRNCVKLGINAGIDMVMVPFK 326
Query: 123 FTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHRKL 182
+ +FI D+T LV++ IPM+R++DAV+RIL VKF+ G+FE+P AD SL+G +G ++HR++
Sbjct: 327 YEQFIQDMTDLVESGEIPMARVNDAVERILRVKFVAGLFEHPLADRSLLGTVGCKEHREV 386
Query: 183 AREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSGNN 242
AREAVRKSLVLLKNG +A+ P LPL + +ILV G HA++LG QCGGWT G SG
Sbjct: 387 AREAVRKSLVLLKNGKNADTPFLPLDRNAKRILVVGMHANDLGNQCGGWTKIKSGQSGRI 446
Query: 243 HLEGTTILTAIKNTVDPETTVIYKENPDTE-FVESNGFSYAIVVVGEHPYAEMEGDSMNL 301
+ GTT+L +IK V +T VI+++ P E S+GFSYAIV VGE PYAEM+GD+ L
Sbjct: 447 TI-GTTLLDSIKAAVGDKTEVIFEKTPTKETLASSDGFSYAIVAVGEPPYAEMKGDNSEL 505
Query: 302 TIPSPGPETITNVCEAITCVVIIISGRPLVIEP-YLALIDALVAGWLPGSEGQGVADVLY 360
TIP G IT V E I +VI+ SGRP+V+EP L +ALVA W PG+EGQG++DV++
Sbjct: 506 TIPFNGNNIITAVAEKIPTLVILFSGRPMVLEPTVLEKTEALVAAWFPGTEGQGMSDVIF 565
Query: 361 GDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLST 401
GDY F GKLP +WFK VDQLP+N YDPLFP GFGL++
Sbjct: 566 GDYDFKGKLPVSWFKRVDQLPLNAEANSYDPLFPLGFGLTS 606
>At3g47050 beta-D-glucan exohydrolase - like protein
Length = 612
Score = 422 bits (1085), Expect = e-118
Identities = 218/402 (54%), Positives = 285/402 (70%), Gaps = 5/402 (1%)
Query: 1 EKVIACAKHYVGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSSWNG 60
+ V+ACAKH+VGDGGT I+ NT++ + L R H+ Y IS+GV+T+M SYSSWNG
Sbjct: 204 KNVVACAKHFVGDGGTNKAINEGNTILRYEDLERKHIAPYKKCISQGVSTVMASYSSWNG 263
Query: 61 IKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIP 120
K+H+++ ++T LK L FKG+V+SD+EG+DR++ PP +N+ V+ G++AGIDM M+P
Sbjct: 264 DKLHSHYFLLTEILKQKLGFKGYVVSDWEGLDRLSDPPGSNYRNCVKIGINAGIDMVMVP 323
Query: 121 KFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYLGIQKHR 180
+ +F NDL LV++ + M+R++DAV+RIL VKF+ G+FE P D SL+ +G ++HR
Sbjct: 324 FKYEQFRNDLIDLVESGEVSMARVNDAVERILRVKFVAGLFEFPLTDRSLLPTVGCKEHR 383
Query: 181 KLAREAVRKSLVLLKNGISAEKPILPLTKKVPKILVAGTHADNLGYQCGGWTIEWQGVSG 240
+LAREAVRKSLVLLKNG E LPL +ILV GTHAD+LGYQCGGWT G SG
Sbjct: 384 ELAREAVRKSLVLLKNGRYGE--FLPLNCNAERILVVGTHADDLGYQCGGWTKTMYGQSG 441
Query: 241 NNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESN-GFSYAIVVVGEHPYAEMEGDSM 299
+GTT+L AIK V ET VIY+++P E + S FSYAIV VGE PYAE GD+
Sbjct: 442 -RITDGTTLLDAIKAAVGDETEVIYEKSPSEETLASGYRFSYAIVAVGESPYAETMGDNS 500
Query: 300 NLTIPSPGPETITNVCEAITCVVIIISGRPLVIEP-YLALIDALVAGWLPGSEGQGVADV 358
L IP G E IT V E I +VI+ SGRP+ +EP L +ALVA WLPG+EGQG+ADV
Sbjct: 501 ELVIPFNGSEIITTVAEKIPTLVILFSGRPMFLEPQVLEKAEALVAAWLPGTEGQGIADV 560
Query: 359 LYGDYGFTGKLPRTWFKSVDQLPMNVGDPHYDPLFPFGFGLS 400
++GDY F GKLP TWFK VDQLP+++ Y PLFP GFGL+
Sbjct: 561 IFGDYDFRGKLPATWFKRVDQLPLDIESNGYLPLFPLGFGLN 602
>At5g64570 beta-xylosidase
Length = 784
Score = 100 bits (250), Expect = 1e-21
Identities = 111/413 (26%), Positives = 186/413 (44%), Gaps = 52/413 (12%)
Query: 2 KVIACAKHYVG-DGGTINGID--GNNTVIDRDGLMRVHMPGYFSSISKG-VATIMVSYSS 57
KV AC KHY D G++ N V+ + + + P + S + G VA++M SY+
Sbjct: 219 KVAACCKHYTAYDVDNWKGVERYSFNAVVTQQDMDDTYQPPFKSCVVDGNVASVMCSYNQ 278
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVS--AGID 115
NG + D+++G ++ G+++SD + +D + H T + A +S AG+D
Sbjct: 279 VNGKPTCADPDLLSGVIRGEWKLNGYIVSDCDSVDVLYKNQHYTKTPAEAAAISILAGLD 338
Query: 116 MFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFE-NPFADYSLVGYL 174
+ F + + VK+ L+ + ID A+ +G F+ NP + G L
Sbjct: 339 L-NCGSFLGQHTEE---AVKSGLVNEAAIDKAISNNFLTLMRLGFFDGNP--KNQIYGGL 392
Query: 175 G-----IQKHRKLAREAVRKSLVLLKNGISAEKPILPLT-KKVPKILVAGTHADNLGYQC 228
G +++LA +A R+ +VLLKN LPL+ K + + V G +A+
Sbjct: 393 GPTDVCTSANQELAADAARQGIVLLKN-----TGCLPLSPKSIKTLAVIGPNANVTKTMI 447
Query: 229 GG-------WTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSY 281
G +T QG++G TT L N V T+ + S
Sbjct: 448 GNYEGTPCKYTTPLQGLAGT---VSTTYLPGCSNVACAVADVA----GATKLAATADVS- 499
Query: 282 AIVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVIEPYLAL 338
++V+G E E D ++L +P E + V +A +++I+SG I A
Sbjct: 500 -VLVIGADQSIEAESRDRVDLHLPGQQQELVIQVAKAAKGPVLLVIMSGGGFDIT--FAK 556
Query: 339 IDALVAGWL----PGSEGQ-GVADVLYGDYGFTGKLPRTWFKS--VDQLPMNV 384
D +AG L PG G +AD+++G Y +GKLP TW+ V+++PM +
Sbjct: 557 NDPKIAGILWVGYPGEAGGIAIADIIFGRYNPSGKLPMTWYPQSYVEKVPMTI 609
>At1g02640 beta-xylosidase, putative
Length = 768
Score = 100 bits (249), Expect = 1e-21
Identities = 121/446 (27%), Positives = 198/446 (44%), Gaps = 68/446 (15%)
Query: 2 KVIACAKHYVG-DGGTINGIDG---NNTVIDRDGLMRVHMPGYFSSISKGVATIMVSYSS 57
KV AC KH+ D NG+D N V +D +P VA+IM SY+
Sbjct: 201 KVAACCKHFTAYDLDNWNGVDRFHFNAKVSKQDIEDTFDVPFRMCVKEGNVASIMCSYNQ 260
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEA--GVSAGID 115
NG+ + +++ ++N G+++SD + + + H T A + AG+D
Sbjct: 261 VNGVPTCADPNLLKKTIRNQWGLNGYIVSDCDSVGVLYDTQHYTGTPEEAAADSIKAGLD 320
Query: 116 MFMIPKFFTEFINDLTI-LVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVGYL 174
+ P F+ TI VK L+ S +D+A+ L V+ +G+F+ A G+L
Sbjct: 321 LDCGP-----FLGAHTIDAVKKNLLRESDVDNALINTLTVQMRLGMFDGDIAAQP-YGHL 374
Query: 175 G-----IQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPK-ILVAGTHAD----NL 224
G H+ LA EA ++ +VLLKN S+ LPL+ + + + V G ++D +
Sbjct: 375 GPAHVCTPVHKGLALEAAQQGIVLLKNHGSS----LPLSSQRHRTVAVIGPNSDATVTMI 430
Query: 225 GYQCG---GWTIEWQGVSG---NNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNG 278
G G G+T QG++G H +G + + + + D + G
Sbjct: 431 GNYAGVACGYTSPVQGITGYARTIHQKGCVDVHCMDDRLF-----------DAAVEAARG 479
Query: 279 FSYAIVVVGEHPYAEME-GDSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVIE-- 333
++V+G E E D +L +P E ++ V +A +++++SG P+ I
Sbjct: 480 ADATVLVMGLDQSIEAEFKDRNSLLLPGKQQELVSRVAKAAKGPVILVLMSGGPIDISFA 539
Query: 334 PYLALIDALVAGWLPGSE-GQGVADVLYGDYGFTGKLPRTWFKS--VDQLP---MNVGDP 387
I A+V PG E G +AD+L+G GKLP TW+ + LP M++
Sbjct: 540 EKDRKIPAIVWAGYPGQEGGTAIADILFGSANPGGKLPMTWYPQDYLTNLPMTEMSMRPV 599
Query: 388 H-----------YD--PLFPFGFGLS 400
H YD ++PFG GLS
Sbjct: 600 HSKRIPGRTYRFYDGPVVYPFGHGLS 625
>At5g49360 xylosidase
Length = 774
Score = 97.1 bits (240), Expect = 2e-20
Identities = 111/443 (25%), Positives = 189/443 (42%), Gaps = 66/443 (14%)
Query: 2 KVIACAKHYVG-DGGTINGIDGN--NTVIDRDGLMRVHMPGYFSSISKG-VATIMVSYSS 57
KV AC KHY D NG+D N + + L + + S + +G VA++M SY+
Sbjct: 207 KVAACCKHYTAYDLDNWNGVDRFHFNAKVTQQDLEDTYNVPFKSCVYEGKVASVMCSYNQ 266
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGID------RITSPPHANFTYSVEAGVS 111
NG + +++ ++ G+++SD + +D TS P S++AG+
Sbjct: 267 VNGKPTCADENLLKNTIRGQWRLNGYIVSDCDSVDVFFNQQHYTSTPEEAAARSIKAGLD 326
Query: 112 AGIDMFMIPKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLV 171
F+ FTE VK L+ + I+ A+ L V+ +G+F+ Y+ +
Sbjct: 327 LDCGPFL--AIFTEG------AVKKGLLTENDINLALANTLTVQMRLGMFDGNLGPYANL 378
Query: 172 GYLGI--QKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKIL-----VAGTHADNL 224
G + H+ LA EA + +VLLKN + P+ P + ++ V T N
Sbjct: 379 GPRDVCTPAHKHLALEAAHQGIVLLKNS-ARSLPLSPRRHRTVAVIGPNSDVTETMIGNY 437
Query: 225 GYQCGGWTIEWQGVS---GNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSY 281
+ +T QG+S H G + N +E T
Sbjct: 438 AGKACAYTSPLQGISRYARTLHQAGCAGVACKGNQGFGAAEAAAREADAT---------- 487
Query: 282 AIVVVGEHPYAEME-GDSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVI-----E 333
++V+G E E D L +P + +T V +A +++++SG P+ + +
Sbjct: 488 -VLVMGLDQSIEAETRDRTGLLLPGYQQDLVTRVAQASRGPVILVLMSGGPIDVTFAKND 546
Query: 334 PYLALIDALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMNV----GDP 387
P +A I + AG+ + G +A++++G GKLP TW+ V ++PM V
Sbjct: 547 PRVAAI--IWAGYPGQAGGAAIANIIFGAANPGGKLPMTWYPQDYVAKVPMTVMAMRASG 604
Query: 388 HY----------DPLFPFGFGLS 400
+Y +FPFGFGLS
Sbjct: 605 NYPGRTYRFYKGPVVFPFGFGLS 627
>At5g10560 beta-xylosidase - like protein
Length = 792
Score = 92.8 bits (229), Expect = 3e-19
Identities = 98/390 (25%), Positives = 171/390 (43%), Gaps = 32/390 (8%)
Query: 5 ACAKHYVG----DGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKGVAT-IMVSYSSWN 59
AC KH+ G D N V ++D + + P + + I G A+ +M SY++ N
Sbjct: 224 ACCKHFTAYDLEKWGNFTRYDFNAVVTEQD-MEDTYQPPFETCIRDGKASCLMCSYNAVN 282
Query: 60 GIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMI 119
G+ D++ + F+G++ SD + + I + + +T S E V+ I +
Sbjct: 283 GVPACAQGDLLQK-ARVEWGFEGYITSDCDAVATIFA--YQGYTKSPEEAVADAIKAGVD 339
Query: 120 PKFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFENP--FADYSLVGYLGI- 176
T + ++ + +D A+ + V+ +G+F+ Y +G I
Sbjct: 340 INCGTYMLRHTQSAIEQGKVSEELVDRALLNLFAVQLRLGLFDGDPRRGQYGKLGSNDIC 399
Query: 177 -QKHRKLAREAVRKSLVLLKNGISAEKPILPLTKK-VPKILVAGTHADNLGYQCGGWTIE 234
HRKLA EA R+ +VLLKN + +LPL K V + + G A+N+ G +T
Sbjct: 400 SSDHRKLALEATRQGIVLLKN----DHKLLPLNKNHVSSLAIVGPMANNISNMGGTYT-- 453
Query: 235 WQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVES----NGFSYAIVVVGEHP 290
G T +L +K T + DT F E+ G + IVV G
Sbjct: 454 --GKPCQRKTLFTELLEYVKKTSYASGCSDVSCDSDTGFGEAVAIAKGADFVIVVAGLDL 511
Query: 291 YAEMEG-DSMNLTIPSPGPETITNVCEAITCVVIIISGRPLVIEPYLALIDALVAG--WL 347
E E D ++L++P + +++V VI++ ++ A D + W+
Sbjct: 512 SQETEDKDRVSLSLPGKQKDLVSHVAAVSKKPVILVLTGGGPVDVTFAKNDPRIGSIIWI 571
Query: 348 --PGSEG-QGVADVLYGDYGFTGKLPRTWF 374
PG G Q +A++++GD+ G+LP TW+
Sbjct: 572 GYPGETGGQALAEIIFGDFNPGGRLPTTWY 601
>At3g19620 beta-xylosidase, putative
Length = 781
Score = 91.7 bits (226), Expect = 7e-19
Identities = 118/446 (26%), Positives = 197/446 (43%), Gaps = 67/446 (15%)
Query: 2 KVIACAKHYVG-DGGTINGIDGNN--TVIDRDGLMRVHMPGYFSSISKG-VATIMVSYSS 57
KV +C KHY D GID + + + L + + S + +G V+++M SY+
Sbjct: 202 KVSSCCKHYTAYDLDNWKGIDRFHFDAKVTKQDLEDTYQTPFKSCVEEGDVSSVMCSYNR 261
Query: 58 WNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTY--SVEAGVSAGID 115
NGI + +++ G ++ G+++SD + I + H T +V + AG++
Sbjct: 262 VNGIPTCADPNLLRGVIRGQWRLDGYIVSDCDSIQVYFNDIHYTKTREDAVALALKAGLN 321
Query: 116 MFMIPKFFTEFINDLT-ILVKNKLIPMSRIDDAVKRILWVKFMMGIFEN-----PFADYS 169
M +F+ T VK K + S +D+A+ V +G F+ PF +
Sbjct: 322 MNC-----GDFLGKYTENAVKLKKLNGSDVDEALIYNYIVLMRLGFFDGDPKSLPFGNLG 376
Query: 170 LVGYLGIQKHRKLAREAVRKSLVLLKNGISAEKPILPLTK-KVPKILVAGTHADNLGYQC 228
+ H+ LA EA ++ +VLL+N + LPL K V K+ V G +A+
Sbjct: 377 PSDVCS-KDHQMLALEAAKQGIVLLEN-----RGDLPLPKTTVKKLAVIGPNANATKVMI 430
Query: 229 GGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYA------ 282
+ GV + T+ + ++ V PE V D + + S A
Sbjct: 431 SNYA----GVP----CKYTSPIQGLQKYV-PEKIVYEPGCKDVKCGDQTLISAAVKAVSE 481
Query: 283 ----IVVVGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVIE-- 333
++VVG E EG D +NLT+P + + +V A T V++I+S P+ I
Sbjct: 482 ADVTVLVVGLDQTVEAEGLDRVNLTLPGYQEKLVRDVANAAKKTVVLVIMSAGPIDISFA 541
Query: 334 PYLALIDALVAGWLPG-SEGQGVADVLYGDYGFTGKLPRTWF-----KSVDQLPMNV--- 384
L+ I A++ PG + G +A V++GDY +G+LP TW+ V MN+
Sbjct: 542 KNLSTIRAVLWVGYPGEAGGDAIAQVIFGDYNPSGRLPETWYPQEFADKVAMTDMNMRPN 601
Query: 385 ---GDPHYD-------PLFPFGFGLS 400
G P P++ FG+GLS
Sbjct: 602 STSGFPGRSYRFYTGKPIYKFGYGLS 627
>At5g09730 beta-xylosidase - like protein
Length = 773
Score = 89.4 bits (220), Expect = 3e-18
Identities = 103/406 (25%), Positives = 178/406 (43%), Gaps = 40/406 (9%)
Query: 2 KVIACAKHY----VGDGGTINGIDGNNTVIDRDGLMRVHMPGYFSSISKG-VATIMVSYS 56
KV AC KHY + + +N + N V+++ L P + S + G VA++M SY+
Sbjct: 209 KVAACCKHYTAYDIDNWRNVNRLTFN-AVVNQQDLADTFQPPFKSCVVDGHVASVMCSYN 267
Query: 57 SWNGIKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFT--YSVEAGVSAGI 114
NG + D+++G ++ G+++SD + +D + H T +V + AG+
Sbjct: 268 QVNGKPTCADPDLLSGVIRGQWQLNGYIVSDCDSVDVLFRKQHYAKTPEEAVAKSLLAGL 327
Query: 115 DMFMIPKFFTEFINDLTIL--VKNKLIPMSRIDDAVKRILWVKFMMGIFENPFADYSLVG 172
D+ + N + VK L+ + ID A+ +G F+ L G
Sbjct: 328 DL------NCDHFNGQHAMGAVKAGLVNETAIDKAISNNFATLMRLGFFDGD-PKKQLYG 380
Query: 173 YLG-----IQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKIL-VAGTHADNLGY 226
LG +++LAR+ R+ +VLLKN + LPL+ K L V G +A+
Sbjct: 381 GLGPKDVCTADNQELARDGARQGIVLLKNSAGS----LPLSPSAIKTLAVIGPNANATET 436
Query: 227 QCGGWTIEWQGVSGNNHLEGTTILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIV-V 285
G + GV + + +T V + V+ + A+V V
Sbjct: 437 MIG----NYHGVPCKYTTPLQGLAETVSSTYQLGCNVACVDADIGSAVDLAASADAVVLV 492
Query: 286 VGEHPYAEMEG-DSMNLTIPSPGPETITNVCEAI--TCVVIIISGRPLVI---EPYLALI 339
VG E EG D ++L +P E +T V A V++I+SG I + +
Sbjct: 493 VGADQSIEREGHDRVDLYLPGKQQELVTRVAMAARGPVVLVIMSGGGFDITFAKNDKKIT 552
Query: 340 DALVAGWLPGSEGQGVADVLYGDYGFTGKLPRTWFKS--VDQLPMN 383
+ G+ + G +ADV++G + +G LP TW+ V+++PM+
Sbjct: 553 SIMWVGYPGEAGGLAIADVIFGRHNPSGNLPMTWYPQSYVEKVPMS 598
>At1g78060 putative protein
Length = 767
Score = 74.3 bits (181), Expect = 1e-13
Identities = 89/390 (22%), Positives = 167/390 (42%), Gaps = 32/390 (8%)
Query: 5 ACAKHYVG-DGGTINGIDGN--NTVIDRDGLMRVHMPGYFSSISKGVAT-IMVSYSSWNG 60
AC KH+ D GI N + L + P + I +G A+ IM +Y+ NG
Sbjct: 206 ACCKHFTAYDLDRWKGITRYVFNAQVSLADLAETYQPPFKKCIEEGRASGIMCAYNRVNG 265
Query: 61 IKMHTNHDMITGFLKNTLHFKGFVISDFEGIDRITSPPHANFTYSVEAGVSAGIDMFMIP 120
I + +++T + F+G++ SD + + I + S E V+ + M
Sbjct: 266 IPSCADPNLLTRTARGQWAFRGYITSDCDAVSIIYDAQ--GYAKSPEDAVADVLKAGMDV 323
Query: 121 KFFTEFINDLTILVKNKLIPMSRIDDAVKRILWVKFMMGIFEN-----PFADYSLVGYLG 175
+ ++ K + + ID A+ + V+ +G+F P+ + S +
Sbjct: 324 NCGSYLQKHTKSALQQKKVSETDIDRALLNLFSVRIRLGLFNGDPTKLPYGNIS-PNEVC 382
Query: 176 IQKHRKLAREAVRKSLVLLKNGISAEKPILPLTKK-VPKILVAGTHADNLGYQCGGWTIE 234
H+ LA +A R +VLLKN + +LP +K+ V + V G +A + G +
Sbjct: 383 SPAHQALALDAARNGIVLLKNNLK----LLPFSKRSVSSLAVIGPNAHVVKTLLGNYA-- 436
Query: 235 WQGVSGNNHLEGTTILTAIKNTV---DPETTVIYKENPDTEFVESNGFSYAIVVVGEHPY 291
G + + +KN V ++ D + + ++++G
Sbjct: 437 --GPPCKTVTPLDALRSYVKNAVYHQGCDSVACSNAAIDQAVAIAKNADHVVLIMGLDQT 494
Query: 292 AEMEG-DSMNLTIPSPGPETITNVCEAIT--CVVIIISGRPLVIEPYLA---LIDALVAG 345
E E D ++L++P E IT+V A V+++I G P+ I + A I +++
Sbjct: 495 QEKEDFDRVDLSLPGKQQELITSVANAAKKPVVLVLICGGPVDIS-FAANNNKIGSIIWA 553
Query: 346 WLPGSEGQ-GVADVLYGDYGFTGKLPRTWF 374
PG G ++++++GD+ G+LP TW+
Sbjct: 554 GYPGEAGGIAISEIIFGDHNPGGRLPVTWY 583
>At5g09700 beta-glucosidase - like protein
Length = 411
Score = 51.6 bits (122), Expect = 8e-07
Identities = 53/215 (24%), Positives = 97/215 (44%), Gaps = 16/215 (7%)
Query: 178 KHRKLAREAVRKSLVLLKNGISAEKPILPLTKKVPKIL-VAGTHADNLGYQCGGWTIEWQ 236
++R+LA E R+ +VLLKN + LPL+ K L V G +A+ G + E
Sbjct: 27 ENRELAVETARQGIVLLKNSAGS----LPLSPSAIKTLAVIGPNANVTKTMIGNY--EGV 80
Query: 237 GVSGNNHLEGT--TILTAIKNTVDPETTVIYKENPDTEFVESNGFSYAIVVVGEHPYAEM 294
L+G T+LT + T + + + ++ + +V+ + +
Sbjct: 81 ACKYTTPLQGLERTVLTTKYHRGCFNVTCTEADLDSAKTLAASADATVLVMGADQTIEKE 140
Query: 295 EGDSMNLTIPSPGPETITNVCEAITCVVIII----SGRPLVIEPYLALIDALVAGWLPGS 350
D ++L +P E +T V +A V+++ G + I +++ PG
Sbjct: 141 TLDRIDLNLPGKQQELVTQVAKAARGPVVLVIMSGGGFDITFAKNDEKITSIMWVGYPGE 200
Query: 351 EGQ-GVADVLYGDYGFTGKLPRTWFKS--VDQLPM 382
G +ADV++G + +GKLP TW+ V+++PM
Sbjct: 201 AGGIAIADVIFGRHNPSGKLPMTWYPQSYVEKVPM 235
>At1g23200 putative pectinesterase
Length = 554
Score = 30.0 bits (66), Expect = 2.4
Identities = 12/24 (50%), Positives = 16/24 (66%)
Query: 344 AGWLPGSEGQGVADVLYGDYGFTG 367
AGWLP S ++ + YG+YG TG
Sbjct: 482 AGWLPWSGSFALSTLYYGEYGNTG 505
>At2g43050 putative pectinesterase
Length = 518
Score = 29.6 bits (65), Expect = 3.1
Identities = 11/24 (45%), Positives = 16/24 (65%)
Query: 344 AGWLPGSEGQGVADVLYGDYGFTG 367
+GW P S G G+ + YG+YG +G
Sbjct: 444 SGWSPWSGGFGLKSLFYGEYGNSG 467
>At2g39850 putative serine protease
Length = 783
Score = 29.3 bits (64), Expect = 4.1
Identities = 21/67 (31%), Positives = 30/67 (44%), Gaps = 10/67 (14%)
Query: 262 TVIYKENPDTEFVESNGFSYAIVVVGEHPYAEMEGDSMNLTIPSPGPETITNVCEAITCV 321
T+ + NP+ E + A++ G PY+E+ G SP P N CE ITC
Sbjct: 116 TLKAERNPENE----SDLVVAVIDSGIWPYSELFGSD------SPPPPGWENKCENITCN 165
Query: 322 VIIISGR 328
I+ R
Sbjct: 166 NKIVGAR 172
>At5g62180 putative protein
Length = 327
Score = 28.9 bits (63), Expect = 5.4
Identities = 22/81 (27%), Positives = 36/81 (44%), Gaps = 7/81 (8%)
Query: 110 VSAGIDMFMIPKFFTEFINDLTILVKN---KLIPMSRI----DDAVKRILWVKFMMGIFE 162
+ +DM + F +E DL +V + +L P R+ DD V+ + W+K +
Sbjct: 92 ILCSVDMQLFHDFCSEVARDLNAIVVSPSYRLAPEHRLPAAYDDGVEALDWIKTSDDEWI 151
Query: 163 NPFADYSLVGYLGIQKHRKLA 183
AD+S V +G LA
Sbjct: 152 KSHADFSNVFLMGTSAGGNLA 172
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.140 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,849,089
Number of Sequences: 26719
Number of extensions: 435724
Number of successful extensions: 979
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 928
Number of HSP's gapped (non-prelim): 24
length of query: 409
length of database: 11,318,596
effective HSP length: 102
effective length of query: 307
effective length of database: 8,593,258
effective search space: 2638130206
effective search space used: 2638130206
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0371a.1


